Abstract
Cancer is a deadly disease primarily because of the ability of tumor cells to spread from the primary tumor, to invade into the connective tissue, and to form metastases at distant sites. In contrast to cell migration on a planar surface where large cell tractions and contractile forces are not essential, tractions and forces are thought to be crucial for overcoming the resistance and steric hindrance of a dense 3-dimensional connective tissue matrix. In this review, we describe recently developed biophysical tools including 2-D and 3-D traction microscopy to measure contractile forces of cells. We discuss evidence indicating that tumor cell invasiveness is associated with increased contractile force generation.
Cell migration and invasion
The main reason for the malignancy of cancer is the ability of tumor cells to form secondary tumors and metastasize in distant organs. To form metastases, cancer cells need to take multiple steps: First, they separate from the primary tumor and invade through the tissue and the extracellular matrix. Next, they enter a nearby blood and lymph vessel where they get transported to distant sites. The subsequent steps are in dispute, but a likely scenario is that the cancer cells adhere onto the endothelium of the vessel, transmigrate through the endothelium and, once more, migrate through the tissue. Regardless of whether extravasation takes place, however, the migration through connective tissue (subsequently called invasion) is a prerequisite for metastasis formation.
Although cell invasion is foremost a mechanical process, cancer research has focused largely on gene regulation and signaling that underlie uncontrolled cell growth. More recently, the genes and signals involved in the invasion and transendothelial migration of cancer cells, such as the role of adhesion molecules and matrix-degrading enzymes, have become the focus of research (Paszek et al., 2005; Rolli et al., 2003; Wolf et al., 2003). However, the mechanical processes themselves that control cancer cell invasion, such as cell adhesion, changes of cell shape, cell movements and motility, and the generation of forces, are currently not well understood (Friedl and Brocker, 2000; Ridley et al., 2003; Zaman et al., 2006). In particular, some of the most elementary questions regarding the forces during cancer cell invasion have not yet been answered: Do cells push against the tissue to propel themselves forward, or do they grab tissue matrix in front of them and then pull? How hard do they push or pull? How strong do they adhere to the matrix? What size holes are they able to squeeze through, and what are the forces during amoeboid versus mesenchymal invasion strategies?
Forces in cell migration on 2-D substrates
Most of what we know today about cell migration, mechanical tensions and forces is derived from studies of cells cultured on planar substrates (e.g. tissue culture plastic or glass, or polyacrylamide hydrogels). Methods to visualize traction forces during cell migration in 2-D culture systems have been used for several decades (Harris et al., 1980; Pelham and Wang, 1997) and were more recently developed into quantitative tools (Butler et al., 2002; Dembo and Wang, 1999; Raupach et al., 2007; Sabass et al., 2008). The principal idea behind these methods is the measurement of the deformations of an elastic substrate with known elastic modulus (such as polyacrylamide) on which adherent cells are plated. As the cells adhere and spread, they generate tractions and thereby deform the substrate. The tractions can then be computed from the substrate displacements using continuum mechanics theory. Measurement of the displacement field is accomplished by tracking small fluorescent beads that are embedded near the surface of the substrate gel. The elastic modulus of the polyacrylamide substrate can be adjusted within a wide range by changing the acrylamide and bis-acrylamide cross-linker density (Pelham and Wang, 1997; Yeung et al., 2005). The spatial resolution of the traction map obtained with this method approaches 1 µm under ideal conditions, which is sufficient to resolve the forces from individual focal adhesions (Sabass et al., 2008).
2-D traction microscopy has brought a wealth of new insights into the mechano-biology of cells and cancer cell migration in particular (Mierke et al., 2007a, b; Raupach et al., 2007; Runz et al., 2008). For example, cells feel and respond to the stiffness of their extracellular matrix by a dynamic regulation of adhesion receptor (integrin) clustering, focal adhesion complex formation, and cytoskeletal architecture remodeling (Discher et al., 2005). As a consequence, contractile force generation and cell migration are strongly influenced by the mechanical properties of the matrix (Pelham and Wang, 1997).
Forces in cell invasion through 3-D connective tissue
How such a force feedback mechanism plays out in a 3-D environment is currently not well understood; force generation, migratory behavior, cell adhesion, focal adhesion formation, cytoskeletal organization, and dynamics of cancer cells in 2-D culture have been shown to substantially differ from those observed in a 3-D environment where cells are embedded in a flexible, degradable 3-D extracellular matrix (Cukierman et al., 2001; Zaman et al., 2006).
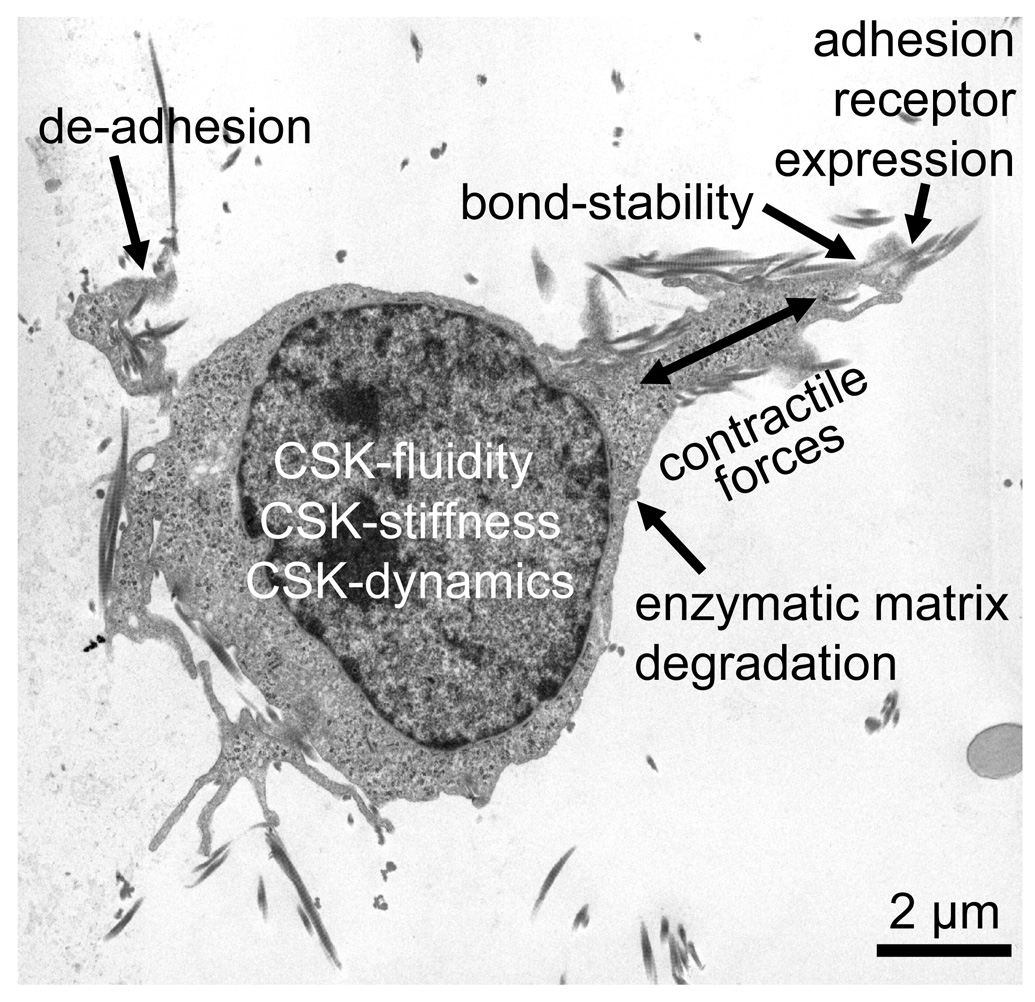
The speed of 3-D cell migration, regardless of cell type, is governed by the balance between four biophysical processes (Zaman et al., 2006) (Fig. 1): 1) Contractile forces need to be generated that help the cell to pull itself through a dense matrix network. 2) These contractile forces need to be transmitted to the surrounding extracellular matrix via cell adhesions, such as integrins. Moreover, the adhesive bonds need to be sufficiently strong under the load imposed by the contractile forces, but they also need to de-adhere in time in order not to permanently hold the cell back. 3) As the cell squeezes through the matrix network, the cell’s resisting elastic and frictional forces against cell shape changes need to be sufficiently small, or else the cytoskeleton needs to be able to dynamically remodel itself to accommodate the necessary shape changes. 4) The resisting (mostly elastic) forces imposed by the matrix when it is deformed as the cell wedges itself through the matrix network need to be sufficiently small, or else the cell needs to degrade the matrix network enzymatically to decrease matrix resistance and steric hindrance. Matrix resistance plays no role in 2-D migration where cell adhesion, de-adhesion and the ability to remodel cytoskeletal structures are the only important mechanical parameters that influence migration speed, whereas the forces needed to overcome the viscous drag imposed by the liquid environment are negligible.
Fig. 1.
Prerequisites for efficient cell invasion. TEM image of an MDA-MB-231 breast carcinoma cell invading into a 3-D collagen matrix. The ability to invade through connective tissue is governed by the balance between four biophysical processes: 1) contractile force generation, 2) transmission of contractile forces via cell-matrix adhesions (integrin expression, adhesion bond stability, de-adhesion), 3) resisting forces of the cell against cell shape changes (cytoskeletal (CSK) fluidity, dynamics, stiffness)), and 4) resisting forces imposed by the matrix against deformations (enzymatic matrix degradation).
3-D force assays
To estimate the forces that cells exert in a 3-D environment, gel contraction assays have been developed and used in numerous studies (Bell et al., 1979; Berendsen et al., 2006; Cooke et al., 2000; Smith et al., 2006): Cells are mixed with collagen prior to gelation into a disk. The gel disk has free boundaries (or is only loosely attached to the wall of the culture dish) and shrinks when the cells exert contractile forces. From the gel shrinkage (assessed from the gel diameter), a qualitative estimate of the contractile forces can be obtained. Theoretically, a quantitative estimate of the average forces generated by the cells inside the gels could also be obtained if the cell number and the viscoelastic gel properties are known and if the spatial cell density distribution and cell orientation is homogenous and isotropic throughout the gel. In practice, however, these prerequisites are not satisfied. Moreover, cells can remodel their extracellular matrix by compacting the matrix, by secretion of matrix-degrading enzymes or by secretion of new matrix proteins, and as a consequence, the local and global viscoelastic properties of the gels can significantly change over time (Bell et al., 1979; Leung et al., 2007). Although the matrix remodeling can be continuously monitored by twisting micrometer-scale ferrimagnetic beads embedded throughout the gel and measuring the angular bead rotation, those measurements do not have the sensitivity and spatial resolution needed for a quantitative estimate of cell forces (Leung et al., 2007). To further complicate matters, cells modify their biomechanical properties in response to the rheological properties (e.g., stiffness), the structural properties (e.g., mesh size, fiber orientation) and the biochemical properties (adhesive ligands) of the extracellular matrix (Discher et al., 2005; Paszek et al., 2005; Pelham and Wang, 1997). Current computational models of cell migration in 3-D matrices are limited because they largely ignore such active cell responses (Zaman et al., 2005, 2006).
3-D traction microscopy
For a quantitative estimate of cell tractions and contractile forces of single cells in a 3-D extracellular matrix, it seems feasible, at least in principle, to extend the 2-D traction microscopy method described above to the third dimension. The following modifications of the 2-D method described above would be necessary. First, instead of a polyacrylamide hydrogel, a reconstituted connective tissue matrix (e.g., collagen or Matrigel) can be used. Invasive cancer cells are able to spontaneously invade deep into such gels. Second, instead of a single layer of fluorescent beads at the gel surface, they need to be randomly dispersed throughout the matrix. By taking multiple images at different focal depth of the matrix gel, one can then determine the x, y, z position of the fluorescent beads. Third, from changes in the bead position (either measured over time, or after cell treatment with drugs relaxing the contractile forces of cells and inducing cell detachment), certain measures of contractile force generation such as the elastic strain energy can be computed.
There are numerous challenges ahead, however, before such an approach will become a routine tool. First, measurements of the deformation field in 3-D are considerably more difficult compared to a 2-D situation. Image stacks with a z-focus distance of 2 µm between adjacent images over a total z-height of approximately 500 µm need to be recorded. The need for sufficient data storage capacity and long image acquisition time (> 1 min) for a single image stack, and the potentially harmful light exposure, will restrict the measurement time. Computing the x, y and z position for each of the typically far more than 10,000 fluorescent beads within the image stack with an accuracy of better than 30 nm poses additional challenges. Because at least two image stacks are needed to measure cell tractions – one stack each before and after the addition of traction-releasing drugs such as trypsin/EDTA, ML-7, cytochalasin D, or latrunculin A (see Fig. 3) – it is necessary to identify the same bead in two image stacks, which can be difficult when large deformations have occurred.
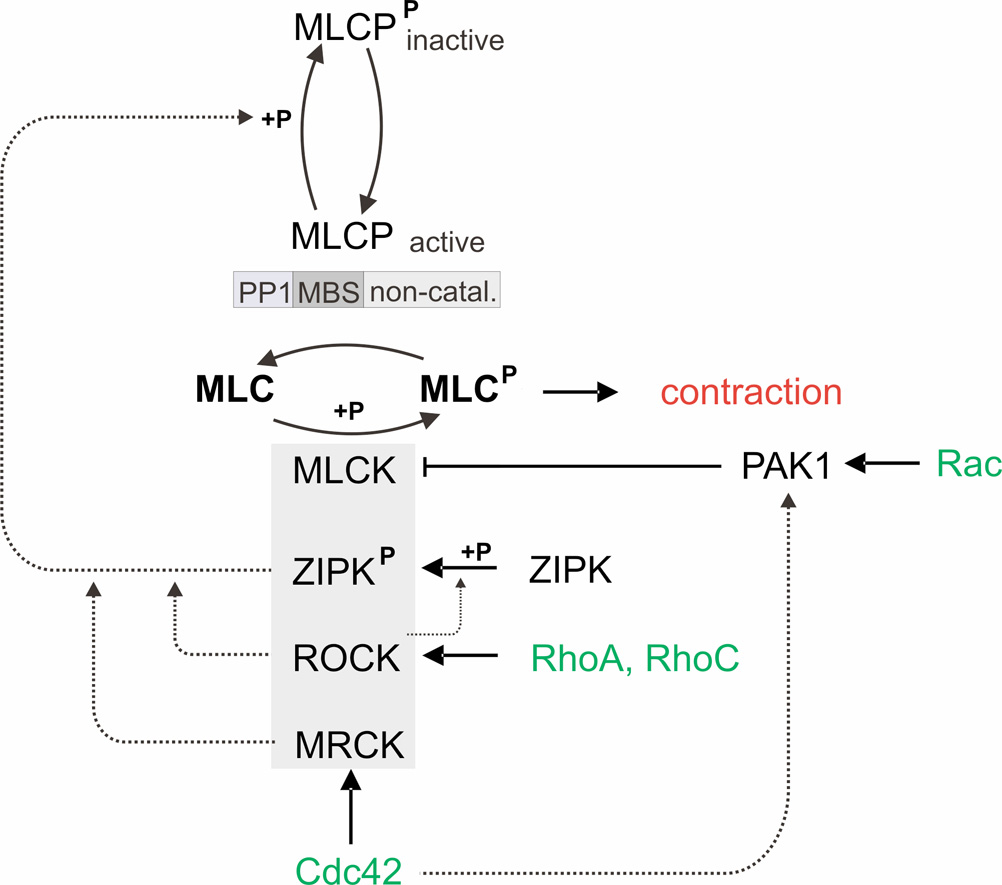
Fig. 3.
Regulation of contractile forces. Contractile forces are controlled by myosin light chain (MLC) phosphorylation, which in turn depends on the balanced activities of the myosin light chain kinase (MLCK) and myosin light chain phosphatase (MLCP). Force up-regulation: MLCP can be inhibited by phosphorylation either through Rho-associated kinase (ROCK), through zipper-interacting protein kinase (ZIPK), or through myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK) (dotted lines). ROCK has been shown to indirectly inhibit MLCP via ZIRP phosphorylation. ROCK is activated by the small GTPases RhoA and RhoC, and MRCK is activated by the small GTPase Cdc42. ZIPK, ROCK and MRCK have also been shown to directly phosphorylate MLC. Force down-regulation: p21-activated protein kinase 1 (PAK1) inhibits MLCK and is activated by the small GTPases Rac and Cdc42.
The second challenge is the measurement of the elastic modulus of the 3-D matrix gels. Unlike polyacrylamide gels, collagen gels are not elastic but viscoelastic, they are not linear but stiffen with increasing strain, and they are not amorphous but filamentous. Because of the filamentous nature of collagen gels, it is not even clear to what degree the macroscopic rheological properties that one can measure with a plate rheometer reflect the microscopic properties experienced by the cell.
The third challenge is the computation of the traction field. In the case of 2-D traction microscopy, the gel can be approximated as a semi-infinite solid, and the gel displacements that result from a point traction on the gel surface are described by an analytically known Green’s function (Butler et al., 2002). In the case of a 3-D gel, the boundary conditions such as the free upper gel surface (usually overgrown with cells that have not yet invaded) and the fixed lower surface, as well as the non-linear rheological properties of the gel need to be considered, and the Green’s function under such conditions is unknown. Moreover, the invaded cell may have generated a path through the gel by secreting matrix-degrading enzymes. Finally, the deformation field of the gel is known only at the locations of the embedded fluorescent beads, which are so sparse (approximately 10–15 µm apart) that the traction reconstruction would severely underestimate the true tractions, although the latter problem may be eased by using the matrix filaments themselves to track the deformation field, as shown in Figure 2.
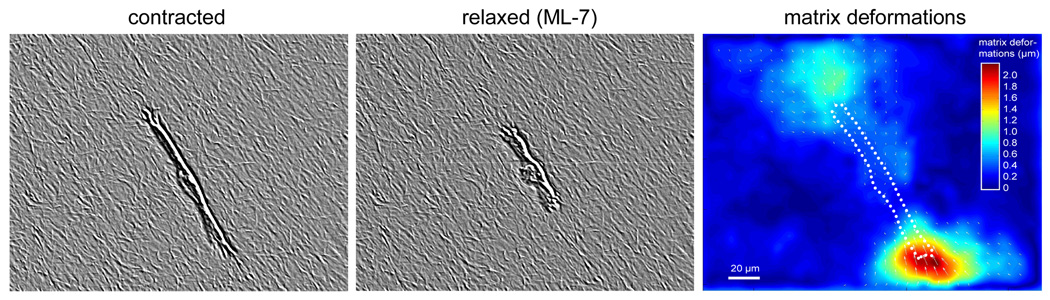
Fig. 2.
Collagen matrix deformations due to contractile forces. An MDA-MD-231 breast carcinoma cell was allowed to invade into a collagen gel for two days and had reached a depth of approximately 200 µm below the gel surface. The left image shows the structure of the collagen matrix surrounding the cell, recorded with a modulation contrast imaging mode (in addition, the contrast was digitally enhanced to better visualize the collagen matrix). The cell was then treated with an over-dose of 250 µM ML-7 (a myosin light chain kinase inhibitor) to release cytoskeletal tension stored in the actin cytoskeleton and to detach the cell from the matrix. Within less than 2 minutes, the collagen matrix returned to its undeformed, relaxed state, and a second image was recorded (middle). The matrix deformations (right) were computed using a difference-with-interpolation method from the left (contracted) and the middle (relaxed) image (Raupach et al., 2007).
One way of overcoming these problems is to use the elastic strain energy stored in the matrix as a robust estimate of contractile cell forces (Butler et al., 2002). The elastic strain energy can be obtained from the local matrix strain between adjacent fluorescent beads, and only the matrix rheological properties but not the boundary conditions need to be known. Wyckoff et al. (2006) used glass microneedles to calibrate the gel deformations that arise from a point source, and they estimated that the total traction forces of rat mammary adenocarcinoma cells (MTLn3) are between 10–20 nN. Calibration measurements of the deformation field in collagen gels that result from point forces (generated with magnetic tweezers (Kollmannsberger and Fabry, 2007)), and measurements in MDA-MB-231 human breast carcinoma cells (Fig. 2) show forces that are at least one order of magnitude higher (unpublished). But even 10–20 nN is a substantial force that can significantly enhance the ability of cells to migrate through a dense connective tissue matrix. As such, it is a tenable hypothesis that cancer cells can become more invasive by becoming more contractile. Indeed, this hypothesis is supported by a recent report (Mierke et al., 2007b). It is, then, also conceivable that pharmacological interventions that alter the contractile properties of cancer cells may offer new therapeutic strategies to reduce cancer cell invasiveness. In the following section, we give a brief outline of the signal transduction pathways that regulate contractile forces in cancer cells (Fig. 3).
Signal transduction pathways involved in contractile regulation
Generation of contractile force is essential for important biological processes such as cytokinesis, chemotaxis, tissue remodeling, and invasiveness (Wyckoff et al., 2006; Yee et al., 2001). In both smooth muscle and non-muscle cells, contraction is powered by the myosin II motor protein complex, which is activated when the myosin II regulatory light chain (MLC) is phosphorylated. Phosphorylation of the MLC of the myosin II motor complex induces its interaction with actin, which thereby activates the myosin ATPase, resulting in enhanced cell contractility. The rapid generation of contractile forces is predominantly governed by the Ca2+/calmodulin-stimulated myosin light chain kinase (MLCK); at a longer time scale, contractile forces are modulated by kinases associated with the Rho-family GTPases (Yee et al., 2001).
The MLC phosphorylation status depends mainly on the balance in the activities of MLCK and myosin light chain phosphatase (MLCP). MLCP is composed of three subunits: a catalytic subunit – protein phosphatase type 1 (PP1), a myosin-binding subunit (MBS), and a small non-catalytic subunit. Rho-family GTPases RhoA, RhoC, Cdc42, and Rac1 modulate MLC phosphorylation and acto-myosin contractility through their associated kinases (Zhao and Manser, 2005).
The monomeric GTPases RhoA and RhoC act through their downstream effector, Rho-associated kinase (ROCK) (Ishizaki et al., 1996; Sahai and Marshall, 2002). ROCK can phosphorylate MLC directly at Ser-19 both in vitro (Amano et al., 1996) and in vivo (Totsukawa et al., 2000; Wyckoff et al., 2006), but the main effect of ROCK on MLC phosphorylation is its ability to prevent the dephosphorylation of MLC via MLCP inhibition (Kimura et al., 1996; Somlyo and Somlyo, 2000; Sward et al., 2000). ROCK phosphorylates MLCP at the inhibitory site Thr-695/696, and at Thr-850/853 at the PP1 subunit where it induces its dissociation from myosin (Ito et al., 2004; Tan et al., 2001; Velasco et al., 2002).
ROCK-dependent phosphorylation of MLC was found to be crucial for the localization and correct organization of MLC at the cell cortex, and has been implied in force generation in invading tumor cells with amoeboid morphology (Wyckoff et al., 2006). Recently, it has been shown that ROCK also phosphorylates and activates zipper-interacting protein kinase (ZIPK). ZIPK can regulate MLC phosphorylation through either phosphorylation and inhibition of MLCP, or direct phosphorylation of MLC (Hagerty et al., 2007).
In contrast to Rho, the Rac effector p21-activated kinase 1 (PAK1) inhibits MLCK (Sanders et al., 1999). Down-regulation of MLC phosphorylation through Rac results in dynamic morphological changes that are associated with 2-D cell migration, in particular the extension of membrane ruffles into membrane protrusions.
Cdc42 can both activate and inhibit MLC phosphorylation. The activation of MLC phosphorylation is mediated by the Cdc42 effector, myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK) (Leung et al., 1998). MRCK cooperates with ROCK to control MLC phosphorylation through the inhibitory phosphorylation of the MBS of MLCP (Amano et al., 1996; Kimura et al., 1996; Tan et al., 2001; Wilkinson et al., 2005). Conversely, Cdc42 can interact with PAK1 (Manser et al., 1995; Sanders et al., 1999), and in smooth muscle, Cdc42 was shown to inhibit MLC phosphorylation (Murthy et al., 2003). Taken together, the small GTPases Rho, Rac and Cdc42, can both inhibit or increase MLC phosphorylation. The net effect and the integrated cellular responses that follow the activation of Rho, Cdc42 and Rac may depend on the precise temporal coordination of GTPase activation, as well as the intracellular localization and extent of MLC phosphorylation (Sanders et al., 1999).
Mechanisms of amoeboid motility in 3-D
Amoeboid cell migration results from more dynamic, transient and less defined cell-substrate contacts associated with amoeboid cell shape (Gunzer et al., 2000). Recently, amoeboid cell migration was identified as a mechanism of tumor cells to invade connective tissue without extracellular matrix degradation (Wolf et al., 2003). Two different modes of amoeboid migration through the extracellular matrix can be distinguished.
HT-1080 fibrosarcoma cells, upon treatment with a protease inhibitor cocktail (to prevent the structural remodeling of collagen by the cells), switched from a mesenchymal migration strategy to an amoeboid migration/invasion mode (Wolf et al., 2003). This protease-independent migration is characterized by an adaptation and alignment of the cell body along collagen fiber strands and consecutive migratory guidance along fibrillar scaffolds. To overcome narrow regions in the collagen network, cells form pseudopods through matrix gaps. This step is followed by constriction and propulsion of the cell body to “squeeze” through the confinements imposed by matrix fibers. These constrictions persist until the cell squeezes or pulls itself out of the confinement (Wolf et al., 2003). The entire process occurs without apparent deformations of the matrix.
In contrast, it has been reported that MTLn3E breast carcinoma cells are able to deform collagen fibers during invasion (Wyckoff et al., 2006). Unlike HT-1080 fibrosarcoma cells, the MTLn3E breast carcinoma cells naturally have an amoeboid morphology and show an amoeboid-type migration through the extracellular matrix also in the absence of a protease inhibitor cocktail. During migration, the MTLn3E cells appeared to push away the collagen at the front, while no consistent pattern of collagen deformation was observed at the rear end of the cells. In cases where cells extend a protrusion that is retracted without leading to translation of the cell body, collagen is pulled toward the cell, indicative of contractile forces. The forces needed to deform the collagen matrix are thought to be generated by actomyosin interactions (as described in (Meshel et al., 2005)). Consistent with this hypothesis, it has been reported that blebbistatin, a non-muscle myosin ATPase inhibitor, dramatically reduced the extent of collagen deformation (Wyckoff et al., 2006). MLC is organized in MTLn3E cells perpendicularly to the direction of movement behind the invading edge. Both the organization and phosphorylation of MLC at the leading edge and the force generation are dependent upon ROCK function.
The following model was proposed for amoeboid invasion of MTLn3E breast carcinoma cells (Wyckoff et al., 2006): As a first step, an F-actin-rich protrusion is extended. Subsequently, cell-matrix adhesions form to connect the F-actin network with the extracellular matrix. ROCK localizes to the invading protrusions, promotes MLC phosphorylation, and enables actomyosin interactions. This leads to the formation of a dense, tensed actin mesh behind the invading cell edge. The cell-matrix adhesions are formed predominantly in front of this contractile zone; therefore, the net effect of the actomyosin contraction is to move the whole cell body toward the adhesions at the protrusion front. Actin and MLC co-localize throughout the cell cortex and drive the cell forward, analogous to the dynamic network model that was proposed for migrating fish keratocytes (Svitkina et al., 1997; Wyckoff et al., 2006). The role of ROCK in this process is particularly important; besides regulating MLC phosphorylation, ROCK also regulates the localization of MLC to the cell cortex, and the density and turn-over dynamics of the cortical actin network. However, it remains elusive how the cell orchestrates cytoskeletal tension, dynamics and reorganization, protrusive forces, and contractile traction forces during migration through the extracellular matrix.
Concluding remarks
Different cell types employ diverse migrating strategies. It is generally thought that more invasive tumor cells express more adhesion receptors and secrete more proteolytic enzymes compared to non-invasive cells (Rolli et al., 2003), but exceptions are possible. For example, after blocking pericellular proteolysis, cells change their migration strategy from a mesenchymal to an amoeboid pattern (Wolf et al., 2003). No single factor alone can explain the large differences seen in the invasive behavior between different cancer cell types. Apart from the secretion of proteolytic enzymes, the expression of integrins, their adhesive bond stability, and the speed of cytoskeletal remodeling, traction forces appear to be particularly important for cell migration. While traction forces have been studied in 2-D assays, these results cannot be generalized to cell migration in 3-D. Similarly, other parameters that affect cell migration in 2-D systems, such as matrix composition or matrix stiffness, need to be reevaluated in 3-D culture systems.
Acknowledgements
This work was supported by grants from the Czech Republic (301/06/1147), the Ministry of Education of the Czech Republic (research centre grant LC06061), Deutsche Krebshilfe (107384), DFG (FA336/2-1), and NIH (HL65960).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J. Biol. Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- Bell E, Ivarsson B, Merrill C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc. Natl. Acad. Sci. USA. 1979;76:1274–1278. doi: 10.1073/pnas.76.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen AD, Bronckers AL, Smit TH, Walboomers XF, Everts V. Collagen type V enhances matrix contraction by human periodontal ligament fibroblasts seeded in three-dimensional collagen gels. Matrix Biol. 2006;25:515–522. doi: 10.1016/j.matbio.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- Cooke ME, Sakai T, Mosher DF. Contraction of collagen matrices mediated by alpha2beta1A and alpha(v)beta3 integrins. J. Cell Sci. 2000;113:2375–2383. doi: 10.1242/jcs.113.13.2375. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Friedl P, Brocker EB. The biology of cell locomotion within three-dimensional extracellular matrix. Cell. Mol. Life Sci. 2000;57:41–64. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzer M, Friedl P, Niggemann B, Brocker EB, Kampgen E, Zanker KS. Migration of dendritic cells within 3-D collagen lattices is dependent on tissue origin, state of maturation, and matrix structure and is maintained by proinflammatory cytokines. J. Leukoc. Biol. 2000;67:622–629. doi: 10.1002/jlb.67.5.622. [DOI] [PubMed] [Google Scholar]
- Hagerty L, Weitzel DH, Chambers J, Fortner CN, Brush MH, Loiselle D, Hosoya H, Haystead TA. ROCK1 phosphorylates and activates zipper-interacting protein kinase. J. Biol. Chem. 2007;282:4884–4893. doi: 10.1074/jbc.M609990200. [DOI] [PubMed] [Google Scholar]
- Harris AK, Wild P, Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, Narumiya S. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- Ito M, Nakano T, Erdodi F, Hartshorne DJ. Myosin phosphatase: structure, regulation and function. Mol. Cell. Biochem. 2004;259:197–209. doi: 10.1023/b:mcbi.0000021373.14288.00. [DOI] [PubMed] [Google Scholar]
- Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Kollmannsberger P, Fabry B. High-force magnetic tweezers with force feedback for biological applications. Rev. Sci. Instrum. 2007;78:114301–114306. doi: 10.1063/1.2804771. [DOI] [PubMed] [Google Scholar]
- Leung LY, Tian D, Brangwynne CP, Weitz DA, Tschumperlin DJ. A new microrheometric approach reveals individual and cooperative roles for TGF-beta1 and IL-1beta in fibroblast-mediated stiffening of collagen gels. FASEB J. 2007;21:1–10. doi: 10.1096/fj.06-7510com. [DOI] [PubMed] [Google Scholar]
- Leung T, Chen XQ, Tan I, Manser E, Lim L. Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol. Cell. Biol. 1998;18:130–140. doi: 10.1128/mcb.18.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E, Chong C, Zhao ZS, Leung T, Michael G, Hall C, Lim L. Molecular cloning of a new member of the p21-Cdc42/Rac-activated kinase (PAK) family. J. Biol. Chem. 1995;270:25070–25078. doi: 10.1074/jbc.270.42.25070. [DOI] [PubMed] [Google Scholar]
- Meshel AS, Wei Q, Adelstein RS, Sheetz MP. Basic mechanism of three-dimensional collagen fibre transport by fibroblasts. Nat. Cell Biol. 2005;7:157–164. doi: 10.1038/ncb1216. [DOI] [PubMed] [Google Scholar]
- Mierke CT, Kollmannsberger P, Paranhos-Zitterbart D, Smith J, Fabry B, Goldmann WH. Mechano-coupling and regulation of contractility by the vinculin tail domain. Biophys. J. 2007a;94:661–670. doi: 10.1529/biophysj.107.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke CT, Paranhos-Zitterbart D, Kollmannsberger P, Raupach C, Schlötzer-Schrehardt U, Goecke TW, Behrens J, Fabry B. Breakdown of the endothelial barrier function in tumor cell transmigration. Biophys. J. 2007b;94 doi: 10.1529/biophysj.107.113613. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KS, Zhou H, Grider JR, Brautigan DL, Eto M, Makhlouf GM. Differential signalling by muscarinic receptors in smooth muscle: m2-mediated inactivation of myosin light chain kinase via Gi3, Cdc42/Rac1 and p21-activated kinase 1 pathway, and m3-mediated MLC20 (20 kDa regulatory light chain of myosin II) phosphorylation via Rho-associated kinase/myosin phosphatase targeting subunit 1 and protein kinase C/CPI-17 pathway. Biochem. J. 2003;374:145–155. doi: 10.1042/BJ20021274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raupach C, Zitterbart DP, Mierke CT, Metzner C, Muller FA, Fabry B. Stress fluctuations and motion of cytoskeletal-bound markers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;76:011918. doi: 10.1103/PhysRevE.76.011918. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rolli M, Fransvea E, Pilch J, Saven A, Felding-Habermann B. Activated integrin alphavbeta3 cooperates with metalloproteinase MMP-9 in regulating migration of metastatic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:9482–9487. doi: 10.1073/pnas.1633689100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runz S, Mierke CT, Joumaa S, Behrens J, Fabry B, Altevogt P. CD24 induces localization of beta1 integrin to lipid raft domains. Biochem. Biophys. Res. Commun. 2008;365:35–41. doi: 10.1016/j.bbrc.2007.10.139. [DOI] [PubMed] [Google Scholar]
- Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophys J. 2008;94:207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Marshall CJ. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat. Cell Biol. 2002;4:408–415. doi: 10.1038/ncb796. [DOI] [PubMed] [Google Scholar]
- Sanders LC, Matsumura F, Bokoch GM, de Lanerolle P. Inhibition of myosin light chain kinase by p21-activated kinase. Science. 1999;283:2083–2085. doi: 10.1126/science.283.5410.2083. [DOI] [PubMed] [Google Scholar]
- Smith KD, Wells A, Lauffenburger DA. Multiple signaling pathways mediate compaction of collagen matrices by EGF-stimulated fibroblasts. Exp. Cell Res. 2006;312:1970–1982. doi: 10.1016/j.yexcr.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Somlyo AP, Somlyo AV. Signal transduction by G-proteins, rho-kinase and protein phosphatase to smooth muscle and non-muscle myosin II. J. Physiol. 2000;522:177–185. doi: 10.1111/j.1469-7793.2000.t01-2-00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sward K, Dreja K, Susnjar M, Hellstrand P, Hartshorne DJ, Walsh MP. Inhibition of Rho-associated kinase blocks agonist-induced Ca2+ sensitization of myosin phosphorylation and force in guinea-pig ileum. J, Physiol. 2000;522:33–49. doi: 10.1111/j.1469-7793.2000.0033m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan I, Ng CH, Lim L, Leung T. Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J. Biol. Chem. 2001;276:21209–21216. doi: 10.1074/jbc.M102615200. [DOI] [PubMed] [Google Scholar]
- Totsukawa G, Yamakita Y, Yamashiro S, Hartshorne DJ, Sasaki Y, Matsumura F. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell Biol. 2000;150:797–806. doi: 10.1083/jcb.150.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco G, Armstrong C, Morrice N, Frame S, Cohen P. Phosphorylation of the regulatory subunit of smooth muscle protein phosphatase 1M at Thr850 induces its dissociation from myosin. FEBS Lett. 2002;527:101–104. doi: 10.1016/s0014-5793(02)03175-7. [DOI] [PubMed] [Google Scholar]
- Wilkinson S, Paterson HF, Marshall CJ. Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat. Cell Biol. 2005;7:255–261. doi: 10.1038/ncb1230. [DOI] [PubMed] [Google Scholar]
- Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr. Biol. 2006;16:1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- Yee HF, Jr, Melton AC, Tran BN. RhoA/rho-associated kinase mediates fibroblast contractile force generation. Biochem. Biophys. Res. Commun. 2001;280:1340–1345. doi: 10.1006/bbrc.2001.4291. [DOI] [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Zaman MH, Kamm RD, Matsudaira P, Lauffenburger DA. Computational model for cell migration in three-dimensional matrices. Biophys. J. 2005;89:1389–1397. doi: 10.1529/biophysj.105.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman MH, Trapani LM, Siemeski A, Mackellar D, Gong H, Kamm RD, Wells A, Lauffenburger DA, Matsudaira P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZS, Manser E. PAK and other Rho-associated kinases – effectors with surprisingly diverse mechanisms of regulation. Biochem. J. 2005;386:201–214. doi: 10.1042/BJ20041638. [DOI] [PMC free article] [PubMed] [Google Scholar]