Abstract
Conotoxin ι-RXIA, from the fish-hunting species Conus radiatus, is a member of the recently characterized I1-superfamily, which contains eight cysteine residues arranged in a −C-C-CC-CC-C-C- pattern. ι-RXIA (formerly designated r11a) is one of three characterized I1 peptides in which the third last residue is post-translationally isomerized to the d- configuration. Naturally occurring ι-RXIA with d-Phe44 is significantly more active as an excitotoxin than the l-Phe analogue both in vitro and in vivo. We have determined the solution structures of both forms by NMR spectroscopy, the first for an I1-superfamily member. The disulfide connectivities were determined from structure calculations and confirmed chemically as 5-19, 12-22, 18-27, and 21-38, suggesting that ι-RXIA has an ICK structural motif with one additional disulfide (21-38). Indeed, apart from the first few residues, the structure is well defined up to around residue 35 and does adopt an ICK structure. The C-terminal region, including Phe44, is disordered. Comparison of the d-Phe44 and l-Phe44 forms indicates that the switch from one enantiomer to the other has very little effect on the structure, even though it is clearly important for receptor interaction based on activity data. Finally, we identify the target of ι-RXIA as a voltage-gated sodium channel; ι-RXIA is an agonist, shifting the voltage dependence of activation of mouse NaV1.6 expressed in Xenopus oocytes to more hyperpolarized potentials. Thus, there is a convergence of structure and function in ι-RXIA, as its disulfide pairing and structure resemble those of funnel web spider toxins that also target sodium channels.
Through adaptive evolution, marine cone snails of the genus Conus have generated more than 70,000 different venom peptides (conopeptides) (1-3). These highly diverse peptides can be organized into several structural classes, with peptides in the same structural class generally belonging to the same gene superfamily. The peptides in a given superfamily share a highly conserved precursor signal sequence and a disulfide scaffold with a characteristic number, pattern, and pairing of cysteine residues in the mature toxin (1-3). Thus, despite hypermutation of amino acids between half-cystines, the disulfide framework remains conserved within a superfamily. In addition to the accelerated evolution of conopeptide sequences, further molecular diversity is introduced through post-translational modifications. The occurrence of diverse modifications in Conus peptides is now well established (4) but the functional consequences of most post-translational modifications remain unknown, and mechanistic insights require detailed structure/function analyses, which have not been performed in most cases.
Recently, we described the I-superfamily of Conus peptides, defined by framework #11, with eight cysteine residues arranged in two pairs of adjacent Cys flanked by nonadjacent Cys residues (-C-C-CC-CC-C-C-)(5-7). A preliminary analysis of Conus cDNA clones reveals that framework #11 peptides are found broadly across the genus and an extreme diversity of sequences is observed (7, 8). For example, seventeen different cDNA clones were identified in the venom of Conus radiatus alone. These varied toxins, though sharing the same cysteine arrangement, can be divided into two distinct genetic groups, designated the I1- and I2-superfamilies (7). Members of the I1-class have been isolated from the fish-hunting species Conus radiatus (5), and members of the I2-class from worm-hunting species such as Conus betulinus (9) and Conus virgo (10). I1-superfamily conotoxins are excitatory (5-7), and, as we show in this report, the most extensively studied of these, ι-RXIA1 (formerly known as r11a) (11), targets the Nav1.6 subtype of voltage-gated sodium channels (VGSC), but had no effect on KCNQ2/3 and several members of the KV1 family of potassium channels tested. In contrast, two I2-superfamily peptides have been shown to target specific K+ channels, namely the Ca2+- and voltage-sensitive BK channels (9), and the Kv1.1 and Kv1.3 channel subtypes of the Shaker subfamily (10).
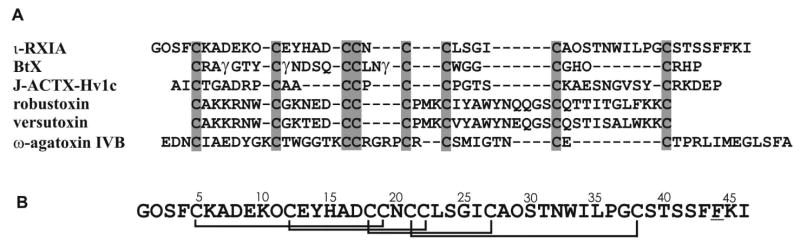
Although framework #11 defines the I-superfamily of Conus peptides, peptides with the same pattern of half-cystines have also been found in the venoms of Australian funnel-web spiders (12). Figure 1A compares the sequences of three framework #11 peptides, two from Conus and one from a spider. Three other spider toxins with four disulfide bridges, although not in a framework #11 pattern, are also shown as they are discussed below. These toxins target a range of ion channels, including Na+, Ca2+ and possibly K+ channels (12-16).
FIGURE 1.
Amino acid sequence and disulfide connectivities. (A) Comparison of various toxin sequences containing four disulfide bridges; the first three have a framework #11 pattern of half-cystine spacings (-C-C-CC-CC-C-C-). SwissProt accession numbers are as follows: ι-RXIA, Q7Z094; BtX, Q9U3Z3; J-atracotoxin Hv1c, P82228; robustoxin, P01478; versutoxin, P13494; ω-agatoxin IVB, P37045. In the I2-superfamily conotoxin BtX, all three Glu residues in the mature toxin sequence are γ-carboxyGlu (9). (B) Amino acid sequence of ι-RXIA showing disulfide connectivities determined in this work by NMR and chemical mapping. The underlined residue, Phe44, has a d- configuration.
For ι-RXIA, a clear demonstration of the functional importance of one post-translationally modified amino acid was reported recently (6). This 46-residue conotoxin contains a d-Phe residue in position 44, three residues from the C-terminus. The naturally occurring peptide and its synthetic counterpart (with d-Phe44) were equipotent in eliciting repetitive action potentials in axons, whereas the l-Phe44-containing analogue was inactive in this assay and 10 to 20-fold less active than ι-RXIA in an in-vivo assay (6). In order to explore the relationship between structure and activity of ι-RXIA, we have determined the solution structure of the l-Phe-44-containing peptide and compared it with the d-Phe44 form. The structure was similar to those of funnel web spider toxins that target Na+ channels. We then showed that ι-RXIA was an agonist of the NaV1.6 sodium channel, shifting its voltage dependence of activation to more hyperpolarized potentials, but had no effect on several members of the KV1 family of potassium channels tested.
Experimental Procedures
Peptide Synthesis and Oxidative Folding
Peptides, ι-RXIA and ι-RXIA[l-Phe44] were synthesized on solid supports by an automated peptide synthesizer using N-Fmoc (N-(9-fluorenyl)methoxycarbonyl)-protected amino acids, 2-(-1H-benzo-triazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and diisopropylethylamine (DIEA), courtesy of Dr. Robert Schackmann of the DNA/Peptide Facility, University of Utah. All cysteine residues were trityl-protected; the coupling time was 1 h. Peptide cleavage/deprotection was accomplished with reagent K (82.5% TFA (trifluoroacetic acid): 5% phenol: 5% H2O: 5% thioanisole: 2.5% 1,2-ethanedithiol) for 2 h at room temperature. Soluble crude peptides were precipitated with cold methyl t-butyl ether (MTBE) and centrifuged. The pellet was washed with methyl t-butyl ether, centrifuged again, then dissolved in 25% acetonitrile in 0.1% TFA and lyophilized. The linear peptides were purified on a Vydac C18 semi-preparative HPLC column with a gradient of 30-60% solvent B (0.1% TFA in 90% acetonitrile; solvent A was 0.1% TFA in water) over 30 min at a flow rate of 3 ml/min.
Oxidative folding of the peptides was carried out in a solution of 0.1 M Tris/HCl, pH 8.7, containing 1 mM EDTA, 1 mM oxidized glutathione and 1 mM reduced glutathione at room temperature for 3 h. The reaction was initiated by adding the linear synthetic peptide to a final concentration of 20 μM then quenched by adding formic acid to a final concentration of 8%. The oxidized peptide was purified on a Vydac C18 semipreparative HPLC column with a gradient of 15-60% solvent B over 40 min at a flow rate of 3 ml/min.
Disulfide Mapping of ι-RXIA
The disulfide-bonding framework of ι-RXIA was determined by the partial reduction (17) and stepwise alkylation method. Ten nmol of oxidized peptide was dissolved in 100 μl of 20% acetonitrile in water and partially reduced by addition of 120 μl of 20 mM TCEP in 0.2 M sodium citrate, pH 3.0 and incubation for 5 min at room temperature. The reaction was stopped by immediate injection on Vydac C18 analytical HPLC column equilibrated at 45 °C, and the partially reduced intermediates were eluted with a shallow gradient of 32-38% solvent B over 1 h at a flow rate of 1 ml/min. Individual partially reduced species were collected manually and stored at −20°C for no longer than 2 h before the first alkylation. Typically, the fractions from two independent runs were pooled and contained about 0.5-1 nmol of peptide in 2 ml of solvent.
For alkylation with NEM (N-ethylmaleimide) an equivalent volume (2 ml) of 60 mM NEM in 0.2 M sodium citrate, pH 3.0, was added directly to the fraction containing partially reduced peptide. After incubation for 1 h at room temperature, the mixture was applied to a Vydac C18 analytical HPLC column, equilibrated at 45°C, and eluted with 30% solvent B for 15 min. followed by gradient of 30-55% solvent B for 25 min then by gradient of 55-100% solvent B for 5 min. at a flow rate of 1 ml/min. Collected fractions were dried and stored at −20°C for no longer than overnight. The first alkylation yielded between 0.25 and 1 nmol of a single species of NEM-labeled peptide.
For the complete reduction and alkylation with IAA (iodoacetic acid), the NEM-labeled peptide (∼1 nmol) was dissolved in 40 μl of 50% acetonitrile in 0.1% TFA, and reduced by addition of 10 μl of reducing buffer (0.5 M Tris/acetate, pH 8.0, containing 5 mM EDTA and 100 mM dithiothreitol) and incubation at 65 °C for 30 min. The peptide was then alkylated with 40 μl of 0.5 M IAA in 0.5 M Tris/acetate, pH 8.0, containing 5 mM EDTA in the dark at room temperature for 30 min. The reaction mixture was applied to a Vydac C18 analytical HPLC column, equilibrated at 45 °C, and eluted as described above for the first-step alkylation.
The completed partial reduction and stepwise alkylation yielded between 0.1 and 0.3 nmol of a given species of NEM- and IAA-double-labeled product, starting from 80-100 nmol of oxidized peptide. The doubly alkylated variants of ι-RXIA were sequenced by standard automated Edman degradation performed with an Applied Biosystem Model 492 Sequenator, courtesy of Dr. Robert Schackmann of the DNA/Peptide Synthesis Facility, University of Utah.
Mass Spectrometry
Matrix-assisted laser desorption ionization (MALDI) mass spectra of the native and synthetic peptides were obtained using a Bruker REFLEX time-of-flight mass spectrometer (Bruker Daltonics) fitted with gridless reflectron, an N2 laser and a 100 MHz digitizer at the Salk Institute for Biological Studies (La Jolla, CA, USA).
Mass spectrometry of partially reduced forms of ι-RXIA was performed with a Sciex API-III electrospray ionization (ESI) quadruple mass spectrometer, courtesy of Dr. Chad Nelson of the Mass Spectrometry and Proteomics Core Facility, University of Utah. Molecular masses of all variants were within 1.0 amu of calculated values.
NMR Spectroscopy
Samples were prepared for NMR by dissolving lyophilized peptide in 95% H2O/ 5% 2H2O to a final peptide concentration of 1.1 mM for ι-RXIA[l-Phe44], and 0.8 mM for ι-RXIA. At 5°C and 25°C, a series of one-dimensional spectra was collected at pH 2.8, 5.9 and 6.9, respectively, for both peptides. Two-dimensional homonuclear TOCSY with a spin-lock time of 70-ms, DQF-COSY and NOESY spectra were recorded at pH 5.9 on Bruker DRX-600 and Avance 800 spectrometers. NOESY spectra were recorded at mixing times of 50, 100, 250 and 300 ms. A 250-ms NOESY spectrum was used for resonance assignments and NOE intensity measurements. To monitor backbone amide exchange, a series of 1D spectra was recorded at various time intervals for the sample in 100% 2H2O at 278 K on the 600 MHz spectrometer. For ι-RXIA, the translational diffusion coefficient was measured at 5 °C using a PFG longitudinal eddy-current delay pulse sequence (18, 19), as implemented by Yao et al. (20). Spectra were referenced to an impurity peak at 0.15 ppm which has been referenced previously against the internal standard dioxane (3.75 ppm) and does not vary with solution conditions over the pH and temperature range used in this study. Spectra were processed using TopSpin, version 1.3, and analysed using XEASY, version 1.3.13 (21). 1H-13C HSQC spectra for the assignment of 13C chemical shifts and a 1H-15N HSQC spectrum for the assignment of 15N chemical shifts (22, 23) were collected at 25 °C on a Bruker Avance 500 spectrometer equipped with a cryoprobe. 15N and 13C chemical shifts are presented in Tables S1 and S2 in Supporting Information.
Structure Calculations
Distance constraints were taken from the volumes of NOESY cross-peaks at 25 °C, pH 5.9 and 800 MHz. NOE buildup rates were monitored for a number of crosspeaks at 25 °C, pH 5.9 and 600 MHz, as shown in Figure S1 in Supporting Information. 3JHNHα values were measured from a DQF-COSY spectrum and converted to ϕ dihedral restraints as follows: 3JHNHα > 8 Hz, ϕ = -120 ± 40°; 3JHNHα <6 Hz, ϕ = -60 ±30°. For residues with 6<3JHNHα<8 Hz, if NOE data showed they did not have positive ϕ angles, restraints with ϕ = -90±90° were applied in the structure calculation. χ1 angles for residues 5, 6, 8, 10, 12, 14, 17, 18, 21, 27 were determined by analysing NOESY spectra with a short mixing time of 50 ms. Intensities of NOE cross-peaks measured in XEASY were calibrated using the CALIBA macro from the program CYANA (24). Initial structures were calculated using torsion angle dynamics and simulated annealing protocols in CYANA and structures were optimized for a low target function. These structures were used to identify hydrogen bond partners for backbone amide protons that were found to be in slow or intermediate exchange with solvent; where such an amide formed a bond with the same partner in ≥ 80% of the CYANA structures this hydrogen bond was included as a restraint in the final round of structure calculations. For each hydrogen bond constraint, upper limits of 2.4 and 3.3 Å were used for the distances from proton to acceptor and donor nitrogen atom to acceptor, respectively. The final constraint set was then used to calculate a new family of 200 structures using XPLOR-NIH (25). The 50 lowest energy structures were selected for energy minimization in a box of water. A final family of 20 lowest energy structures was chosen for analysis using PROCHECK-NMR (26) and MOLMOL (27). The thirty lowest energy structures before water refinement had no experimental distance violations > 0.3 Ǻ or dihedral angle violations > 5°. For the final 20 structures after water refinement, no distance violations > 0.3 or dihedral angle violations > 5° were present. Structural figures were prepared using MOLMOL (27), PyMol (28) and GRASP (29).
Electrophysiology
The clone for the mouse NaV1.6 channel subtype was kindly provided by Alan Goldin and cRNA was prepared from it by Layla Azam. Clones for human KCNQ2/3 and KV1.2 through 1.6 were purchased from PROTINAC GmbH, Hamburg, Germany, and corresponding cRNAs were produced using the mMessage mMachine RNA trancription kit from Ambion (Austin, TX). Each oocyte was injected with 1.5 ng NaV1.6, 2.5 ng KCNQ2 plus 2.5 ng KCNQ3, or 5 ng (KV1.2, 1.3, 1.4, 1.5 or 1.6) of cRNA in 50 nl of distilled water and incubated at 16 °C in ND96 (composition in mM: 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, 5 HEPES, pH ∼7.3) supplemented with Pen/Strep, Septra and Amikacin for 2 to 6 days. The recording chamber consisted of a cylindrical well (4 mm diameter, total volume ∼ 30 μl) in a wafer made of Sylgard (Dow Corning, Midland, MI). Oocytes were two-electrode voltage clamped with a Warner OC-725C amplifier (Warner Instruments, Hamden, CT) using glass microelectrodes filled with 3 M KCl (0.1 to 0.5 MΩ resistance). The chamber was perfused with ND96 for NaV1.6 or KCM 4.1.1 (96 mM NaCl, 4 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM HEPES) for KCNQ2/3, or frog Ringer's (115 mM NaCl, 2.5 mM KCl, 1.8 mM CaCl2, 10 mM HEPES, pH 7.4) for KV1 channels. Both solutions also contained 0.1 mg/ml bovine serum albumin. For NaV1.6, the membrane potential was held at −100 mV, and VGSCs were activated by 60 ms depolarising steps applied every 20 s; the protocol for inactivation measurements was essentially the same except a 30 ms test pulse to −10 mV was applied immediately after a 500 ms conditioning pulse of varying amplitude. For KCNQ2/3, the membrane potential was held at -80 mV and channels were activated by a 2 s step to 0 mV applied every 10 s. After each pulse, deactivating (‘tail’) currents were recorded at a potential of -60 mV. For KV1 channels, the membrane potential was held at −90 mV and channels were activated by a 150 ms step to +20 mV applied every 60 s. Current signals were low-pass filtered at 2 kHz (NaV1.6) or 1 kHz (potassium channels), digitized at a sampling frequency of 10 kHz, and leak-subtracted by a P/6 protocol using in-house software written in LabVIEW (National Instruments, Austin, Texas). Voltage dependencies of activation and inactivation data were curve-fit to the Bolzmann equation using Graphpad Prism (GraphPad Software, San Diego, CA). ι-RXIA was dissolved in ND96 (for NaV1.6), KCM 4.1.1 (for KCNQ2/3) or Ringer's (for KV1 channels) with 0.1 mg/ml bovine serum albumin and applied by halting the perfusion, injecting 3 μl of peptide solution (at ten times the final concentration) into the 30 μl bath with a pipettor, and manually stirring the bath for about 5 s by gently aspirating and expelling ∼ 5 μl of bath fluid several times with the pipettor. Responses in the presence of peptide, as well as control responses, were acquired in static baths to conserve peptide. All experiments were conducted at room temperature (∼ 22 °C).
Results
Peptide Synthesis
Two variants of ι-RXIA, with l- or d-Phe44, where synthesized on a solid support using a standard Fmoc protocol. Peptides were folded in the presence of oxidized and reduced glutathione as described in Experimental Procedures. The HPLC profiles of the folding reactions for the l- or d-Phe44 forms of ι-RXIA were identical, and the yield of oxidation was about 80% in both cases. As shown previously by HPLC co-elution experiments, the fully oxidized form of the peptide containing d-Phe44 was identical to the natural toxin isolated from the venom of Conus radiatus (6). In this paper we refer to the d-Phe44 version of the peptide simply as ι-RXIA and the l-Phe44 version as ι-RXIA[l-Phe44]. Both synthetic variants of ι-RXIA were used to determine and compare the solution structures, while the cysteine bridging pattern was determined chemically for the biologically-active, d-Phe44-containing variant.
NMR Spectroscopy
One-dimensional 1H NMR spectra of ι-RXIA[l-Phe44] at pH 2.8, 5.9 and 6.9 indicated that the toxin was partially denatured at the lower pH but well structured at both pH 5.9 and 6.9 (Figure S2 in Supporting Information). One- and two-dimensional spectra recorded at 5 and 25 °C showed that only one conformation was present in aqueous solution, Although ι-RXIA contains one proline and three hydroxyproline residues, there was no evidence of cis-trans isomerisation in either ι-RXIA or ι-RXIA[l-Phe44].
Sequence-specific chemical shift assignments for backbone and side-chain protons were made at 600 and 800 MHz by analysing standard two-dimensional homonuclear NMR spectra. Assignments at 25 °C and pH 5.9 for ι-RXIA[l-Phe44] and ι-RXIA are tabulated in Tables S1 and S2 in Supporting Information and deposited in BioMagResBank (30) with accession numbers 15174 and 15175, respectively. Figure 2 shows the deviations of the backbone NH and CαH chemical shifts from random coil values (31) at 25 °C for ι-RXIA[l-Phe44] (Fig. 2A) and ι-RXIA (Figure 2B). For ι-RXIA, the translational diffusion coefficient at 5 °C was 1.17± 0.04 × 10-10 m2s-1. This is lower than the corresponding value of 1.38 ± 0.03 × 10-10 m2s-1 at 5 °C for Magi 5, a 29-residue spider toxin with a compact ICK motif structure (32), reflecting the larger mass of ι-RXIA and its more extended C-terminal tail (described below).
FIGURE 2.
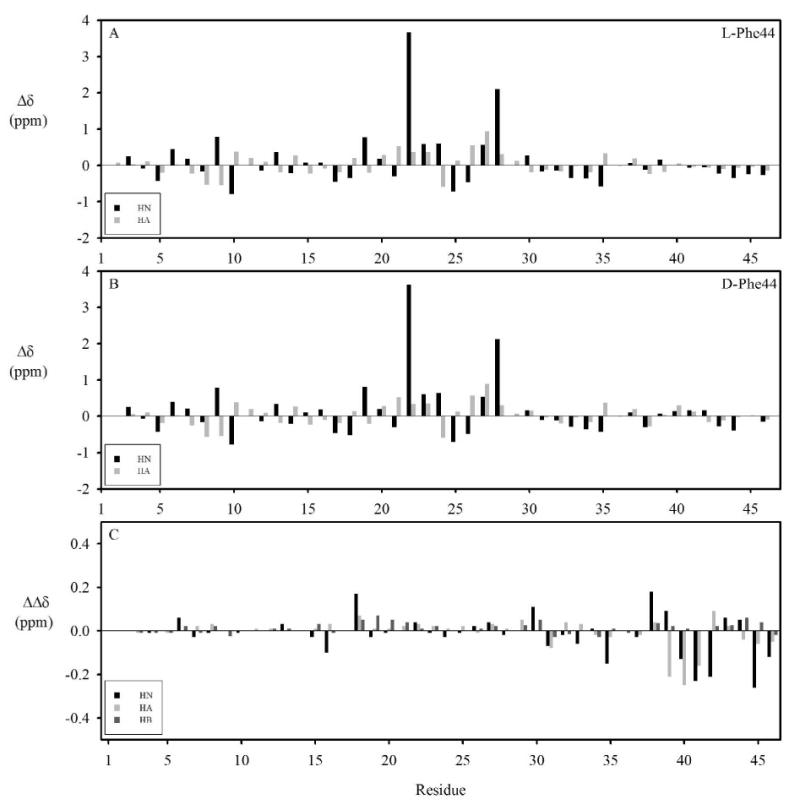
Deviations of ι-RXIA[l-Phe44] (A) and ι-RXIA (B) chemical shifts from random coil values at 298 K. The NH and CαH random coil chemical shifts are from (31). (C) Chemical shift differences of HN, CαH and CβH resonances between ι-RXIA[l-Phe44] and ι-RXIA.
Structural Restraints
We determined the structures of ι-RXIA and ι-RXIA[l-Phe44], but the ι-RXIA[l-Phe44] structure is described first because a larger quantity was available, yielding better spectra. Analysis of a DQF-COSY spectrum showed that 3JHNHα coupling constants of Cys18, Asn20, Cys21, Leu23, Ser24, Ala28, Thr31, Asn32, Trp33 were > 8 Hz and there were none < 6 Hz. Amide exchange experiments of ι-RXIA[l-Phe44] conducted at 5 °C and pH 5.9 indicated that the amide protons of Cys12, Asn20, Cys21, Leu23, Ile26, Cys27 and Ala28 were the slowest to exchange, those of Glu9, Cys19, Cys18, Tyr14, Gly25 and Lys10 had intermediate exchange residues, and the rest exchanged with solvent within minutes of dissolution of the polypeptide in 2H2O.
The disulfide bond pairings were determined by preliminary structure calculations in CYANA. First, a family of 20 structures was determined in CYANA based only on distance constraints. Distances were then measured between pairs of Cys S atoms and pairs of Cys CβH protons. As a result, it was obvious that Cys5 and Cys19 were much closer to one another than to other Cys residues, as were Cys21 and Cys38. To determine the other two disulfide bond pairings, structure calculations, including the above two disulfide connectivities, were repeated with six possible combinations among Cys12, Cys22, Cys18, Cys27. It was found that the combination of Cys12-Cys22 and Cys18-Cys27 had the lowest energies and fewest NMR restraint violations compared with other possible combinations. This pattern suggests that ι-RXIA[l-Phe44] has an ICK motif structure (33-35) with one additional disulfide (Cys21-Cys38), as shown in Figure 1B.
Disulfide Mapping of ι-RXIA
The disulfide connectivities were confirmed by partial reduction of the peptide with TCEP followed by stepwise alkylation and microsequencing of the final product. Key features of the strategy were optimization of the reaction to yield the highest ratio of partially reduced disulfide species relative to fully reduced peptide, and proper choice of alkylating agent. The rearrangement of disulfide bonds was avoided by performing the first alkylation with N-ethylmaleimide (NEM) at an acidic pH directly after HPLC separation of the partially reduced forms. The second labeling was done with iodoacetic acid (IAA), as NEM- and IAA-labeled cysteines are readily distinguishable during microsequencing.
Figure 3A shows the HPLC elution of partially reduced ι-RXIA. Four out of five major partially reduced forms were separated and analyzed by ESI mass spectrometry. Their molecular masses were determined to be 4976.5, 4978.1, 4978.2 and 4979.9 Da, corresponding to species containing three, two, two, and one remaining disulfide bond, respectively. We refer to these partially reduced intermediates as 3S-S, 2S-S1, 2S-S2 and 1S-S species. The first alkylation of each intermediate gave one main product, which was repurified by HPLC (Figure 3B). Each alkylated variant was then lyophilized and re-suspended in the pH 8.0 buffer, and the remaining disulfides were reduced and the free cysteines labeled with IAA. The double-labeled species appeared as one major peak during purification with RP-HPLC (Figure 3C).
FIGURE 3.
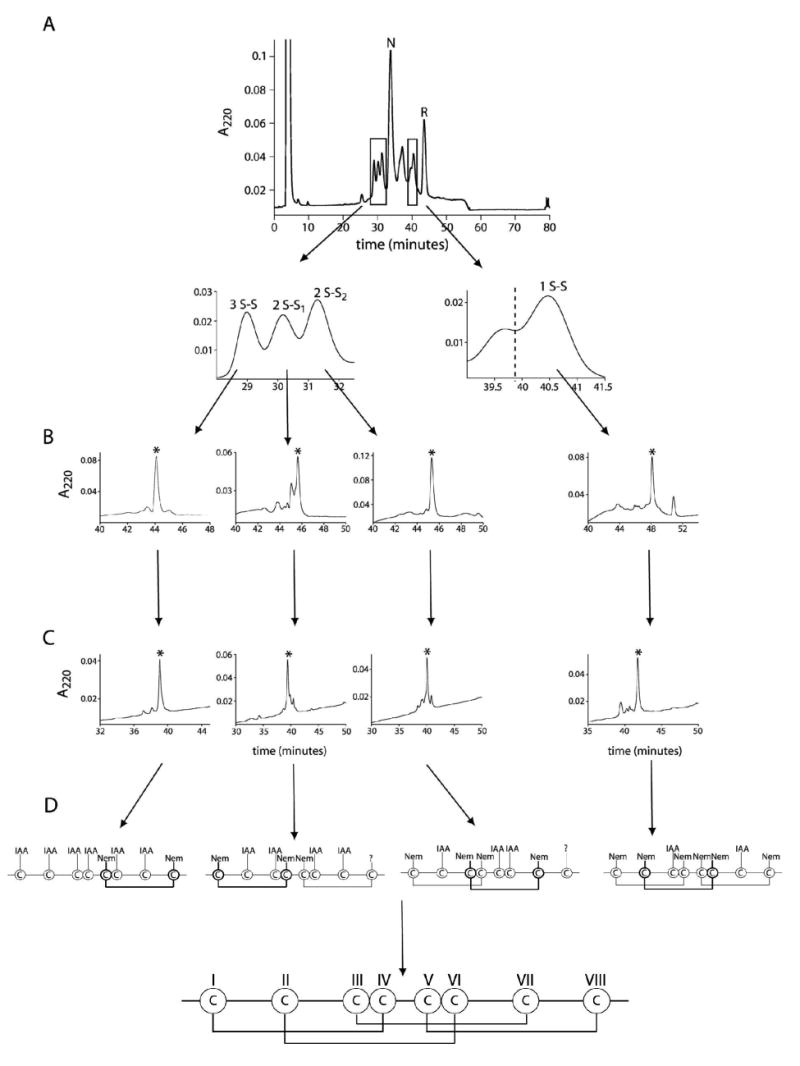
Disulfide mapping of ι-RXIA. (A) HPLC trace for TCEP partial reduction of ι-RXIA. The reaction was carried out with 20 mM TCEP in 0.2 M sodium citrate, pH 3.0 for 5 min at room temperature. N and R denote native and reduced form, respectively. Four major partially reduced forms, which correspond to the species containing three (3S-S), two (2S-S1 and 2S-S2) and one (1S-S) remaining disulfide bond, were separated. (B) HPLC traces for purification of NEM-alkylated variants. The reaction was performed with 60 mM NEM in 0.2 M sodium citrate, pH 3.0 for 1 h at room temperature directly after HPLC separation of partially reduced forms. (C) HPLC purification of double-labeled forms. The NEM-alkylated variants were completely reduced and then alkylated with 0.2 M IAA in 0.2 M Tris/acetate, 0.2 mM EDTA, pH 8.0, for 30 min at room temperature. Each proper product is marked with an asterisk. (D) Schematic representation of a labeling pattern of cysteine residues resulting from the microsequencing data and disulfide connectivity of ι-RXIA.
Four different alkylated variants of ι-RXIA (3S-S, 2S-S1, 2S-S2 and 1S-S) were sequenced to identify the labeling pattern of cysteine residues. Although the microsequencing data were clear, a few factors made the analysis challenging. First, ι-RXIA is a large, rather hydrophobic peptide, and after double alkylation and lyophilization it remained almost insoluble. Thus, the sequencing was performed directly after HPLC with 0.5 - 1 ml fractions containing about 300 pmol of the peptide loaded overnight. The other challenge was to identify the IAA-labeling when Cys was in very close proximity to Ser as those two signals could not be separated. Furthermore, in the case of species with two remaining disulfides (2S-S1, 2S-S2) the signal for the last cysteine residue (position 38 in the sequence) was too low to identify the labeling. Figure 3D shows the cysteine-labeling pattern resulting from microsequencing data and the surmised disulfide connectivity of ι-RXIA. This was in perfect agreement with the disulfide connectivities determined by NMR for ι-RXIA[l-Phe44], showing that both forms of this polypeptide fold in the same way.
Solution Structure of ι-RXIA [L-Phe44]
Parameters characterizing the final 20 structures in XPLOR and structural statistics for ι-RXIA[l-Phe44] are summarized in Table 1, and stereo views of the family of structures superimposed over the backbone heavy atoms are shown in Figure 4A. Numerous medium- and long-range NOEs (i-j>4) were observed for residues 5-30 (Figure S3 in Supporting Information), implying that this region has a well-defined structure. By contrast, very few medium- or long-range NOEs were observed for the N- and C-terminal regions, indicating that both termini lacked a well-defined structure in solution.
Table 1.
Experimental Constraints and Structural Statistics for ι-RXIA[l-Phe44] and ι-RXIA.
| ι-RXIA[l-Phe44] | ι-RXIA | |
|---|---|---|
| No. of distance restraints | ||
| total | 482 | 448 |
| intraresidue (i =j) | 223 | 198 |
| sequential (|i-j| = 1) | 156 | 132 |
| medium-range (1 < |i-j| < 5) | 38 | 22 |
| long-range (|i-j| > 4) | 65 | 96 |
| Hydrogen bond restraints | 9 | 7 |
| No. of dihedral restraints | 49 | 39 |
| RMS deviations from experimental data | ||
| NOEs (Å) | 0.021±0.004 | 0.027±0.004 |
| dihedrals (deg) | 0.560±0.2 | 0.299±0.2 |
| Deviations from ideal a | ||
| angles (deg) | 0.675±0.03 | 0.707±0.04 |
| bonds (Å) | 0.0046±0.0006 | 0.0048±0.0006 |
| impropers (deg) | 0.486±0.03 | 0.486±0.03 |
| RMS deviations b | ||
| residues 5-30 | ||
| all heavy atoms | 1.07± 0.17 | 1.17± 0.21 |
| backbone heavy atoms (N, Cα, C) | 0.49± 0.15 | 0.56± 0.13 |
| residues 5-35 | ||
| all heavy atoms | 2.28± 1.10 | 2.17± 0.43 |
| backbone heavy atoms (N, Cα, C) | 1.64± 1.04 | 1.39± 0.38 |
| Ramachandran plot b | ||
| most favored (%) | 50.0 | 52.4 |
| allowed (%) | 42.3 | 38.5 |
| additionally allowed (%) | 4.4 | 5.9 |
| disallowed (%) | 3.3 | 3.2 |
The values for the bonds, angles, and impropers show the deviations from ideal values based on perfect stereochemistry.
As determined by the program PROCHECK-NMR for all residues. The high percentage of residues in the disallowed residue of the Ramachandran plot reflects the positive ϕ angle for Glu9. All violations, energies, and RMS difference are given as the mean ± standard deviation.
FIGURE 4.
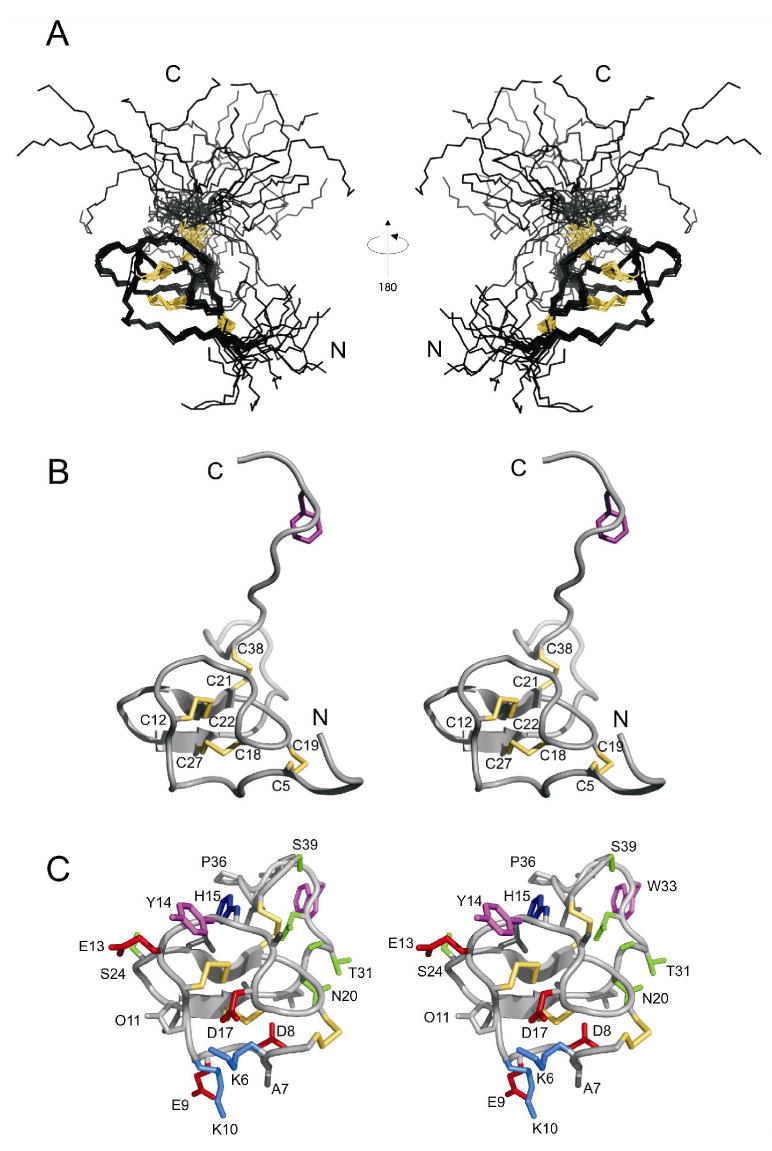
Structure of ι-RXIA[l-Phe44]. (A) Family of 20 structures, superimposed over backbone heavy atoms of residues 5-30. The disulfide bond connectivities are in yellow. (B) Stereo view of ribbon diagram of the closest-to-average structure, four disulfide bonds (5-19, 12-22, 18-27, 21-38) are in yellow and Phe44 is in magenta. C, Stereo view showing only residues 5-39 of the closest-to-average structure, but including all side chains (most of which are labelled with single-letter code and residue number). Side chains are colored as follows: Lys, marine; His, blue; Asp, Glu, red; Trp, Tyr, magenta; Thr, Ser, Asn, green. This figure was prepared using PyMol.
The angular order parameters S for the ϕ, ψ and χ1 angles in the final family of 20 structures (Figure S3 in Supporting Information) show that the structured region of ι-RXIA[l-Phe44] encompasses residues 5-35. The RMSD over the backbone heavy atoms was 0.49 Ǻ for Cys5-Ser30 and 1.64 Ǻ for Cys5-Leu35. The closest-to-average structure is shown in Figure 4B and C. The structures of ι-RXIA[l-Phe44] have been deposited with the RCSB Protein Data Bank (36) with accession number 2P4L.
Of the four disulfide bridges (Cys5-Cys19, Cys12-Cys22, Cys18-Cys27, and Cys21-Cys38), the first three are located in the well-defined region of the structure. The separation of the half-cystines in this region, -CX6CX5CCX2CX4C-, where X is any amino acid, fits the original consensus for an ICK motif structure, -CX3-7CX3-6CX0-5CX1-4CX4-13C- (33, 34). Secondary structure analysis using PROCHECK (26) showed that ι-RXIA contained a small, two-stranded, anti-parallel β-sheet (Cys21-Cys22, Cys27-Ala28). The fourth disulfide, Cys21-Cys38, links the ICK motif to the C-terminal region. An examination of amide exchange data as well as the ensemble of structures suggested that hydrogen bonds exist between the following pairs of residues: Cys27 and Glu9, Cys21 and Ala28, Leu23 and Ile26, Cys19 and Lys6, Cys12 and Cys22, Lys10 and Asp8. These are all part of, or adjacent to, the two-stranded β-sheet that forms the core of the structure. An examination of backbone ϕ and ψ angles as well as hydrogen bonds in the final family of structures indicated that residues 8-10 form a γ-turn, with Glu9 having a positive ϕ angle. An interesting feature of the structured region of ι-RXIA[l-Phe44] is the abundance of charged side chains (Lys, Asp and Glu) in the N-terminal region (Figure 4C and Figure 5); the C-terminal region, including the disordered tail, contains only one charged side chain, Lys45.
FIGURE 5.
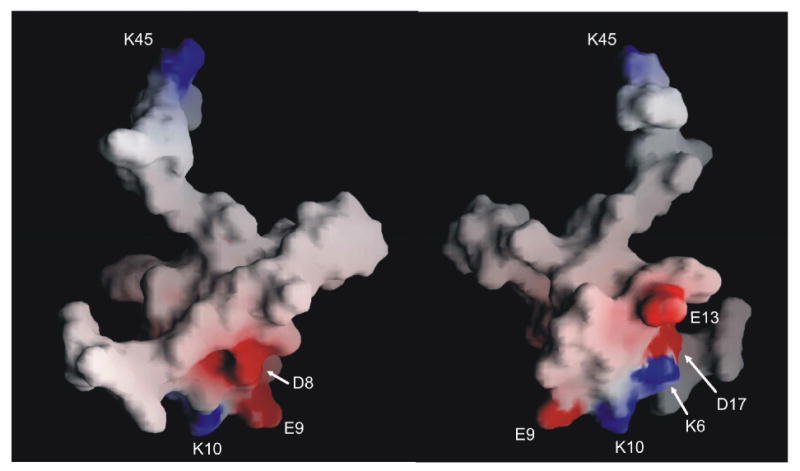
Surface representation of the closest-to-average structure of ι-RXIA[L-Phe44]. The surface is colored, with basic residues in blue and acidic residues in red. The two views are related by a 180° rotation around the vertical axis. This figure was prepared using GRASP (29).
Structural comparison of ι-RXIA[l-Phe44] using Dali (37) revealed that there are no other known structures with a Dali Z-score larger than 2, over either the whole 46 residues (Table S3 in Supporting Information) or just the well-defined Cys5-Leu35 region. The toxin versutoxin (16) (RCSB accession number 1VTX), which contains a triple-stranded antiparallel β sheet and has the same pattern of disulfide connectivities as ι-RXIA[l-Phe44], is the closest match, with a Dali Z-score of 0.6. ω-agatoxin IVB(38), ω-agatoxin IVA(39) and ι-RXIA[l-Phe44] contain four disulfide bridges, but in the case of ω-agatoxins IVA and IVB the fourth disulfide is located within one of the loops, whereas in ι-RXIA[l-Phe44] it connects one of the loops to the C-terminus. It is interesting to note that there are similarities between ι-RXIA and ω-agatoxin IVB (RCSB accession number 1AGG), which has d-Ser at position 46 (38, 40, 41) (note that in reference 35, ω-agatoxin IVB is referred to as ω-agatoxin TK), and an unstructured C-terminal region (residues 37-48).
Figure S4 in Supporting Information shows the superposition of [l-Phe44]ι-RXIA with ω-agatoxin IVB, robustoxin (RCSB accession number 1QDP), versutoxin (RCSB accession number 1VTX), and J-ACTX-Hv1c (RCSB accession number 1DLO). The pairwise RMSDs between [l-Phe44]ι-RXIA and the above four molecules, superimposed over N, Cα, C of the β-sheet and Cα, Cβ of the three disulfides of the ICK fold, are 2.80, 3.72, 3.39, and 1.32 Å, respectively.
Comparison of ι-RXIA[L-Phe44] with ι-RXIA
Figure 2 shows that ι-RXIA[l-Phe44] and ι-RXIA have a similar pattern of backbone NH and CαH chemical shift deviations from random coil values at pH 5.9 and 25 °C. The differences in backbone CαH and NH and sidechain CβH chemical shifts between ι-RXIA[l-Phe44] and ι-RXIA (Figure 2C) are very small, the largest being < 0.3 ppm. The main differences occur adjacent to Phe44 (residues 38-46), although the NH of Leu35 is perturbed and there are some small changes more distant from Phe44, notably for Cys18 and Ser30. Comparison of 800 MHz NOESY spectra of the two peptides showed that there were very few NOE differences between them (see comparison in Figure S3 in Supporting Information). The same is true for the backbone 3JHNNα coupling constants (Tables S1 and S2 in Supporting Information), although small differences are evident around residue 44.
In order to confirm our inference from comparison of chemical shifts, backbone coupling constants and NOE distribution that the solution structures of ι-RXIA[l-Phe44] and ι-RXIA were very similar we have calculated the structure of ι-RXIA using the procedure described above for ι-RXIA[l-Phe44]. The structural statistics for ι-RXIA are compared with those for ι-RXIA[l-Phe44] in Table 1 and the structures are compared in Figure 6. The families of structures are virtually superimposable over the well-defined regions (residues 5-30), as shown in Figure 6 and from RMSD comparisons shown in Figure S5 of the Supporting Information. The N- and C-terminal regions are poorly defined, consistent with the chemical shifts (Figure 2) and lack of medium- and long-range NOEs (Figure S3 in Supporting Information). Thus, even though the regions of conformational space sampled by the families of structures of ι-RXIA and ι-RXIA[l-Phe44] differ slightly in Figure 6, no conclusions can be drawn about the solution conformations of the C-terminal tail containing Phe44 other than that they are disordered in both cases. Comparison of the local structures in the vicinity of Phe44 also reveals no significant differences; the families of structures are similar when superimposed over the backbone heavy atoms of residues 41-45, and the angular averages for the backbone dihedral angles are nearly identical for these residues.
FIGURE 6.
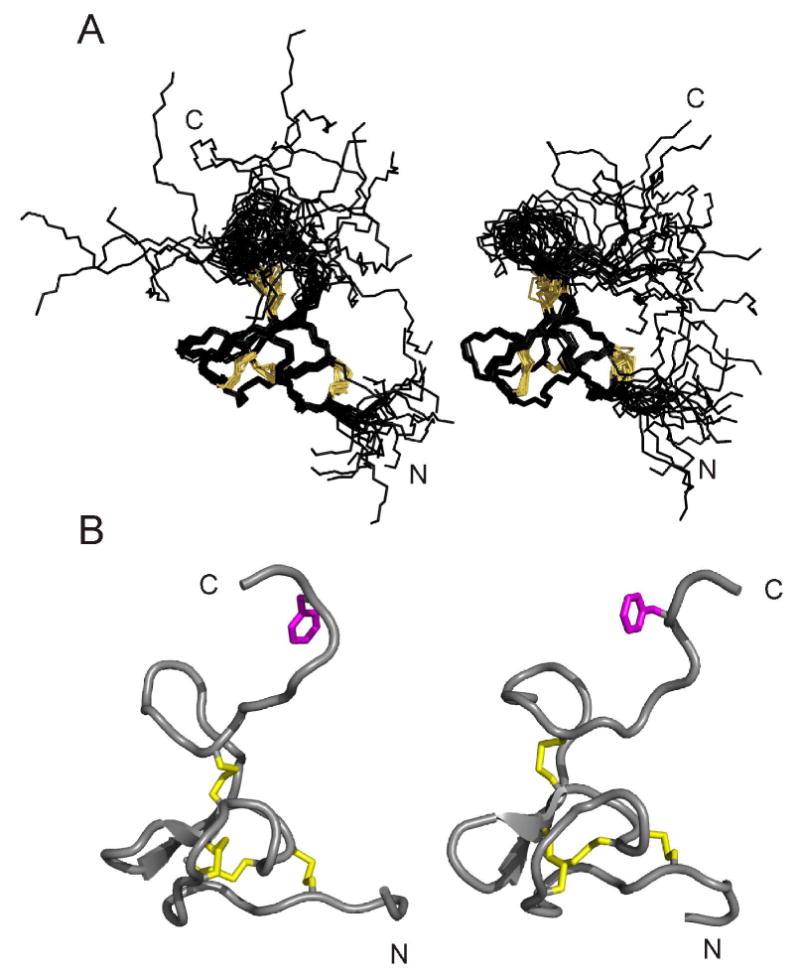
Comparison of structures of ι-RXIA and ι-RXIA[l-Phe44]. (A) Families of 20 structures of ι-RXIA (left) and ι-RXIA[l-Phe44] (right), superimposed over backbone heavy atoms of residues 5-30 in each case. The disulfide bond connectivities are in orange. (B) Ribbon diagram of the closest-to-average structures of ι-RXIA (left) and ι-RXIA[l-Phe44] (right); these are the closest-to-average structures in each case following superposition of each family over the entire backbone (residues 1-46). The four disulfide bonds (5-19, 12-22, 18-27, 21-38) are in yellow and Phe44 is in magenta.
Activity of ι-RXIA on NaV1.6, KCNQ2/3 and KV1.2 through 1.6 expressed in Xenopus oocytes
In previous experiments with frog preparations (5, 6), we observed that ι-RXIA induced repetitive action potentials in motor axons which have KV1.2 (in frogs) and KCNQ2 (and possibly KCNQ3) and NaV1.6 (in mammals) at their nodes of Ranvier as shown by immunohistochemistry (42-44). The heteromeric human KCNQ (Q2 and Q3), and homomeric human KV1.2, 1.3, 1.4, 1.5 and 1.6 K channels were expressed in Xenopus oocytes and tested for their sensitivities to ι-RXIA as described in Experimental Procedures. None of these channels was affected by 100 μM ι-RXIA (not illustrated). In contrast, when ι-RXIA was tested on mouse NaV1.6 expressed in Xenopus oocytes, the voltage-gated sodium current (INa) was affected profoundly. Figure 7 shows representative current traces before (panel A) and after (panel B) ∼10 min exposure to 5 μM ι-RXIA For a given voltage step, the peptide increased the peak INa. To facilitate comparison, the responses to a −30 mV voltage step in the absence and presence of ι-RXIA are superimposed in Figure 7C. Figure 7D shows that ι-RXIA displaced the voltage dependence of activation to more hyperpolarized potentials (V0.5 shifted by 12.5 mV with no significant change in slope factor). In contrast, the toxin affected the voltage dependence of fast inactivation minimally, if at all (Figure 7E). These results indicate that ι-RXIA acted as an agonist of the sodium channel. The effects of ι-RXIA were reversed when the peptide was washed out (not illustrated).
FIGURE 7.
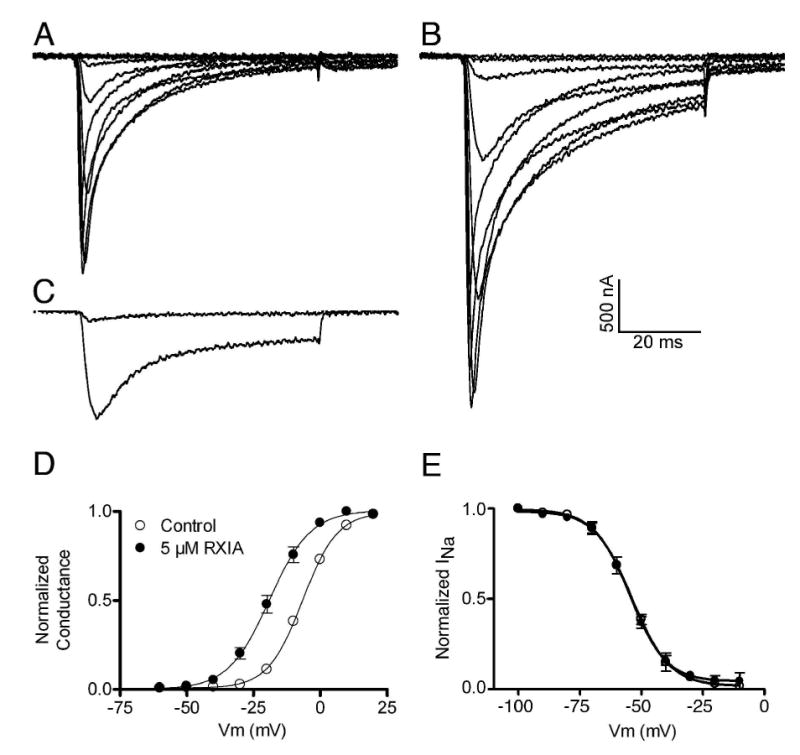
ι-RXIA promotes the activation of NaV1.6 without significantly affecting channel inactivation. Xenopus oocytes expressing mouse NaV1.6 were prepared and used as described in Experimental Procedures. Oocytes were clamped at −100 mV, and 60 ms depolarizing steps were applied in 10 mV increments from −60 to +20 mV. Representative sodium currents (INa) in control solution (A) and after the effects of 5 μM ι-RXIA had reached steady-state in about 10 min. (B). The peak amplitudes of INa were increased by the peptide. (C) One trace each from A and B representing the responses to a −30 mV voltage step are superimposed and illustrate the dramatic effect of ι-RXIA on INa at this potential. When their peaks were normalized, the traces essentially overlapped (not illustrated), indicating that toxin treatment does not alter the time course of INa. (D) The voltage dependence of activation was shifted to more hyperpolarized potentials by ι-RXIA. Solid lines represent Boltzmann fits: V0.5 values were −6.64 ± 0.27 mV (control) and −19.09 ± 0.74 mV (with peptide); respective slope factors were 6.50 ± 0.24 and 7.72 ± 0.69, which were not significantly different from one another (p < 0.5, Student's t-test). (E) Voltage dependence of inactivation was essentially unaltered by 5 μM ι-RXIA. Pulse protocol for inactivation was as described in Experimental Procedures. Solid lines are Boltzmann fits: inactivation V0.5 values were −54.02 ± 0.74 mV (control) and − 54.49 ± 0.69 mV (with toxin); respective slope factors were 7.60 ± 0.67 and 7.25 ± 0.63. Symbol legend in (D) applies to both (D) and (E), and data points in (D) and (E) represent mean ± s.e. (N = 3 oocytes).
Discussion
The disulfide bond connectivities of ι-RXIA and ι-RXIA[l-Phe44] have been determined, respectively, by chemical mapping and analysis of structures calculated using NMR-derived restraints. The solution structure shows that ι-RXIA forms an ICK-fold motif stabilized by the three disulfide bridges Cys5-Cys19, Cys12-Cys22, and Cys18-Cys27, with one additional disulfide (Cys21-Cys38) linking the ICK motif to the C-terminus. This is the same pattern seen in the Sydney funnel-web spider toxin, robustoxin, ie I-IV/II-VI/III-VII/V-VIII (15). However, in a different spider toxin, J-atracotoxin, which shares an identical cysteine framework with I-conotoxins (Figure 1), NMR studies, combined with a partial reduction/alkylation technique, showed that the cysteine connectivities were I-VI, II-VII, III-IV and V-VIII, as illustrated in Figure 8. An unusual feature of J-atracotoxin is the presence of a rare vicinal disulfide bridge (CysIII-CysIV), which appears to be critical for the insecticidal activity of this polypeptide (12). Thus, apparently conserved cysteine frameworks do not lead to the same disulfide connectivities. ι-RXIA is more similar to J-atracotoxin (in terms of spacing between cysteine residues), than to some other I-superfamily members characterized from Conus. It remains to be seen what the disulfide linkages are in other framework #11 Conus peptides, such as the I2-superfamily conotoxin BtX (Figure 1).
FIGURE 8.
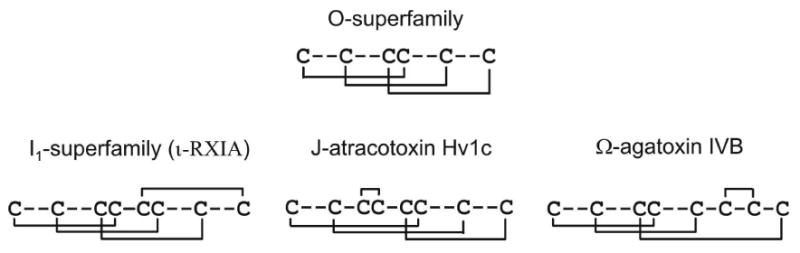
Disulfide patterns in toxins containing three and four disulfide bridges. The O-conotoxins (framework #6) have the disulfide pattern I-IV/II-V/III-VI. The I-conotoxins (framework #11), including ι-RXIA, have the disulfide pattern I-IV/II-VI/III-VII/V-VIII, as do robustoxin and versutoxin. The spider toxin, J-atracotoxin, which shares an identical cysteine framework with the I-conotoxins, has the disulfide pattern I-VI/II-VII/III-IV/V-VIII. Yet another pattern of connectivities of eight half-cystines is found in ω-agatoxin IVB. The disulfide bridges above each of the amino acid sequences shows the different ways in which the fourth bridge could have been added to the ICK pattern of three bridges in the course of evolution.
Robustoxin and versutoxin have the same pattern of disulfide connectivities as ι-RXIA (I-IV/II-VI/III-VII/V-VIII) even though they do not have the −C-C-CC-CC-C-C- pattern characteristic of framework #11. They also adopt ICK motif structures (15, 16), with the fourth disulfide linking this motif to the C-terminus (15). Structurally, therefore, they are more similar to ι-RXIA than is J-atracotoxin, even though the latter has a framework #11 arrangement of cysteine residues. A possible reason for this difference is the presence of Pro between the CC doublets in the J-atracotoxins (12), in contrast to the Asn present in ι-RXIA (Figure 1A) and related peptides from C. radiatus (5, 7), although other conserved non-Cys residues may also have important roles as folding determinants for this disulfide framework. Thus, different patterns of half-cystines but the same disulfide connectitivies (as in ι-RXIA and robustoxin) can produce similar structures, but the same pattern of half-cystines (as in ι-RXIA and J-atracotoxin) does not necessarily lead to the same pattern of disulfide connectitivies (Figure 8). A remarkable feature of all of these molecules, however, is the prevalence of the ICK structural motif (33, 34), or minor variants thereof, as the basic structural scaffold. It is possible that both Conus and spider peptides may have arisen from peptides with a disulfide connectivity characteristic of the ICK motif, and that the fourth disulfide bond was added subsequently (Figure 8). The identical structural motif of knotted cystines is characteristic of another superfamily of Conus peptides, the O-superfamily (45, 46). The I-conotoxins (framework #11) and O-conotoxins (defined by framework #6, C-C-CC-C-C) belong to distinct gene superfamilies and there is no evident homology between their precursor sequences (7, 47), reflecting a long period of evolutionary divergence.
Comparison of the chemical shifts, backbone coupling constants and NOEs of ι-RXIA with those of ι-RXIA[l-Phe44] suggests that post-translational modification of residue 44 from l- to d- has very little effect on the structure. This is readily accounted for by the structure described in this paper, as the C-terminal region beyond the last disulfide bridge (Cys21-Cy38) is largely disordered in both structures. As the naturally occurring d-Phe44 form of ι-RXIA is significantly more active as an excitotoxin than the l-Phe44 analogue both in vitro and in vivo (6), it is likely that the C-terminal tail of ι-RXIA undergoes a transition to a more ordered conformation when in complex with its receptor. In this conformation the switch from d to l configuration at Phe44 may be expected to have a significant effect on affinity and/or activity. The more structured region of ι-RXIA, the ICK motif, probably plays a common role in both ι-RXIA and ι-RXIA[l-Phe44] of mediating peptide binding to the appropriate site on the target in such a way as to allow the C-terminal tail to interact with neighboring residues.
The functional importance of epimerization of a single amino acid near the C-terminus was also observed for ω-agatoxin IVB, which has a similar combination of an ICK motif core and an unstructured tail (13, 38, 40, 41). In this 48-residue peptide, d-Ser46 was crucial for the inhibition of P-type calcium channels, whereas the l-Ser enantiomer was about 90-fold less active (40).
As with the funnel web spider toxins robustoxin and versutoxin, with which it shares a common disulfide pattern and structures, ι-RXIA proves to be active against sodium channels. The agonistic action of ι-RXIA on NaV1.6 (Figure 7) explains how the peptide renders motor axons hyperexcitable, as it effectively lowers the threshold for the generation of action potentials by shifting the voltage-sensitivity of the sodium channels. The effect of ι-RXIA in shifting the voltage dependence of activation in a hyperpolarizing direction is reminiscent of δ-conopeptides PVIA and SVIE (48). However, unlike the δ-conopeptides, ι-RXIA does not shift the voltage dependence of steady-state inactivation appreciably, if at all (Figure 7E). Further experiments are in progress to examine ι-RXIA's VGSC subtype-selectivity as well as its mechanism of action in greater detail. However, it is noteworthy that there is a convergence of structure and function in ι-RXIA, as its structure resembles those of funnel web spider toxins that also target sodium channels. Their effects on these channels, however, are not identical, and it will be of interest to define which residues in ι-RXIA differentiate its activity on the sodium channel from those of robustoxin and versutoxin, and what the respective roles of the ICK core and the C-terminal tail containing Phe44 are in their activities.
Supplementary Material
NMR data and structure comparisons referred to in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank Shenggen Yao for help with the NMR experiments, Zhi-Ping Feng and Jennifer Sabo for help with the structure calculations, Alan Goldin for the mouse NaV1.6 clone, Layla Azam for preparing the corresponding cRNA, Michael Sanguinetti for help with testing KCNQ clones, and Minmin Zhang for helpful discussions.
Footnotes
This work was supported in part by N.I.H. grant GM 48677 (to B.M.O.) from the National Institute of General Medical Sciences. R.S.N. acknowledges support from the Australian National Health and Medical research Council.
Chemical shift assignments for ι-RXIA[l-Phe44] and ι-RXIA have been deposited in BioMagResBank with accession numbers 15174 and 15175, respectively. The families of structures of ι-RXIA[l-Phe44] and ι-RXIA have been deposited in the Protein Data Bank with accession numbers 2P4L and 2JRY, respectively.
Abbreviations used are: IAA, iodoacetic acid; INa, sodium current; ι-RXIA, iota-conotoxin RXIA (previously designated r11a); Kv, voltage-gated potassium channel(s); Nav or VGSC, voltage-gated sodium channel; NEM, N-ethylmaleimide; NMR, nuclear magnetic resonance; NOE, nuclear Overhauser effect; TCEP, tris(2-caroxyethyl)-phosphine hydrochloride; TFA, trifluoroacetic acid.
References
- 1.Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 2.Norton RS, Olivera BM. Conotoxins down under. Toxicon. 2006;48:780–798. doi: 10.1016/j.toxicon.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 3.Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- 4.Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jimenez EC, Shetty RP, Lirazan M, Rivier J, Walker C, Abogadie FC, Yoshikami D, Cruz LJ, Olivera BM. Novel excitatory Conus peptides define a new conotoxin superfamily. J Neurochem. 2003;85:610–621. doi: 10.1046/j.1471-4159.2003.01685.x. [DOI] [PubMed] [Google Scholar]
- 6.Buczek O, Yoshikami D, Bulaj G, Jimenez EC, Olivera BM. Post-translational amino acid isomerization: a functionally important D-amino acid in an excitatory peptide. J Biol Chem. 2005;280:4247–4253. doi: 10.1074/jbc.M405835200. [DOI] [PubMed] [Google Scholar]
- 7.Buczek O, Yoshikami D, Watkins M, Bulaj G, Jimenez EC, Olivera BM. Characterization of d-amino-acid-containing excitatory conotoxins and redefinition of the I-conotoxin superfamily. FEBS J. 2005;272:4178–4188. doi: 10.1111/j.1742-4658.2005.04830.x. [DOI] [PubMed] [Google Scholar]
- 8.Kauferstein S, Huys I, Kuch U, Melaun C, Tytgat J, Mebs D. Novel conopeptides of the I-superfamily occur in several clades of cone snails. Toxicon. 2004;44:539–548. doi: 10.1016/j.toxicon.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Fan CX, Chen XK, Zhang C, Wang LX, Duan KL, He LL, Cao Y, Liu SY, Zhong MN, Ulens C, Tytgat J, Chen JS, Chi CW, Zhou Z. A novel conotoxin from Conus betulinus, κ-BtX, unique in cysteine pattern and in function as a specific BK channel modulator. J Biol Chem. 2003;278:12624–12633. doi: 10.1074/jbc.M210200200. [DOI] [PubMed] [Google Scholar]
- 10.Kauferstein S, Huys I, Lamthanh H, Stocklin R, Sotto F, Menez A, Tytgat J, Mebs D. A novel conotoxin inhibiting vertebrate voltage-sensitive potassium channels. Toxicon. 2003;42:43–52. doi: 10.1016/s0041-0101(03)00099-0. [DOI] [PubMed] [Google Scholar]
- 11.Olivera BM, Cruz LJ. Conotoxins, in retrospect. Toxicon. 2001;39:7–14. doi: 10.1016/s0041-0101(00)00157-4. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Connor M, Smith R, Maciejewski MW, Howden ME, Nicholson GM, Christie MJ, King GF. Discovery and characterization of a family of insecticidal neurotoxins with a rare vicinal disulfide bridge. Nat Struct Biol. 2000;7:505–513. doi: 10.1038/75921. [DOI] [PubMed] [Google Scholar]
- 13.Adams ME, Mintz IM, Reily MD, Thanabal V, Bean BP. Structure and properties of ω-agatoxin IVB, a new antagonist of P-type calcium channels. Mol Pharmacol. 1993;44:681–688. [PubMed] [Google Scholar]
- 14.Maggio F, King GF. Scanning mutagenesis of a Janus-faced atracotoxin reveals a bipartite surface patch that is essential for neurotoxic function. J Biol Chem. 2002;277:22806–22813. doi: 10.1074/jbc.M202297200. [DOI] [PubMed] [Google Scholar]
- 15.Pallaghy PK, Alewood D, Alewood PF, Norton RS. Solution structure of robustoxin, the lethal neurotoxin from the funnel-web spider Atrax robustus. FEBS Lett. 1997;419:191–196. doi: 10.1016/s0014-5793(97)01452-x. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher JI, Chapman BE, Mackay JP, Howden ME, King GF. The structure of versutoxin (δ-atracotoxin-Hv1) provides insights into the binding of site 3 neurotoxins to the voltage-gated sodium channel. Structure. 1997;5:1525–1535. doi: 10.1016/s0969-2126(97)00301-8. [DOI] [PubMed] [Google Scholar]
- 17.Gray WR. Disulfide structures of highly bridged peptides: a new strategy for analysis. Protein Sci. 1993;2:1732–1748. doi: 10.1002/pro.5560021017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbs SJ, Johnson CS. A PFG-NMR experiment for accurate diffusion and flow studies in the presence of eddy currents. J Magn Reson. 1991;93:395–402. [Google Scholar]
- 19.Dingley AJ, Mackay JP, Chapman BE, Morris MB, Kuchel PW, Hambly BD, King GF. Measuring protein self-association using pulsed-field-gradient NMR spectroscopy: application to myosin light chain 2. J Biomol NMR. 1995;6:321–328. doi: 10.1007/BF00197813. [DOI] [PubMed] [Google Scholar]
- 20.Yao S, Howlett GJ, Norton RS. Peptide self-association in aqueous trifluoroethanol monitored by pulsed field gradient NMR diffusion measurements. J Biomol NMR. 2000;16:109–119. doi: 10.1023/a:1008382624724. [DOI] [PubMed] [Google Scholar]
- 21.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 22.Palmer AG, Cavanagh J, Wright PE, Rance M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J Magn Reson. 1991;93:151–170. [Google Scholar]
- 23.Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 24.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 25.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 27.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–5. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMol Molecular Graphics System. DeLano Scientific; San Carlos CA USA: 2002. http://www.pymol.org. [Google Scholar]
- 29.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–96. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 30.Seavey BR, Farr EA, Westler WM, Markley JL. A relational database for sequence-specific protein NMR data. J Biomol NMR. 1991;1:217–236. doi: 10.1007/BF01875516. [DOI] [PubMed] [Google Scholar]
- 31.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 32.Corzo G, Sabo JK, Bosmans F, Billen B, Villegas E, Tytgat J, Norton RS. Solution structure and alanine scan of a spider toxin that affects the activation of mammalian voltage-gated sodium channels. J Biol Chem. 2007;282:4643–4652. doi: 10.1074/jbc.M605403200. [DOI] [PubMed] [Google Scholar]
- 33.Pallaghy PK, Nielsen KJ, Craik DJ, Norton RS. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994;3:1833–1839. doi: 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norton RS, Pallaghy PK. The cystine knot structure of ion channel toxins and related polypeptides. Toxicon. 1998;36:1573–1583. doi: 10.1016/s0041-0101(98)00149-4. [DOI] [PubMed] [Google Scholar]
- 35.Miles LA, Dy CY, Nielsen J, Barnham KJ, Hinds MG, Olivera BM, Bulaj G, Norton RS. Structure of a novel P-superfamily spasmodic conotoxin reveals an inhibitory cystine knot motif. J Biol Chem. 2002;277:43033–43040. doi: 10.1074/jbc.M206690200. [DOI] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H, Rosen MK, Saccomano NA, Phillips D, Volkmann RA, Schreiber SL. Sequential assignment and structure determination of spider toxin ω-Aga-IVB. Biochemistry. 1993;32:13123–13129. doi: 10.1021/bi00211a022. [DOI] [PubMed] [Google Scholar]
- 39.Kim JI, Konishi S, Iwai H, Kohno T, Gouda H, Shimada I, Sato K, Arata Y. Three-dimensional solution structure of the calcium channel antagonist ω-agatoxin IVA: consensus molecular folding of calcium channel blockers. J Mol Biol. 1995;250:659–671. doi: 10.1006/jmbi.1995.0406. [DOI] [PubMed] [Google Scholar]
- 40.Kuwada M, Teramoto T, Kumagaye KY, Nakajima K, Watanabe T, Kawai T, Kawakami Y, Niidome T, Sawada K, Nishizawa Y, et al. ω-agatoxin-TK containing d at position 46, but not synthetic ω-[l-Ser46]agatoxin-TK, exerts blockade of P-type calcium channels in cerebellar Purkinje neurons. Mol Pharmacol. 1994;46:587–593. [PubMed] [Google Scholar]
- 41.Reily MD, Thanabal V, Adams ME. The solution structure of ω-Aga-IVB, a P-type calcium channel antagonist from venom of the funnel web spider, Agelenopsis aperta. J Biomol NMR. 1995;5:122–132. doi: 10.1007/BF00208803. [DOI] [PubMed] [Google Scholar]
- 42.Caldwell JH, Schaller KL, Lasher RS, Peles E, Levinson SR. Sodium channel NaV1.6 is localized at nodes of Ranvier, dendrites, and synapses. Proc Nat Acad Sci. 2000;97:5616–5620. doi: 10.1073/pnas.090034797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasband MN. It's “juxta” potassium channel! J Neurosci Res. 2004;76:749–757. doi: 10.1002/jnr.20073. [DOI] [PubMed] [Google Scholar]
- 44.Schwarz JR, Glassmeier G, Cooper EC, Kao TC, Nodera H, Tabuena D, Kaji R, Bostock H. KCNQ channels mediate IKs, a slow K+ current regulating excitability in the rat node of Ranvier. J Physiol. 2006;573:17–34. doi: 10.1113/jphysiol.2006.106815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McIntosh JM, Hasson A, Spira ME, Gray WR, Li W, Marsh M, Hillyard DR, Olivera BM. A new family of conotoxins that blocks voltage-gated sodium channels. J Biol Chem. 1995;270:16796–16802. doi: 10.1074/jbc.270.28.16796. [DOI] [PubMed] [Google Scholar]
- 46.Shon KJ, Stocker M, Terlau H, Stuhmer W, Jacobsen R, Walker C, Grilley M, Watkins M, Hillyard DR, Gray WR, Olivera BM. κ-Conotoxin PVIIA is a peptide inhibiting the shaker K+ channel. J Biol Chem. 1998;273:33–38. doi: 10.1074/jbc.273.1.33. [DOI] [PubMed] [Google Scholar]
- 47.Kauferstein S, Melaun C, Mebs D. Direct cDNA cloning of novel conopeptide precursors of the O-superfamily. Peptides. 2005;26:361–367. doi: 10.1016/j.peptides.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 48.West PJ, Bulaj G, Yoshikami D. Effects of δ-conotoxins PVIA and SVIE on sodium channels in the amphibian sympathetic nervous system. J Neurophysiol. 2005;94:3916–3924. doi: 10.1152/jn.01304.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NMR data and structure comparisons referred to in the text. This material is available free of charge via the Internet at http://pubs.acs.org.