Abstract
Cdc42Hs(F28L) is a single-point mutant of Cdc42Hs, a member of the Ras superfamily of GTP-binding proteins, that facilitates cellular transformation brought about by an increased rate of cycling between GTP and GDP (Lin et al. (1997) Curr. Biol. 7, 794−797). Dynamics studies of Cdc42Hs(F28L)-GDP have shown increased flexibility for several residues at the nucleotide-binding site (Adams et al. (2004) Biochemistry 43, 9968−9977). The solution structure of Cdc42Hs-GDP (wildtype) has previously been solved by NMR spectroscopy (Feltham et al. (1997) Biochemistry 36, 8755−8766). Here, we describe the solution structure of Cdc42Hs(F28L)-GDP, which provides insight into the structural basis for the change in affinity for GDP. Heteronuclear NMR experiments were performed to assign resonances in the protein, and distance, hydrogen-bonding, residual dipolar coupling, and dihedral angle constraints were used to calculate a set of low energy structures using distance geometry and simulated annealing refinement protocols. The overall structure of Cdc42Hs(F28L)-GDP is very similar to Cdc42Hs wildtype, consisting of a centrally located six-stranded β-sheet structure surrounding the C-terminal α-helix (Feltham et al. (1997) Biochemistry 36, 8755−8766). In addition, the same three regions in wildtype Cdc42Hs that show structural disorder (Switch I, II and the insert region) are disordered in F28L as well. Although the structure of Cdc42Hs(F28L)-GDP is very similar to wildtype, interactions with the nucleotide and hydrogen bonding within the nucleotide binding site are altered, and the region surrounding L28 is substantially more disordered.
The Ras superfamily of GTP-binding proteins is involved in a variety of cellular processes such as cell growth, protein trafficking, cytoskeletal organization, and secretion. The proteins are controlled by cycling between the biologically active GTP form and the inactive GDP-bound form (1, 2). GTP is hydrolyzed to GDP by the intrinsic GTPase activity of the protein; however, the cycle of binding, hydrolysis and rebinding of the nucleotide is controlled by a number of regulatory and effector proteins (3-5). The Ras proteins, especially the Rho-subfamily, have been extensively investigated due to their potential roles in cell transformation. Oncogenic Ras proteins transform cells either due to a defect in GTPase activity, which leads to a terminally active state or to an increased rate of GTP/GDP cycling. In either case, the activity of downstream kinases is increased. Point mutations in Ras that inhibit the GTPase activity (e.g., G12V; Q61L) have been implicated in several types of cancer (6-8). In addition, the F28L mutation in Cdc42Hs (a member of the Rho-subfamily) results in cell transformation due to the increased rate of cycling between the active and inactive forms of the protein. The mutation-induced cell transformation by Cdc42Hs(F28L) is similar to cell transformation induced by the Dbl oncoprotein, which also catalyzes fast nucleotide exchange in Cdc42Hs (3).
Structural studies of wildtype Cdc42Hs-GDP (9) as well as H-Ras-GDP (10, 11) and Rac-GDP, another member of the Ras superfamily (12), have found several well conserved regions in these proteins. However, unique structural aspects of each of these proteins provide specificity for various effector and/or regulatory proteins. In all three cases, the bound nucleotide interacts with the same regions: P Loop or Loop 1 (residues 10−15), residues 116−119 (residues 115−118 in Cdc42Hs), residues 145−147 (residues 158−160 in Cdc42Hs), and F28. F28 serves as the principal stabilizer of the nucleotide via a strong interaction with the aromatic ring of the bound nucleotide (10, 13, 14). When F28 is mutated to leucine, this interaction is lost. 31P NMR studies have shown chemical shift differences of the bound phosphates in H-Ras p21(F28L) relative to H-Ras wildtype (14). In addition, Cdc42Hs(F28L)-GDP (15) as well as Ras(F28L) (14, 16) show a drastic increase in the GDP dissociation rate in the F28L mutant relative to wildtype suggesting that F28 is an important contributor to the stability of GDP binding. NMR dynamics studies of Cdc42Hs(F28L)-GDP (17) have shown several differences in flexibility at the nucleotide-binding site of Cdc42Hs(F28L)-GDP relative to wildtype (18), while the dynamics of the remainder of the protein are not significantly altered by the mutation.
In this paper, we present the solution structure of the single point mutant, Cdc42Hs(F28L)-GDP. Analysis of the solution structure of Cdc42Hs(F28L)-GDP in comparison to wildtype Cdc42Hs (9), show that, overall, the core of Cdc42Hs(F28L)-GDP is very similar to the wildtype structure. In the mutant protein, however, some of the regions comprising the nucleotide-binding site exhibit differences that may explain the increase in the GDP dissociation rate. Further, the regions that show the most pronounced differences in stabilizing interactions with the nucleotide correlate with the regions of the protein with the greatest changes in dynamics relative to wildtype (17).
Experimental Procedures
Protein Expression and Purification
A Cdc42Hs construct with a C-terminal truncation (19) was used to prepare the F28L mutation. The construct was cloned into a pET-15b vector and subsequently over expressed in E. coli. Five-ml cultures were initially grown to saturation and used to seed 1 or 2-L cultures grown in LB broth for natural abundance samples, and in minimal media containing 15NH4Cl as the sole nitrogen source for 15N-labelled samples. Expression and purification proceeded as outlined previously (17). For 2D-NMR experiments using 15N-labelled GDP, unlabelled Cdc42Hs wildtype and Cdc42Hs(F28L)-GDP samples were prepared as described above except that natural abundance NH4Cl was used, and 0.1 mM 15N-GDP was incorporated in all buffers throughout purification. Expression levels using 13C-glucose were too low to produce 15N,13C-labeled protein for triple resonance and 1H,13C-NOESYHSQC experiments.
NMR Spectroscopy
All protein samples were prepared in an NMR sample buffer solution containing 25 mM NaCl, 10 mM NaH2PO4, 5 mM MgCl2, and 1 mM NaN3 with 8% D2O at pH 5.5 at Cdc42Hs(F28L)-GDP concentrations of 0.2−0.5 mM in volumes of ∼250 μL. NMR spectra were obtained using Varian Inova 600 MHz and 500 MHz spectrometers. The 600 MHz spectrometer was equipped with a triple-resonance pulsed-field gradient probe, and the 500 MHz spectrometer with a triple-resonance cryogenic probe. All NMR spectra were acquired at 25°C in States-TPPI mode for quadrature detection (20). Carrier frequencies for 1H and 15N were 4.77 and 115.9 ppm, respectively. 2D 1H-15N HSQC spectra were acquired on 15N-labelled Cdc42Hs(F28L)-GDP. A 2D NOESY experiment was performed using a natural abundance sample (in 100% D2O) in order to detect side chain-side chain NOE peaks for some residues with relatively isolated methyl resonances. Hydrogen-deuterium exchange was measured by lyophilizing the sample and resuspending in 100% D2O. A series of 1H-15N HSQC experiments were performed at 0, 5, 15, 25, and 60 minutes, and the decrease in the volume and eventual disappearance of 1H-15N crosspeaks were used to determine the extent of exchange of deuterons for protons. 3D 1H-15N-TOCSYHSQC experiments were acquired with mixing times of 50 and 70 ms. 3D 1H-15N-NOESYHSQC spectra were acquired using mixing time of 125 ms.
NMR spectra were processed using NMRPipe, version 1.6 (21). The spectra were zero-filled, then apodized using a Gaussian window function prior to Fourier transformation. After Fourier transformation, a baseline correction was applied. Spectra were visualized and analyzed using the Xeasy program (version 1.3.6) (22). Peaks were integrated using the program Peakint (N. Schaefer, diploma thesis, ETH, Zurich, Switzerland). The backbone proton and nitrogen assignments for Cdc42Hs(F28L)-GDP were reported previously (17). We were able to assign a majority of the Hα, Hβ, Hδ, and Hγ protons by analysis of the 1H,15N-TOCSYHSQC and 1H,15N-NOESYHSQC spectra of Cdc42Hs(F28L)-GDP overlaid with the corresponding spectra of wildtype. Over 90% of the side chain protons had similar chemical shifts. Residues in two stretches T35-N39 (Switch I region) and G60, R65-R68 (Switch II region) in Cdc42Hs(F28L)-GDP were unassigned due to line broadening resulting from conformational dynamics, similar to that observed for wildtype Cdc42Hs (9).
For residual dipolar coupling (RDC) measurements, a 15N-Cdc42(F28L)-GDP sample was partial aligned in the magnetic field (Varian Inova 500) with a nonionic liquid crystalline medium made up of 4% C12E6 (n-dodecyl-hexa(ethylene glycol)) and n-hexanol (23). Alignment was verified using the quadrupolar splitting of the 2H spectrum. RDCs were determined from partially aligned and unaligned samples using TROSY experiments with the phase cycling adjusted to observe the two nitrogen components individually.
Structure Calculation
1H-15N-NOESYHSQC experiments were used to derive distance constraints. A majority of the resonances for Cdc42Hs(F28L)-GDP were similar to the corresponding resonances of wildtype, making the NOE assignment relatively straightforward. The volumes of several well dispersed and isolated sequential dαN and dαN(i,j) NOE peaks (resulting from correlation within and between parallel β-strands, respectively) were calibrated to the distances expected for ideal β-sheets (2.2 Å and 3 Å, respectively). NOESY constraints were classified according to their relative strength in the NOESY spectrum (< 2.4, < 3.4, < 4.0, and < 5.5 Å). Also, 55 hydrogen bond constraints were input into the structure calculations based on H-D exchange studies by NMR, the chemical shift index (indicating α-helix or β-sheet) (24, 25), and expected NOE patterns indicative of hydrogen bonding. For each hydrogen bond, the distance between the amide proton and the carbonyl oxygen for the residue was constrained to be 1.8−2.3 Å, and the amide nitrogen to the carbonyl oxygen distance was constrained to be between 2.5−3.3 Å (9). Dihedral constraints were included for residues in α-helices and β-sheets (α-helix, φ = −80° ± 50°, ψ = −20° ± 50°; β-sheet, φ = −105° ± 65°, ψ = 145° ± 45°). Sixty-eight RDCs from well-resolved resonances were used as additional constraints. All constraints were input into Xplor-NIH (26) to calculate an ensemble of structures for the protein. The fifteen “best” structures from 600 calculated structures were selected based on their overall low energies and minimal constraint violations. The experimental statistics from the structure calculations are given in Table 1. The structures were visualized using Swiss PDBviewer (version 3.7) (27) and MacPyMol (28), and analyzed using AQUA (29), PROCHECK-NMR (29), and the NMR ensemble program, OLDERADO (30). For Cdc42Hs(F28L)-GDP, very few intermolecular NOEs were observed between GDP and the protein (except for the N1 proton of the guanine ring), making it not possible to properly constrain GDP in the protein for the structure calculations. There was also a lack of NOE data for wildtype as only two NOEs were observed between the protein and the bound nucleotide (9), one of which involved the side chain proton of F28 not expected in the mutant protein. The position of the GDP was determined starting with superimposing the Cdc42Hs-GDP crystal structure onto the core Cα atoms of each of the final 15 Cdc42Hs(F28L) structures, similar to that done for GDP in Cdc42Hs wildtype (9). The nucleotide was then copied into the Cdc42Hs(F28L) structure from its position in the crystal structure of Cdc42Hs wildtype. The Cdc42Hs(F28L)-GDP structures were energy minimized to find the most favorable positions of the nucleotide in the protein. After minimization, NOEs that were observed between the N1 proton of the nucleotide ring and various residues of the protein were used to back calculate distances between the residues and the nucleotide ring. These distances between the N1 proton of the ring and other protons were checked in the structures and found to be compatible with observed NOEs.
Table 1.
Statistics and restraint data for the structure calculation of Cdc42Hs(F28L).
| Ensemble of 15 structures | Most representative | ||
|---|---|---|---|
| RMSD from experimental constraints | |||
| NOE distances (Å) | 0.08 ± 0.002 | 0.07 | |
| Dihedral Angles (°) | 0.99 ± 0.15 | 0.83 | |
| RMSD from secondary structure | |||
| Bonds (Å) | 0.003 ± 0.0006 | 0.003 | |
| Angles (°) | 0.56 ± 0.01 | 0.57 | |
| Impropers (°) | 0.48 ± 0.02 | 0.46 | |
| Restraint Data | |||
| Long-Range NOEs (|i-j| > 4) | 461 | ||
| Medium-Range NOEs (4 > |i-j| > 1) | 401 | ||
| Total NOEs | 862 | ||
| Intra | 361 | ||
| Sequential | 255 | ||
| Medium | 111 | ||
| Long | 134 | ||
| Hydrogen Bond | 71 5 2 | ||
| Ψ | 92 | ||
| Φ | 90 | ||
| NH Backbone RDC Restraints | 68 |
Results and Discussion
Overall Structure of Cdc42Hs(F28L)-GDP
We have determined the 3D solution structure of Cdc42Hs(F28L)-GDP in order to characterize: (1) differences in the nucleotide-binding region in Cdc42Hs(F28L)-GDP that may be consistent with the dynamics results, (2) conformational changes outside of the nucleotide-binding site that may be involved in effector binding, and (3) changes resulting from the lost interaction between the nucleotide base and the phenyl ring of F28. A set of 15 superimposed structures of Cdc42Hs(F28L)-GDP is shown in Figure 1A. The core of the protein (atoms in the protein as defined by OLDERADO (30) whose positions are well defined and largely invariant across the ensemble of structures) gave an RMSD to the average structure of 0.87 ± 0.03 Å for backbone non-hydrogen atoms (Figure 1B). The core atoms of the Cdc42Hs(F28L)-GDP structures are residues 3−9, 16−24, 27, 29, 41−47, 49−56, 75−85, 90−103, 108−116, 136, 139−155, 161−177, very similar to the core atoms of the final structures of Cdc42Hs wildtype (9). Distance, hydrogen-bonding, dihedral, and residual dipolar coupling restraints determined were used to calculate the structure of Cdc42Hs(F28L)-GDP. The secondary structure of Cdc42Hs(F28L)-GDP was determined based on analysis of H-D exchange data, Ramachandran plots of the ensemble of structures, characteristic NOE patterns for secondary structure motifs, and the chemical shift index (24, 25). There were no secondary structural differences in F28L relative to wildtype; the mutant protein also consists of a centrally located six-stranded β-sheet structure surrounding the C-terminal helix of the protein (Figure 1C). Other structural features found in both wildtype and Cdc42Hs(F28L)-GDP included helices α3 and α4 flanking the convex surface of the β-sheet, helix α1 lying perpendicular to the β-sheet on the concave side of the sheets, and a short αI (Insert) helix lying adjacent to a loop connecting β4 and α3 in the protein (Figure 3C). Also consistent with wildtype are the three regions in Cdc42Hs(F28L)-GDP that show the most structural disorder: Switch I, Switch II, and the Insert region. The disorder in Switches I and II is key to the docking of effector proteins with Cdc42Hs (18) and other Ras proteins (31).
Figure 1.
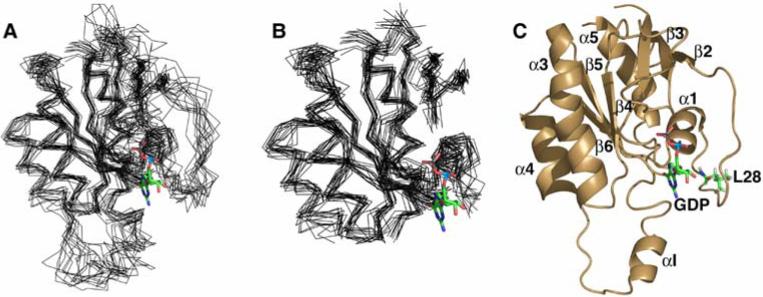
Structure Cdc42Hs(F28L)-GDP. The 15 lowest energy structures were superimposed (using the core residues indicated in the text) on the average structure generated by Xplor-NIH using MacPyMol (28). The nucleotide was superimposed on the structures from the coordinates of crystal structure of wildtype Cdc42Hs-GDP (1AN0). (A) Overlay of the entire structure, (B) overlay with the Switch I and II and the insert helix removed, and (C) a diagram of the most representative structure (defined by OLDERADO, (30)) showing the secondary structure.
Figure 3.
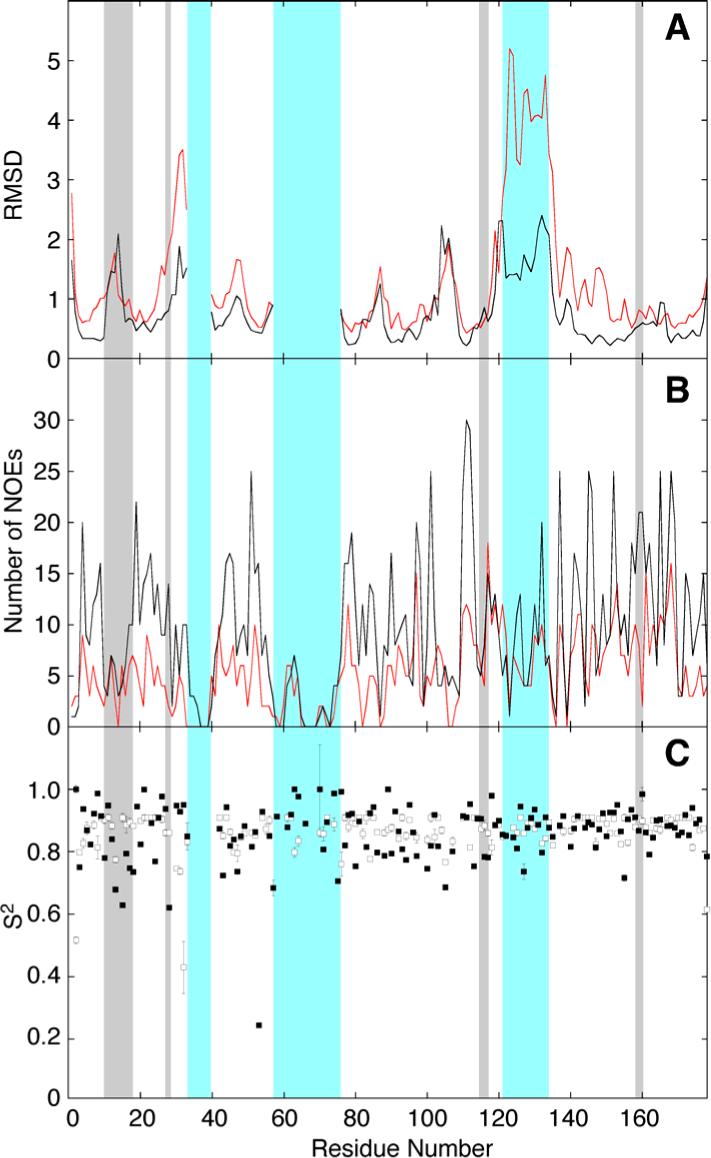
Distribution of (A) RMSDs of the Cα atom, (B) number of interresidue NOEs per residue, and (C) order parameter (17) for Cdc42Hs-GDP and Cdc42Hs(F28L)-GDP. The wildtype data for (A) and (B) were taken from Feltham et al. (9) and are shown in black (the Cdc42(F28L) data is shown in red). The data for (C) was from Adams et al. (17) and the wildtype data is shown with open squares and the Cdc42Hs(F28L) data is shown with closed squares. The gray boxes indicate the positions of the nucleotide binding pocket, and the blue boxes indicate the positions of Switch I, Switch II, and the insert region. The Switch I and II regions are poorly defined and the RMSD values have been removed. The difference between the insert region of wildtype and F28L arises largely due to the unavailability of the 13C-separated NOESY experiment for F28L (see Methods). Although still poorly defined relative to the core of the protein, the structure is somewhat more constrained in wildtype due to several long-range sidechain-sidechain NOEs.
Structure of the Nucleotide-Binding Site in Cdc42Hs(F28L)-GDP
Dynamics studies have shown increased flexibility in residues of the nucleotide-binding site in Cdc42Hs(F28L)-GDP that could affect the signaling processes of Cdc42Hs (17). The nucleotide-binding site of Cdc42Hs consists of several regions that have been described from the crystal structures of Cdc42Hs (1ANO) and H-Ras (31). The side chain hydroxyl group of T35, the main chain NH of G60, and the P Loop residues 10−15 (GXGXXG motif) interact with the phosphates of the nucleotide. Residues 115−118 interact with one side of the guanine ring of the nucleotide, while residues 158−160 and 28 interact with the opposite side of the guanine ring. Resonances for T35 and G60 could not be assigned, presumably because they are located in the flexible Switch regions of the protein. Figure 2 shows the remaining nucleotide-binding site residues of 15 low energy structures of Cdc42Hs(F28L)-GDP with the core atoms of the protein superimposed. Figure 3 shows the sequence-dependent number of interresidue NOEs, RMSDs of the Cα's, and the order parameters (17) of Cdc42Hs(F28L)-GDP compared to wildtype. Because of the differences in the number of NOEs used in the wildtype vs. F28L and the use of RDC restraints for F28L, the RMSD values are not directly comparable. For that reason, specific regions of the each protein are compared to the relatively well-defined core of the protein. The RMSD value for the nucleotide-binding site residues (10−15; 28; 115−118; 158−160) is increased 52% relative to the protein core, in comparison to a 46% increase for the corresponding residues in wildtype (Table 2), with the most striking increase in RMSD surrounding residue 28. Some flexibility in the nucleotide-binding site is expected in order to facilitate proper cycling of GTP to GDP in Cdc42Hs, but the increased RMSD, particularly in the region of L28, may play a role in the increased rate of nucleotide dissociation.
Figure 2.
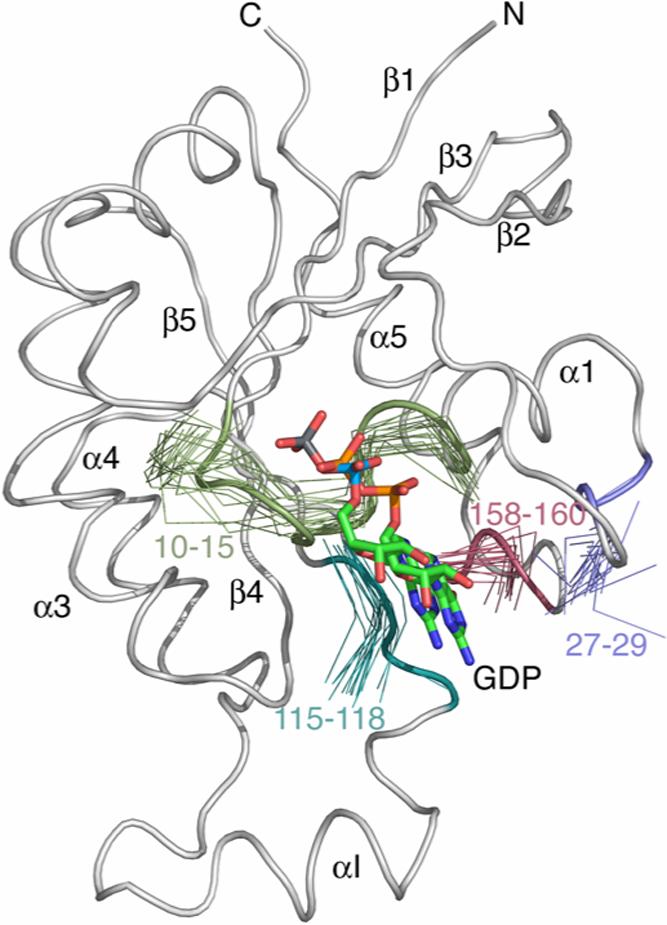
A ribbon diagram of the crystal structure of wildtype Cdc42Hs (1AN0) with segments of the 15 lowest energy structures of Cdc42Hs(F28L)-GDP representing the binding pocket. All structures were superimposed on the average structure computed from the 15 structures shown in Figure 1A.
Table 2.
Percent Increase in Backbone RMSD (%ΔR)a for Regions in Nucleotide-Binding Site Relative to the Average RMSD Value of the Protein coreb.
| Construct |
||
|---|---|---|
| Cdc42Hs-GDP | Cdc42Hs(F28L)-GDP | |
| %ΔR All backbone atoms in nucleotide binding site (10−15;115−118;158−160;28) | 46.0 | 52.0 |
| %ΔR (10−15) | 56.0 | 62.0 |
| %ΔR (115−118) | 47.0 | 65.0 |
| %ΔR (158−160) | 0.0 | 31.0 |
| %ΔR (28) | 11.0 | 84.0 |
Where %ΔR = × 100
Cdc42Hs-GDP RMSDcore=0.59 (9).
The principal stabilizing factor for the nucleotide in its binding site is F28. Studies have shown that mutation of this amino acid to leucine increases the GDP dissociation rate significantly in Ras (14, 16) and Cdc42Hs (15). An examination of this region in the crystal structure of wildtype Ras (10, 14), and Cdc42Hs (1ANO) shows that the phenyl ring of F28 is oriented perpendicularly to the guanine ring of the bound nucleotide and serves to stabilize the nucleotide in the binding pocket, an interaction not possible in Cdc42Hs(F28L)-GDP. The RMSD for the backbone atoms of L28 is 84% greater than the core of the mutant protein whereas the difference in RMSD for F28 relative to its core in wildtype is negligible (11%; Table 2). Dynamics studies also showed that the backbone of L28 in Cdc42Hs(F28L)-GDP exhibits an increased amplitude of local motion relative to the rest of the protein (17), suggestive of conformational disorder for the L28 backbone not seen for F28 in wildtype. Additionally, the crystal structure of Cdc42Hs wildtype shows that a hydrophobic interaction between the side chain of F28 and L160 (another nucleotide-binding site residue) acts to help orient the phenyl side chain (1ANO). As judged from the lack of NOE crosspeaks between the sidechain of L28 and L160, this interaction is lost by the F28L mutation, contributing to the diminished stability of the nucleotide in Cdc42Hs(F28L)-GDP.
The P Loop region (residues 10−15) interacts with the phosphate groups of the bound nucleotide. Both wildtype Cdc42Hs-GDP and Cdc42Hs(F28L)-GDP show some conformational disorder in this region (Table 2). However, dynamics studies have suggested that the P loop residues of Cdc42Hs(F28L)-GDP display more chemical exchange than the corresponding residues in wildtype. Also, the chemical environment of the P Loop residues is different in F28L relative to wildtype, as suggested by significant chemical shift differences for most of these residues (17). Analysis of the wildtype NOESY spectra show that there are several backbone NH NOEs between residues of the P Loop and the α3 helix (G10NH-N92Hε, D11NH-W97Hε, and G12NH-W97Hε) and residues 116−118. These interactions help stabilize the phosphate region of the bound nucleotide (11). Only the D11NH-W97Hε NOE is seen in the NOESY spectrum of F28L. In addition, all six residues of the P Loop undergo H-D exchange in F28L, while only residues 12 and 14 exhibit H-D exchange in wildtype. In addition, hydrogen bonding networks between the backbone NH's of residues 16−18 and the α and β-phosphate groups of the bound nucleotide evident in wildtype Cdc42Hs, which help stabilize the phosphates of the nucleotide (1AN0), are much weaker in Cdc42Hs(F28L)-GDP (17). NOEs observed in wildtype Cdc42Hs between the NH's of T17 and C18 and the Hε protons of G116, a residue in another region directly interacting with the nucleotide, are weak or non-existent in F28L. Although not obvious from the average solution structure, the apparently weaker hydrogen bonds and flexibility on the μs-ms timescale (17) may contribute to a greater degree of conformational freedom for the nucleotide and increased rate of dissociation.
The guanine base of the nucleotide is also stabilized via interactions with residues 115−118 on the side of the nucleotide ring opposite F28 and with residues 158−160 perpendicular to the nucleotide ring on the same side of the nucleotide as F28 (1ANO). Studies of H-Ras have shown that the principal function of residues 116−119 (residues 115−118 in Cdc42Hs) is to provide a link between the other regions of the protein involved in nucleotide-binding: the P Loop residues, and residues 145−147 (158−160 in Cdc42Hs) (31). The chemical shifts for these residues differ little between wildtype and F28L (17), indicating that the chemical environment is similar. Also, the dynamics parameters for these residues are similar in both wildtype and F28L (17). Likewise, little change is seen in the solution structures of the two forms of Cdc42Hs (Figure 2, Table 2). Despite similarities between wildtype and F28L, several NOEs are observed in wildtype Cdc42Hs between the backbone NH of residues 115−118 and other regions that are absent in the F28L spectra (Table 3). The structure of wildtype Cdc42Hs shows that the NH of Q116 has a hydrogen bond with the carbonyl of C157, and D118 is hydrogen-bonded to the hydroxyl side chain of S158 helping to form a tight binding pocket for the nucleotide. In F28L, these two hydrogen bonds are weak or nonexistent as H-D exchange is seen for the backbone NH of Q116 and D118.
Table 3.
NH NOEs for Cdc42Hs (wildtype) and Cdc42Hs(F28L) for residues comprising the nucleotide binding site.
| Cdc42Hs (wildtype) | Cdc42Hs(F28L) |
|---|---|
| 11HN-92Hδ | 11HN-97Hε |
| 11HN-97Hε | 12HN-61Hα |
| 12HN-97Hε | |
| 15HN-85Hγ | |
| 17HN-116Hε | |
| 18HN-28 Hε | |
| 18HN-116Hε | |
| 28HN-160Hδ | 28HN-160Hδ |
| 116HN-158Hα | 116HN-158Hα |
| 116HN-159Hb | |
| 117HN-161Hγ | |
| 117HN-158Hα | 117HN-161HN |
| 117HN-84HN | |
| 118HN-158Hα | |
| 118HN-160Hδ | 118HN-161Hγ |
| 158HN-117Hγ | |
| 158HN-117Hδ | |
| 157HN-117Hγ | |
| 158HN-117Hγ | 158HN-117HN |
| 158HN-117Hδ | 158HN-117Hα |
| 159HN-117Hγ | 159HN-27HN |
| 159HN-117HN | |
| 160HN-28 Hδ | 160HN-28HN |
The structure of F28L in the region of residues 158−160 shows a small increase in the RMSD (%ΔR = 31) while the corresponding region in wildtype shows no change in RMSD relative to its core (%ΔR ≈ 0). The chemical shift difference for these residues between wildtype and F28L is also significant, indicating that these residues lie in a different chemical environment in Cdc42Hs(F28L)-GDP (17). In addition, several more NOEs are observed between the backbone NH of these residues and other residues in the nucleotide-binding site for the wildtype protein than for F28L (Table 3). In the crystal structure of Cdc42Hs (1ANO), the backbone NH of S158 is hydrogen bonded to the carbonyl of K163, while the backbone NHs of A159 and L160 are hydrogen bonded to the carbonyl oxygen of the guanine ring. All three of these hydrogen-bonding interactions are weak or missing in F28L as determined from H-D exchange experiments.
The Nucleotide
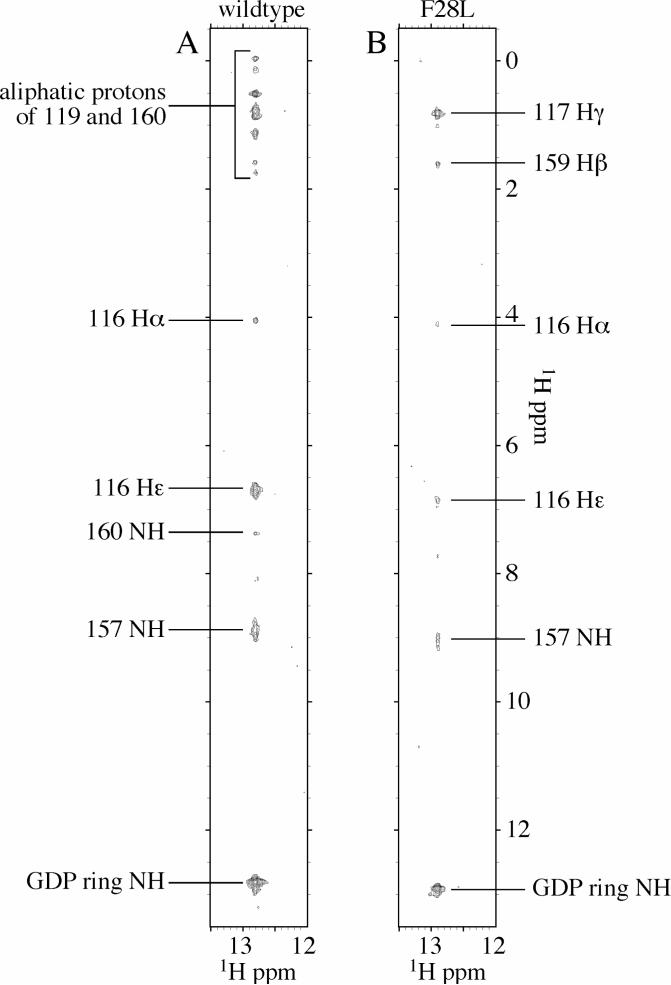
Compelling evidence for conformational differences at the nucleotide-binding site in Cdc42Hs(F28L)-GDP relative to wildtype come from the 2-D NMR spectrum of 15N-labelled-GDP complexed to both proteins (Figure 4). The N1 proton of the guanine ring in the wildtype complex has a chemical shift of 12.81 ppm versus 12.92 ppm in the Cdc42Hs(F28L)-GDP complex. This finding is indicative of a more polar environment for the proton in the F28L complex due to either more solvent exposure or an increased interaction with a more electron-rich group (32, 33). NMR analysis of 15N-GDP-bound Cdc42Hs and Cdc42Hs(F28L)-GDP show that the nucleotide makes more contacts with residues of the nucleotide-binding site in wildtype than in Cdc42Hs(F28L)-GDP. The 1H-15N NOESYHSQC spectra of Cdc42Hs-15N-GDP and Cdc42Hs(F28L)-GDP-15N-GDP show NOE crosspeaks involving the nucleotide N1 proton and residues 116 and 157 in both proteins. However, several crosspeaks involving the nucleotide N1H and residues 119 and 160 in the spectrum of wildtype-15N-GDP are not observed in the corresponding spectrum of Cdc42Hs(F28L)-15N-GDP. Likewise, the 1H-15N-NOESYHSQC spectrum of Cdc42Hs(F28L)-15N-GDP shows two NOE crosspeaks involving residues 117 γ and 159 Hβ that were not evident for wildtype, suggestive of a conformational change involving the nucleotide that brings the N1H of the nucleotide ring into proximity to both the A159 Hβ and I117 Hγ protons in Cdc42Hs(F28L)-GDP. These studies reveal several different contacts between the nucleotide and residues of the wildtype protein versus those in F28L due perhaps to a number of lost interactions in the nucleotide-binding site for Cdc42Hs(F28L)-GDP.
Figure 4.
A 1H,15N-NOESYHSQC spectrum of 15N-GDP bound to wildtype (A) and F28L (B) Cdc42Hs-GDP. The experiment was run in two dimensions, with an INEPT sequence filtering only protons bound to nitrogen prior to detection.
Summary
We have shown previously that this single-point mutation introduces increased flexibility on multiple timescales to most of the residues comprising the nucleotide-binding site as well as the nucleotide itself (17). We show here that although the structural changes are subtle, the interactions within in the nucleotide-binding pocket, both within the protein and between the protein and nucleotide, likely contribute to the decrease in affinity for nucleotide. NMR studies on H-Ras p21(F28L) indicated that the mutation distorted the binding site around the α and β phosphate groups, but the area surrounding the γ phosphate group was undisturbed (34). While this study does not address the γ phosphate of the nucleotide because only the GDP form has been studied here, our results do show that the conformational differences in this mutant are not confined to the site of the mutation, and that the entire nucleotide-binding site of Cdc42Hs(F28L)-GDP is disturbed.
Most oncogenic Ras proteins are locked in a perpetually active state (GTP-like state), and transduce signals that are directly involved in proliferation, transformation, or tumorigenesis (35). Mutations that alter the intrinsic ability of the protein to hydrolyze GTP to GDP and affect proper exchange may facilitate oncogenesis (1, 11, 36). Therefore, these proteins and the signal transduction pathways that help to regulate them are important targets for anticancer therapy. As mentioned previously, Cdc42Hs(F28L)-GDP mimics the fast nucleotide exchange and transformation capability caused by an interaction of wildtype Cdc42Hs with the Dbl oncoprotein (37). These studies describing the structural correlates of the F28L mutation in Cdc42Hs have provided important molecular information about the protein's function, and will be extended to other mutations that affect normal functioning in signaling pathways.
Acknowledgements
We thank the Laboratory of Chemical Physics at the National Institutes of Health and Drs. Dan Garrett and Frank Delaglio for making available the programs PIPP and NMRPipe, which were useful in analyzing our NMR data. We thank Ahmed Ahmed, Adrienne Loh, and Dawit Gizachew for helpful discussions.
Abbreviations
- F28L
mutation of phenylalanine 28 in Cdc42Hs to leucine
- GDP
guanosine-5′-diphosphate
- GTP
guanosine-5′-triphosphate
- GAP
GTPase activating protein
- GEF
guanidine nucleotide exchange factor
- GDI
guanidine nucleotide dissociation inhibitor
- HSQC
heteronuclear single quantum correlation
- IPTG
isopropyl β-D-thiogalactopyranoside
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- RDC
residual dipolar coupling
- P Loop/Loop 1
residues 10−15 on Cdc42Hs
- Switch I
residues 31−40 on Cdc42Hs
- Switch II
residues 57−74 on Cdc42Hs
- Insert region
residues 121−134 on Cdc42Hs
Footnotes
This material is based upon work supported by the National Science Foundation under a grant awarded to PDA in 2002 and by a grant from the National Institutes of Health (R01 GM56223) to REO. The coordinates of Cdc42Hs(F28L)-GDP have been deposited with the Protein Data Bank as 2ASE.
References
- 1.Barbacid M. ras Genes. Ann. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.Bourne HR, A. SD, F. M. The GTPase superfamily: Conserved Structure and Molecular Mechanism. Nature. 1991:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 3.Hart M, Eva A, Evans T, Aaronson S, Cerione R. Catalysis of Guanine Nucleotide Exchange on the Cdc42Hs Protein by the dbl Oncogene Product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 4.Horii Y, Beeler JF, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signaling pathways. Embo J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonard D, Hart MJ, Platko JV, Eva A, Henzel W, Evans T, Cerione RA. The identification and characterization of a GDP-dissociation inhibitor (GDI) for the CDC42Hs protein. J Biol Chem. 1992;267:22860–22868. [PubMed] [Google Scholar]
- 6.Farr CJ, Saiki RK, Erlich HA, McCormick F, Marshall CJ. Analysis of RAS gene mutations in acute myeloid leukemia by polymerase chain reaction and oligonucleotide probes. Proc Natl Acad Sci U S A. 1988;85:1629–1633. doi: 10.1073/pnas.85.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiaris H, Ergazaki M, Spandidos DA. Instability at the H-ras minisatellite is associated with the spontaneous abortion of the embryo. Biochem Biophys Res Commun. 1995;214:788–792. doi: 10.1006/bbrc.1995.2355. [DOI] [PubMed] [Google Scholar]
- 8.Shinohara N, Koyanagi T. Ras signal transduction in carcinogenesis and progression of bladder cancer: molecular target for treatment? Urol Res. 2002;30:273–281. doi: 10.1007/s00240-002-0275-0. [DOI] [PubMed] [Google Scholar]
- 9.Feltham JL, Dotsch V, Raza S, Manor D, Cerione RA, Sutcliffe MJ, Wagner G, Oswald RE. Definition of the switch surface in the solution structure of Cdc42Hs. Biochemistry. 1997;36:8755–8766. doi: 10.1021/bi970694x. [DOI] [PubMed] [Google Scholar]
- 10.Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine nucleotide binding domain of the Ha-Ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- 11.Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. refined crystal structure of the triphosphate conformation of h-ras p21 at 1.35 Å resolution: implication for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirshberg M, Stockley RW, Dodson G, Webb MR. The crystal structure of human rac1, a member of the rho-family complexed with a GTP analogue. Nature Struc. Biol. 1997;4:147–152. doi: 10.1038/nsb0297-147. [DOI] [PubMed] [Google Scholar]
- 13.deVos AM, Tong L, Milburn M, Matias P, Jancarik J, Miura K, Noguchi S, Nishimura S, Ohtsuka E, Kim S-H. Science. 1988;239:883–893. doi: 10.1126/science.2448879. [DOI] [PubMed] [Google Scholar]
- 14.Schlichting I, John J, Frech M, Chardin P, Wittinghofer A, Zimmermann H, Roesch P. Proton NMR Studies of Transforming and Nontransforming H-ras p21 Mutants. Biochemistry. 1990;29:504–511. doi: 10.1021/bi00454a026. [DOI] [PubMed] [Google Scholar]
- 15.Lin R, Bagrodia S, Cerione R, Manor D. A novel Cdc42Hs mutant induces cellular transformation. Curr Biol. 1997;7:794–797. doi: 10.1016/s0960-9822(06)00338-1. [DOI] [PubMed] [Google Scholar]
- 16.John J, Frech M, Wittinghofer A. Biochemical properties of Ha-ras encoded p21 mutants and mechanism of the autophosphorylation reaction. J Biol Chem. 1988;263:11792–11799. [PubMed] [Google Scholar]
- 17.Adams PD, Loh AP, Oswald RE. Backbone dynamics of an oncogenic mutant of Cdc42Hs shows increased flexibility at the nucleotide-binding site. Biochemistry. 2004;43:9968–9977. doi: 10.1021/bi0490901. [DOI] [PubMed] [Google Scholar]
- 18.Loh A, Guo W, Nicholson L, Oswald RE. Backbone dynamics of inactive, active, and effector-bound Cdc42Hs from measurements of 15N relaxation parameters at multiple field strengths. Biochemistry. 1999;38:12547–12557. doi: 10.1021/bi9913707. [DOI] [PubMed] [Google Scholar]
- 19.Guo W, Sutcliffe MJ, Cerione RA, Oswald RE. Identification of the binding surface on Cdc42Hs for p21-activated kinase. Biochemistry. 1998;37:14030–14037. doi: 10.1021/bi981352+. [DOI] [PubMed] [Google Scholar]
- 20.States D, Haberkorn R, Ruben D. A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J. Magn. Reson. 1982;48:282–292. [Google Scholar]
- 21.Delaglio F, Grzeseik S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Bartels C, Xia T-H, Billeter M, Guntert T, Wuthrich K. Journal of Biomolecular NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 23.Otting G, Rückert M. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 2000;122:7793–7797. [Google Scholar]
- 24.Wishart DS, Sykes BD, Richards RM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 25.Wishart DS, Sykes BD, Richards FM. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 26.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 27.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 28.DeLano WL. The PyMol molecular graphics system. 2002 http://www.pymol.org.
- 29.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 30.Kelley LA, Sutcliffe MJ. OLDERADO: on-line database of ensemble representatives and domains. On Line Database of Ensemble Representatives And DOmains. Protein Sci. 1997;6:2628–2630. doi: 10.1002/pro.5560061215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wittinghofer A, Pai E. The Structure of Ras protein: a Model for a Universal Molecular Switch. Trends Biomed. Sci. 1991;16:382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]
- 32.Redfield AG, Papastavros MZ. NMR Study of the Phosphoryl Binding Loop in Purine Nucleotide Proteins: Evidence for Strong Hydrogen Bonding in Human N-ras p21. Biochemistry. 1990;29:3509–3514. doi: 10.1021/bi00466a013. [DOI] [PubMed] [Google Scholar]
- 33.Wuthrich K. NMR of Proteins and Nucleic Acids. John Wiley and Sons; New York: 1986. [Google Scholar]
- 34.Reinstein J, Schlichting I, Frech M, Goody R, Wittinghofer A. p21 with a Phenylalanine 28 to Leucine Mutation Reacts Normally with the GTPase Activating Protein GAP but Nevertheless Has Transforming Properties. J. Biol. Chem. 1991;266:17700–17706. [PubMed] [Google Scholar]
- 35.Ramirez de Molina A, Rodriguez-Gonzalez A, Lacal JC. From Ras signalling to ChoK inhibitors: a further advance in anticancer drug design. Cancer Lett. 2004;206:137–148. doi: 10.1016/j.canlet.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 36.Bos JL, Toksoz D, Marshall CJ, Verlaan-de Vries M, Veeneman GH, van der Eb AJ, van Boom JH, Janssen JW, Steenvoorden AC. Amino-acid substitutions at codon 13 of the N-ras oncogene in human acute myeloid leukaemia. Nature. 1985;315:726–730. doi: 10.1038/315726a0. [DOI] [PubMed] [Google Scholar]
- 37.Hart MJ, Eva A, Zangrilli D, Aaronson SA, Evans T, Cerione RA, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]