Abstract
Cancer cells treated with the cyclooxygenase-2 inhibitor celecoxib show growth inhibition and induced apoptosis. This study was conducted to determine if the same processes are relevant to celecoxib’s effects on human colorectal adenocarcinomas treated in vivo. A cohort of 23 patients with primary colorectal adenocarcinomas was randomized to receive a 7-day course of celecoxib (400 mg b.i.d.) or no drug prior to surgical resection. Gene expression profiling was performed on resected adenocarcinomas from the cohort of patients. Using fold change (>1.5) and p-value (<0.05) cut-offs, 190 genes were differentially expressed between adenocarcinomas from patients receiving celecoxib and those that did not. The celecoxib pre-treated samples showed decreased expression levels in multiple genes involved in cellular lipid and glutathione metabolism; changes associated with diminished cellular proliferation. Celecoxib pre-treatment for 7 days in vivo is associated with alterations in colorectal adenocarcinoma gene expression which are suggestive of diminished cellular proliferation.
Keywords: cDNA microarrays, Colorectal Neoplasms, Celecoxib, Cyclooxygenase 2 inhibitors, Gene expression profiling
Introduction
The inducible form of cyclooxygenase (COX), COX-2 is markedly up-regulated in pathological conditions such as inflammation and cancer (1). For instance, a majority of colorectal adenocarcinomas display increased expression levels of the COX-2 protein (2). Additionally, COX-2 expression levels have been associated with invasion depth (3), liver metastasis (4), and poor clinical outcome (5). Thus, the pharmacological inhibition of COX-2 activity has been explored as a therapeutic option for the management of colorectal cancer. In fact, administration of celecoxib, a selective COX-2 inhibitor, for 6 months resulted in a 28% reduction in the mean number of colorectal polyps in patients with familial adenomatous polyposis (6). Furthermore, clinical trials of colorectal cancer patients are currently underway investigating the addition of celecoxib to conventional chemotherapeutic regimens (7).
Investigations into celecoxib’s mechanism of action have shown that in vitro administration leads to growth arrest and induction of apoptosis in a variety of cancer cells (8–12). Interestingly, celecoxib’s anti-cancer effects appear to be independent of COX-2 inhibition, involving the inhibition of cell cycle progression, induction of apoptosis and inhibition of angiogenesis (Reviewed in (13)). Investigations into celecoxib’s mechanisms of action in vivo have also been conducted. Gene expression analysis of healthy colon tissue from patients with hereditary nonpolyposis colon cancer (HNPCC) receiving celecoxib for 12 months displayed gene expression changes suggestive of alterations in immune response, cell adhesion and transforming growth factor-β signaling (14).
However, the mechanism with which celecoxib exerts its chemotherapeutics effects in colorectal tumors in vivo is currently not known. We conducted a Phase 0 study to examine the effects of celecoxib pre-treatment on primary colorectal adenocarcinoma gene expression to gain insight into the mechanisms underlying celecoxib’s effects in vivo. Our results indicate that celecoxib pre-treatment in vivo is associated with multiple gene expression changes consistent with alterations in cellular lipid metabolism, glutathione metabolism and cell adhesion, in addition to differential expression of select genes involved in the immune response, apoptosis and angiogenesis. These results suggest that celecoxib tips the balance away from cellular proliferation towards growth inhibition in colorectal adenocarcinoma cells.
Materials and Methods
Patients undergoing surgical resection of histologically proven primary colorectal adenocarcinomas were consented for participation in the study, which was approved by the Washington University, School of Medicine Institutional Review Board (Table 1). The patients enrolled in this study were randomized to receive either 400 mg celecoxib two times per day or no COX-2 inhibitor for 7 days prior to surgical resection. This dose of celecoxib (400 mg b.i.d.) was chosen because it elicited a significant decrease in the mean number of colorectal polyps in patients with familial adenomatous polyposis (6). A 7 day treatment regimen allowed the pharmacological evaluation to be performed without disrupting the surgical management of the colorectal adenocarcinomas. Patients in both groups were not allowed to take aspirin or other non-steroidal anti-inflammatory drugs (NSAIDS) before surgical resection to be eligible to participate in this study. Patients had received no prior therapy for their cancer. There were no apparent differences in either tumor cellularity or histology of the resection samples in the two groups.
Table 1.
Patient demographics
| Celecoxib | No Drug | |
|---|---|---|
| Gender: | ||
| Male | 6 | 4 |
| Female | 5 | 8 |
| Tumor Site: | ||
| Ascending Colon | 3 | 2 |
| Cecum | 3 | 4 |
| Descending Colon | 1 | 0 |
| Hepatic Flexure | 1 | 1 |
| Recto-Sigmoid Colon | 0 | 2 |
| Sigmoid Colon | 3 | 3 |
| Clinical Stage: | ||
| I | 2 | 2 |
| II | 3 | 6 |
| III | 2 | 2 |
| IV | 4 | 2 |
| Pathological Grade: | ||
| Poorly Differentiated | 2 | 2 |
| Moderately Differentiated | 7 | 9 |
| Well Differentiated | 2 | 1 |
Total RNA was isolated from surgically resected, histologically confirmed colorectal primary adenocarcinomas using a TRIzol RNA isolation kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommended protocol at the Siteman Cancer Center Tissue Procurement Core (St. Louis, MO). Total RNA (5 ug) from each sample was converted to double stranded cDNA using a dT-T7 promoter primer. The double stranded cDNA was then used as a template to synthesize biotinylated RNA, which was fragmented and hybridized to the Affymetrix HG_U95av2 microarray chip (Santa Clara, CA) using Affymetrix’s labeling and hybridization protocol within the Siteman Cancer Center Multiplex Gene Expression Core (St. Louis, MO).
The array data was imported into GeneSpring GX 7.3 (Santa Clara, CA) and normalized using GC-RMA. Genes with normalized expression standard deviations < 0.04 across all samples, essentially unchanging genes, were excluded from further analysis. Unsupervised, hierarchical clustering was performed on all samples and all changing genes (n = 10,083 genes) using uncentered correlation as the similarity matrix with complete linkage clustering was performed with Eisen’s clustering program (15). Using a volcano plot in GeneSpring, 190 genes were identified with an absolute fold change greater than 1.5 and non-parametric p-value < 0.05 between adenocarcinomas from patients receiving celecoxib pre-treatment and adenocarcinomas from patients receiving no drug. Hierarchical clustering (15) was performed on all samples using the 190 differentially expressed genes identified in the volcano plot, using uncentered correlation as the similarity matrix with complete linkage clustering. The resulting heatmap and dendrogram were visualized using Java TreeView (16). Gene expression enrichment analysis was performed using the analytical tools available at the DAVID website: http://david.abcc.ncifcrf.gov/ (17). The data discussed in this publication have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE11237.
Results
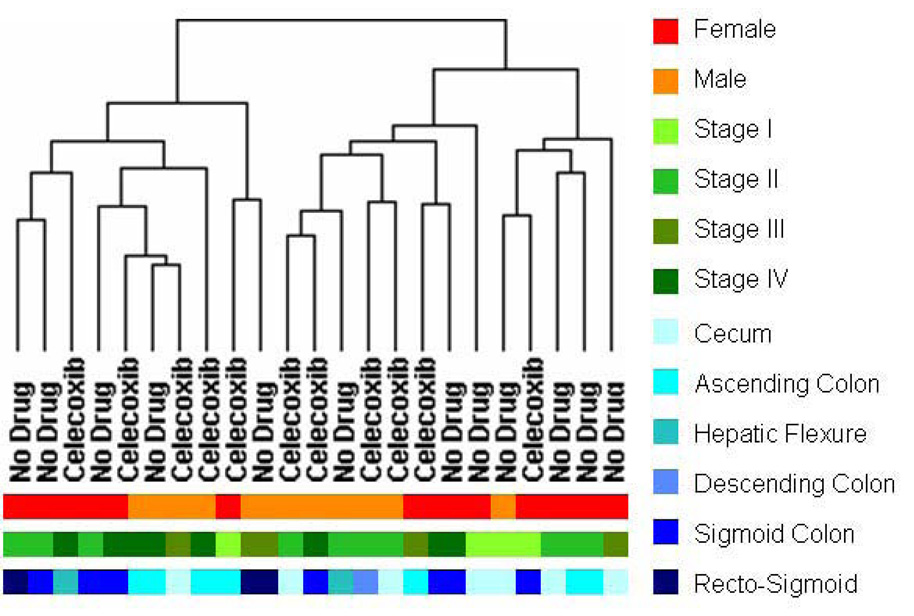
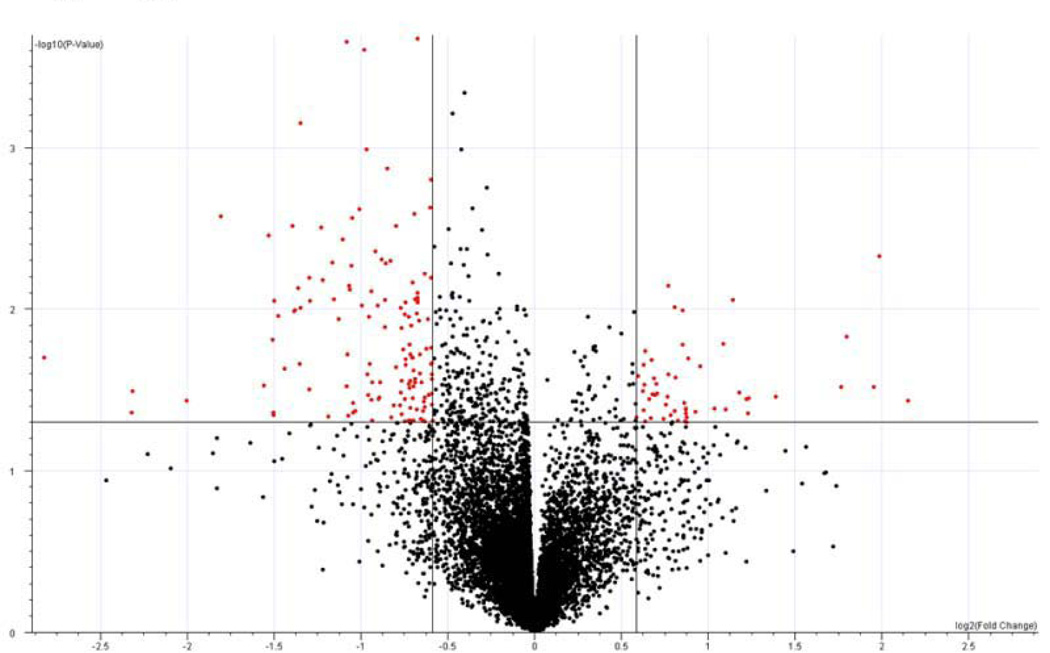
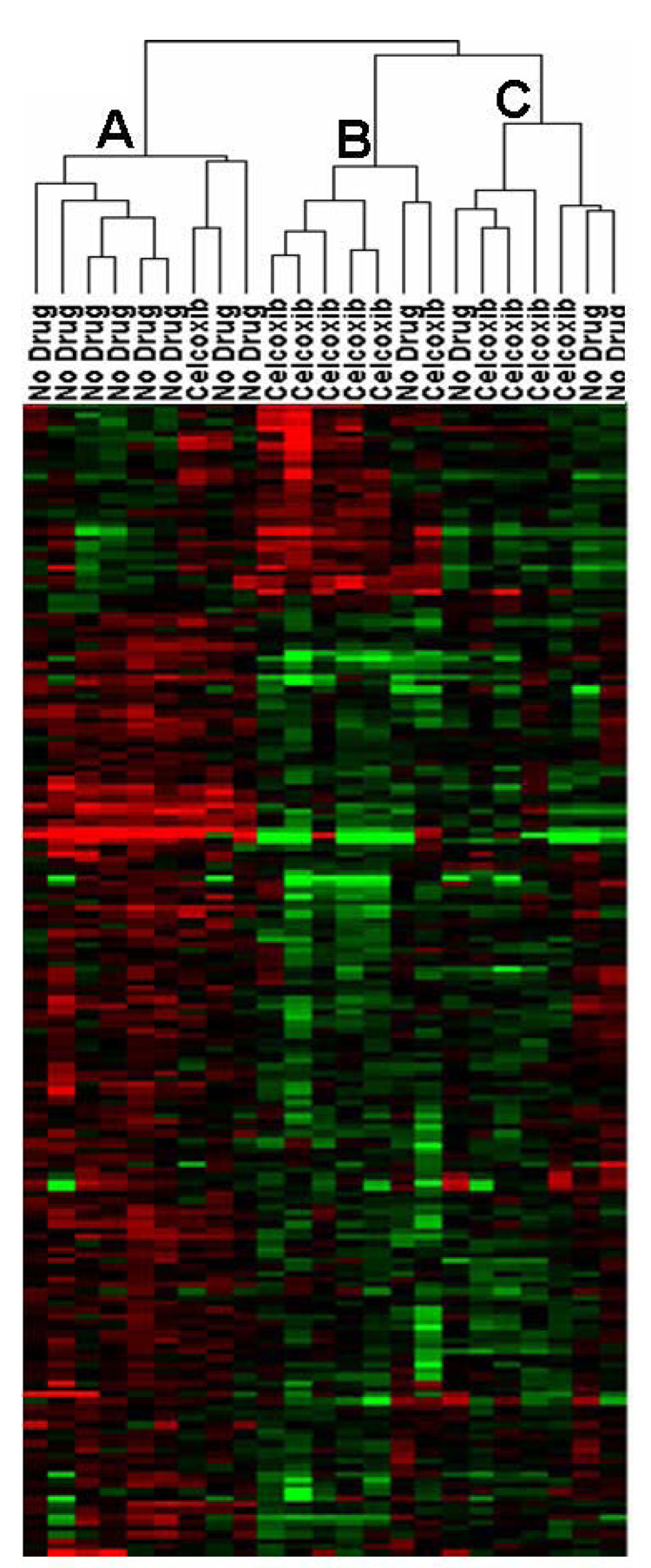
Unsupervised hierarchical clustering using all genes with differing levels of expression across all samples (n = 10,083 genes) showed no obvious separations based on: gender, tumor site, clinical stage or celecoxib pre-treatment status (Figure 1). Differentially expressed genes associated with celecoxib pre-treatment were identified by comparing gene expression intensities in celecoxib-treated patients and patients receiving no drug using a volcano plot. Using a > 1.5 absolute fold change and p<0.05 cut-offs, 35 genes displayed higher expression values and 155 genes displayed lower expression values in adenocarcinomas from patients receiving celecoxib relative to adenocarcinomas from patients receiving no drug (Figure 2A and Supplemental Table 1). Hierarchical clustering of the adenocarcinoma samples across the 199 differentially expressed genes separated the samples into 3 main sub-clusters (Figure 2B). In sub-cluster A, 8 of 9 tumor samples were from patients receiving no celecoxib pre-treatment. In sub-cluster B, 6 of 7 tumor samples were from patients receiving celecoxib pre-treatment. Sub-cluster C contains tumor samples from 4 patients receiving celecoxib and 3 patients receiving no drug.
Figure 1. Clustering of gene expression in colorectal adenocarcinoma patients.
Unsupervised hierarchical clustering was performed on all samples using 10,083 genes with a normalized expression standard deviations greater than 0.04. The dendrogram indicates that the samples do not cluster based solely on any of the recorded phenotypic characteristics.
Figure 2. Differentially expressed genes associated with celecoxib pre-treatment.
2A: Volcano plot of gene expression changes between colorectal adenocarcinomas from patients pre-treated and not pre-treated with celecoxib. 2B: Clustering using the 190 differentially expressed genes. Red indicates increased expression; green indicates decreased expression and black indicates no difference.
Gene annotation enrichment analysis was performed to examine which biological processes were altered by celecoxib pre-treatment. Celecoxib pre-treatment for 7 days was associated with the alteration of 23 genes involved in cellular lipid metabolism, 4 genes involved in glutathione metabolism and 19 genes involved in cell adhesion (Table 2). In addition to these significantly altered biological processes, tumor samples from patients receiving celecoxib pre-treatment displayed changes in expression levels for genes associated with growth arrest (growth arrest-specific 6, 1.7 fold change), apoptosis (BCL2-associated × protein, −1.9 fold change), angiogenesis (angiogenin, −2.8 fold change; kallikrein-1, −2.6 fold change) and inflammatory/immune response (interleukin-6, 4.4 fold change; S100 calcium binding protein A8, 2.4 fold change).
Table 2.
Differentially expressed genes in three overrepresented biological processes associated with celecoxib pre-treatment
| Symbol | Gene Name | Fold Change |
|---|---|---|
| Cellular Lipid Metabolism: p<0.000002* | ||
| ACAA2 | acetyl-Coenzyme A acyltransferase 2 | −1.86 |
| ACADSB | acyl- Coenzyme A dehydrogenase, short/branched chain | −1.63 |
| AGPAT2 | 1-acylglycerol-3-phosphate O-acyltransferase 2 | −1.65 |
| AKR1C1/AKR1C2 | aldo-keto reductase family 1, member C | 1.72 |
| C14ORF1 | chromosome 14 open reading frame 1 | −1.58 |
| CDS1 | CDP-diacylglycerol synthase 1 | −2.89 |
| CYP2J2 | cytochrome P450, family 2, subfamily J, polypeptide 2 | −2.15 |
| DGAT1 | diacylglycerol O-acyltransferase homolog 1 (mouse) | −2.04 |
| DHCR24 | 24-dehydrocholesterol reductase | −1.55 |
| ECH1 | enoyl Coenzyme A hydratase 1, peroxisomal | −1.51 |
| FABP1 | fatty acid binding protein 1, liver | −2.82 |
| FDFT1 | farnesyl-diphosphate farnesyltransferase 1 | −1.58 |
| HADHSC | L-3-hydroxyacyl-Coenzyme A dehydrogenase, short chain | −2.33 |
| HMGCR | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | −1.81 |
| HSD17B2 | hydroxysteroid (17-beta) dehydrogenase 2 | −2.11 |
| HSD17B4 | hydroxysteroid (17-beta) dehydrogenase 4 | −1.66 |
| PCCA | propionyl Coenzyme A carboxylase, alpha polypeptide | −2.60 |
| PCCB | propionyl Coenzyme A carboxylase, beta polypeptide | −2.25 |
| PIGF | phosphatidylinositol glycan anchor biosynthesis, class F | −1.60 |
| RARRES2 | retinoic acid receptor responder (tazarotene induced) 2 | 1.75 |
| SLC27A2 | solute carrier family 27 (fatty acid transporter), member 2 | −2.54 |
| UGT1A1 - A10 | UDP glucuronosyltransferase 1 family | −2.84 |
| UGT8 | UDP glycosyltransferase 8 | −2.54 |
| Glutathione Metabolism: p<0.001* | ||
| GGT1 | gamma-glutamyltransferase 1 | −1.54 |
| GGT2 | gamma-glutamyltransferase 2 | −1.80 |
| GGTL4 | gamma-glutamyltransferase-like 4 | −2.12 |
| GSR | glutathione reductase | −1.67 |
| Cell Adhesion: p<0.02* | ||
| CCL2 | chemokine (C-C motif) ligand 2 | 2.62 |
| CIB1 | calcium and integrin binding 1 (calmyrin) | −1.64 |
| CLDN7 | claudin 7 | −1.71 |
| COL16A1 | collagen, type XVI, alpha 1 | 2.34 |
| CTNNA1 | catenin (cadherin-associated protein), alpha 1, 102kDa | −1.68 |
| FCGBP | Fc fragment of IgG binding protein | −7.09 |
| GMDS | GDP-mannose 4,6-dehydratase | −2.07 |
| ITGA6 | integrin, alpha 6 | −1.58 |
| LAMA4 | laminin, alpha 4 | 1.81 |
| LGALS4 | lectin, galactoside-binding, soluble, 4 (galectin 4) | −2.57 |
| LOXL2 | lysyl oxidase-like 2 | 1.52 |
| MAEA | macrophage erythroblast attacher | −1.51 |
| MUC5AC | mucin 5AC, oligomeric mucus/gel-forming | −2.84 |
| PECAM1 | platelet/endothelial cell adhesion molecule (CD31 antigen) | 2.21 |
| TNC | tenascin C (hexabrachion) | 1.82 |
| TNFAIP6 | tumor necrosis factor, alpha-induced protein 6 | 1.93 |
| TSPAN1 | tetraspanin 1 | −2.61 |
| VCAM1 | vascular cell adhesion molecule 1 | 1.69 |
| VWF | von Willebrand factor | 1.80 |
NOTE: Fold change represents the difference in expression in celecoxib pre-treated patients versus patients receiving no drug pretreatment.
P-value represents the likelihood that the number of differentially expressed genes belonging to the biological process occurred by chance as determined by the Fischer’s exact test.
Discussion
This first analysis of gene expression alterations associated with celecoxib pre-treatment in patients with primary colorectal adenocarcinoma revealed changes of putative biological relevance. Pre-treatment for only 7 days with pharmacological doses of celecoxib (400 mg b.i.d.), resulted in alterations in numerous genes involved in cellular lipid metabolism, glutathione metabolism and cell adhesion. In general, most of the genes involved in cellular lipid metabolism (>90%) were down-regulated in patients receiving celecoxib pre-treatment relative to patients receiving no drug. Importantly, several transcripts involved in fatty acid β-oxidation (ACAA2, ECH1, HADHSC, HSD17B4; see Table 2 for gene names) were down-regulated, suggesting that the tumors from patients receiving celecoxib pre-treatment have a diminished capacity to generate energy. Since fatty acid metabolism has been shown to be elevated in colorectal adenocarcinomas compared to normal colonic tissue (18), celecoxib may exert its antiproliferative effects, at least in part, through counteracting the increase in fatty acid metabolism associated with colorectal carcinogenesis.
Celecoxib pre-treatment was also associated with a decrease in the relative expression of transcripts involved in glutathione metabolism, including 3 γ-glutamyltransferases (GGT1, GGT2, GGTL4) and glutathione reductase. The decreased relative expression of these transcripts suggests that tumors from patients receiving celecoxib pre-treatment have a reduced capacity to handle oxidative stress. Additionally, increased expression of GGT1 has been linked to the proliferative ability of colon cancer cells in vitro (19) by providing a growth advantage over cells devoid of GGT1 expression (20). Furthermore, the proliferation of colon cell lines is associated with increased activity of glutathione reductase, while decreased cell proliferation is associated with decreased cellular glutathione content (21). The decreased expression of glutathione metabolism related genes in this study therefore suggests a decrease in cellular glutathione content by celecoxib pre-treatment and by extension, perhaps decreasing the proliferative ability of the tumors.
In addition to altering expression of genes involved in lipid and glutathione metabolism, celecoxib pre-treatment affected the expression level of genes involved in cell adhesion. Several genes encoding for extracellular matrix proteins, including collagen, laminin, and von Willebrand factor, showed increased relative expression in celecoxib pre-treated patients. These results are in agreement with previous findings on the effects of celecoxib and other NSAIDs on gene expression. For instance, colorectal cancer cells treated in vitro with aspirin were also associated with the up-regulation of collagens (22). Alterations of genes involved in cell adhesion were also noted in healthy colons from HNPCC patients treated with celecoxib for 12 months (14). Another extracellular matrix protein, tenascin C, showed a relative increase in expression levels following celecoxib pre-treatment. Tenascin C expression has been found to be minimal in normal colonic mucosa, but elevated expression levels are observed in colorectal adenocarcinomas (23).
Other gene expression alterations observed in patients receiving celecoxib pre-treatment suggest a trend towards growth inhibitory influences in the tumors. For instance, the inflammatory modulators, interleukin-6 and CXCL6 (chemokine (C-X-C motif) ligand -6), displayed greater relative expression levels in tumor samples from patients receiving celecoxib pre-treatment. Similar increases in inflammatory modulators, including interleukin-6, CXCL2 and CXCL3, have been noted in a breast epithelial cell line treated with 50 µM celecoxib for 48 hours, a treatment regimen associated with growth inhibition and induced apoptosis (9). Angiogenin, a potent mediator of angiogenesis, has been found to promote proliferation of gastric cancer cells (24) and has also been correlated with focal macrophage infiltration in colorectal cancer (25). In this study, celecoxib pre-treatment was associated with decreased expression of angiogenin, again suggesting that celecoxib diminishes the proliferative ability of colorectal cancer cells.
We must note that the cut-offs for differential expression between the two treatment groups were of relatively low stringency. However, by using low stringency cut-offs for selecting differentially expressed genes, we have achieved the goal of this pilot study by identifying gene expression alterations associated with celecoxib pre-treatment, in vivo. It is promising that even a short course of celecoxib resulted in gene expression changes in colorectal adenocarcinomas. Previously, a 12 month regimen of celecoxib was found to induce expression changes in over 1,000 genes in healthy colonic mucosa from HNPCC gene carriers (14). Many of the expression changes were found in genes involved in immune response, cell adhesion, transforming growth factor-β signaling, cell proliferation and angiogenesis. Similarly, the 7 day regimen of celecoxib employed in this study showed gene expression alterations in colorectal tumor tissue, most notably changes in genes involved in cell proliferation, cell adhesion and immune response. Interestingly, no genes involved in transforming growth factor-β signaling were differentially expressed by the 7 day pre-treatment regimen of celecoxib, possibly indicating that these effects of celecoxib occur after prolonged exposure.
How do the results from this study inform on the usefulness of celecoxib as an option for colorectal cancer chemotherapy? This is an important issue, since the prolonged use of several selective COX-2 inhibitors, including celecoxib, has been found to increase the risk of cardiovascular side effects (26, 27). However, the cardiovascular risks associated with long-term use of celecoxib is rather ambiguous, as no increased cardiovascular risk was observed in Prevention of Spontaneous Adenomatous Polyps (PreSAP) trial (http://www.fda.gov/cder/drug/infopage/celebrex/celebrex-hcp.htm). Celecoxib appears to be safer than other selective COX-2 inhibitors, but care should be taken when given to patients already at risk for cardiovascular events (28). In regards to the therapeutic potential in colorectal cancer, celecoxib given by itself at currently recommended doses (400 mg b.i.d.) appears to lessen cellular proliferation of colorectal tumors, as evidenced by the results of this study. However, the observed gene expression alterations appear to be relatively minor and it still remains to be determined if a longer course of celecoxib treatment would significantly arrest the growth of colorectal cancers. However, combination therapy of celecoxib with other chemotherapeutics does not appear to be as promising. A recent Phase II study looking at the effects of celecoxib treatment in metastatic colorectal cancer patients indicates that celecoxib adds no added benefit to combined 5-fluorouracil/oxaliplatin chemotherapy (29). Additionally, previous in vitro findings suggest that the combined use of celecoxib with 5-fluorouracil could be counterproductive, as the growth arrest elicited by celecoxib interferes with 5-fluorouracil mediated cytotoxicity (29, 30). Further investigation is warranted to determine if celecoxib can by employed as an adjunct to chemotherapeutic regimens that do not rely on enhanced cellular proliferation for cytotoxicity.
In conclusion, our results suggest that celecoxib reduces the proliferative capacity of colorectal adenocarcinomas. As with previous results, this small, short-term study, indicates that celecoxib treatment is associated with gene expression alterations in the tumor itself that are indicative of decreased cellular lipid and glutathione metabolism, along with an increased immune response. These gene expression alterations, taken together, suggest that celecoxib pre-treatment leads to growth inhibitory influences on colorectal tumors. This study suggests possible gene expression alterations mediating the effects of celecoxib treatment on colorectal adenocarcinoma cells in vivo. Additionally, these results should provide the impetus for larger studies in the future, either using celecoxib treatment alone or in combination with other chemotherapeutic regimens.
Supplementary Material
Acknowledgments
We would like to thank the staff of the Tissue Procurement Core and the Multiplexed Gene Analysis Core in the Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, Mo. The efforts of the participating colorectal surgeons (Drs. Birnbaum, Dietz, Fleshman, Kodner, Mutch), Michael Lewis, the Barnes-Jewish Hospital Pharmacy Department (Dr. Stephanie Mayo), and Derek Van Booven is greatly appreciated. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant #P30 CA91842. This work was also supported by NIH grants U01GM63340.
Grant support: NIH grants U01GM63340 and P30 CA091842.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: All of the authors state that they do not have any conflicts of interest.
References
- 1.Church RD, Fleshman JW, McLeod HL. Cyclo-oxygenase 2 inhibition in colorectal cancer therapy. Br J Surg. 2003;90(9):1055–1067. doi: 10.1002/bjs.4297. [DOI] [PubMed] [Google Scholar]
- 2.Masunaga R, Kohno H, Dhar DK, et al. Cyclooxygenase-2 expression correlates with tumor neovascularization and prognosis in human colorectal carcinoma patients. Clin Cancer Res. 2000;6(10):4064–4068. [PubMed] [Google Scholar]
- 3.Fujita T, Matsui M, Takaku K, et al. Size- and invasion-dependent increase in cyclooxygenase 2 levels in human colorectal carcinomas. Cancer Res. 1998;58(21):4823–4826. [PubMed] [Google Scholar]
- 4.Tomozawa S, Tsuno NH, Sunami E, et al. Cyclooxygenase-2 overexpression correlates with tumour recurrence, especially haematogenous metastasis, of colorectal cancer. Br J Cancer. 2000;83(3):324–328. doi: 10.1054/bjoc.2000.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheehan KM, Sheahan K, O'Donoghue DP, et al. The relationship between cyclooxygenase-2 expression and colorectal cancer. JAMA. 1999;282(13):1254–1257. doi: 10.1001/jama.282.13.1254. [DOI] [PubMed] [Google Scholar]
- 6.Steinbach G, Lynch PM, Phillips RKS, et al. The effect of celecoxib, a cyclooxygenase-2 inhibitor, in familial adenomatous polyposis. N Engl J Med. 2000;342(26):1946–1952. doi: 10.1056/NEJM200006293422603. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs C, Marshall J, Mitchell E, et al. A randomized trial of first-line irinotecan/fluoropymidine combinations with or without celecoxib in metastatic colorectal cancer (BICC-C) J Clin Oncol (Meeting Abstracts) 2006;24(18suppl):3506. [Google Scholar]
- 8.Grösch S, Tegeder I, Niederberger E, Bräutigam L, Geisslinger G. COX-2 independent induction of cell cycle arrest and apoptosis in colon cancer cells by the selective COX-2 inhibitor celecoxib. FASEB J. 2001;15:2742–2744. doi: 10.1096/fj.01-0299fje. [DOI] [PubMed] [Google Scholar]
- 9.Levitt RJ, Buckley J, Blouin M-J, Schaub B, Triche TJ, Pollak M. Growth inhibition of breast epithelial cells by celecoxib is associated with upregulation of insulin-like growth factor binding protein-3 expression. Biochem Biophys Res Commun. 2004;316(2):421–428. doi: 10.1016/j.bbrc.2004.02.062. [DOI] [PubMed] [Google Scholar]
- 10.Hsu A-L, Ching T-T, Wang D-S, Song X, Rangnekar VM, Chen C-S. The cyclooxygenase-2 inhibitor celecoxib induces apoptosis by blocking Akt activation in human prostate cancer cells independently of Bcl-2. J Biol Chem. 2000;275(15):11397–11403. doi: 10.1074/jbc.275.15.11397. [DOI] [PubMed] [Google Scholar]
- 11.Arico S, Pattingre S, Bauvy C, et al. Celecoxib induces apoptosis by inhibiting 3-phosphoinositide-dependent protein kinase-1 activity in the human colon cancer HT-29 cell line. J Biol Chem. 2002;277(31):27613–27621. doi: 10.1074/jbc.M201119200. [DOI] [PubMed] [Google Scholar]
- 12.Kim SH, Song SH, Kim SG, et al. Celecoxib induces apoptosis in cervical cancer cells independent of cyclooxygenase using NF-κB as a possible target. J Cancer Res Clin Oncol. 2004;130(9):551–560. doi: 10.1007/s00432-004-0567-6. [DOI] [PubMed] [Google Scholar]
- 13.Grösch S, Maier TJ, Schiffmann S, Geisslinger G. Cyclooxygenase-2 (COX-2)-independent anticarcinogenic effects of selective COX-2 inhibitors. J Natl Cancer Inst. 2006;98(11):736–747. doi: 10.1093/jnci/djj206. [DOI] [PubMed] [Google Scholar]
- 14.Glebov OK, Rodriguez LM, Lynch P, et al. Celecoxib treatment alters the gene expression profile of normal colonic mucosa. Cancer Epidemiol Biomarkers Prev. 2006;15(7):1382–1391. doi: 10.1158/1055-9965.EPI-04-0866. [DOI] [PubMed] [Google Scholar]
- 15.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Nat. Acad. Sci. USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 17.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 18.Yeh C-S, Wang J-Y, Cheng T-L, Juan C-H, Wu C-H, Lin S-R. Fatty acid metabolism pathway play an important role in carcinogenesis of human colorectal cancers by Microarray-Bioinformatics analysis. Cancer Lett. 2006;233(2):297–308. doi: 10.1016/j.canlet.2005.03.050. [DOI] [PubMed] [Google Scholar]
- 19.Hanigan MH, Gallagher BC, Townsend DM, Gabarra V. {gamma}-glutamyl transpeptidase accelerates tumor growth and increases the resistance of tumors to cisplatin in vivo. Carcinogenesis. 1999;20(4):553–559. doi: 10.1093/carcin/20.4.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanigan MH. Expression of gamma-glutamyl transpeptidase provides tumor cells with a selective growth advantage at physiologic concentrations of cyst(e)ine. Carcinogenesis. 1995;16(2):181–185. doi: 10.1093/carcin/16.2.181. [DOI] [PubMed] [Google Scholar]
- 21.Bravard A, Beaumatin J, Dussaulx E, Lesuffleur T, Zweibaum A, Luccioni C. Modifications of the anti-oxidant metabolism during proliferation and differentiation of colon tumor cell lines. Int J Cancer. 1994;59(6):843–847. doi: 10.1002/ijc.2910590622. [DOI] [PubMed] [Google Scholar]
- 22.Iizaka M, Furukawa Y, Tsunoda T, Akashi H, Ogawa M, Nakamura Y. Expression profile analysis of colon cancer cells in response to sulindac or aspirin. Biochem Biophys Res Commun. 2002;292(2):498–512. doi: 10.1006/bbrc.2002.6648. [DOI] [PubMed] [Google Scholar]
- 23.Hanamura N, Yoshida T, Matsumoto E-i, Kawarada Y, Sakakura T. Expression of fibronectin and tenascin-C mRNA by myofibroblasts, vascular cells and epithelial cells in human colon adenomas and carcinomas. Int J Cancer. 1997;73(1):10–15. doi: 10.1002/(sici)1097-0215(19970926)73:1<10::aid-ijc2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Zhang S, Chen YP, Lin JY. Increased expression of angiogenin in gastric carcinoma in correlation with tumor angiogenesis and proliferation. World J Gastroenterol. 2006;12(32):5135–5139. doi: 10.3748/wjg.v12.i32.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Etoh T, Shibuta K, Barnard GF, Kitano S, Mori M. Angiogenin expression in human colorectal cancer: the role of focal macrophage infiltration. Clin Cancer Res. 2000;6(9):3545–3551. [PubMed] [Google Scholar]
- 26.Bresalier RS, Sandler RS, Quan H, et al. Cardiovascular events associated with rofecoxib in a colorectal adenoma chemoprevention trial. N Engl J Med. 2005;352(11):1092–1102. doi: 10.1056/NEJMoa050493. [DOI] [PubMed] [Google Scholar]
- 27.Solomon SD, McMurray JJ, Pfeffer MA, et al. Cardiovascular risk associated with celecoxib in a clinical trial for colorectal adenoma prevention. N Engl J Med. 2005;352(11):1071–1080. doi: 10.1056/NEJMoa050405. [DOI] [PubMed] [Google Scholar]
- 28.Antoniou K, Malamas M, Drosos AA. Clinical pharmacology of celecoxib, a COX-2 selective inhibitor. Expert Opin Pharmacother. 2007;8(11):1719–1732. doi: 10.1517/14656566.8.11.1719. [DOI] [PubMed] [Google Scholar]
- 29.André T, Tournigand C, Mineur L, et al. Phase II study of an optimized 5-fluorouracil-oxaliplatin strategy (OPTIMOX2) with celecoxib in metastatic colorectal cancer: a GERCOR study. Ann Oncol. 2007;18(1):77–81. doi: 10.1093/annonc/mdl336. [DOI] [PubMed] [Google Scholar]
- 30.Lim YJ, Rhee JC, Bae YM, Chun WJ. Celecoxib attenuates 5-fluorouracil-induced apoptosis in HCT-15 and HT-29 human colon cancer cells. World J Gastroenterol. 2007;13(13):1947–1952. doi: 10.3748/wjg.v13.i13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.