Abstract
D-cycloserine (DCS) is only used with multi-drug resistant strains of tuberculosis because of serious side-effects. DCS is known to inhibit cell wall biosynthesis, but the in vivo lethal target is still unknown. We have applied NMR-based metabolomics combined with principal component analysis to monitor the in vivo affect of DCS on M. smegmatis. Our analysis suggests DCS functions by inhibiting multiple protein targets.
Keywords: NMR, Principal Component Analysis, Metabolomics, Tuberculosis, D-cycloserine, Mycobacterium smegmatis, D-alanine racemase
Introduction
Tuberculosis (TB) is a world-wide problem as indicated by 5.1 million new and relapsed cases reported in 2005 with South East Asia and Africa accounting for 35% and 23% of these cases, respectively.1, 2 The TB case rate has increased dramatically in the past decade due to the HIV/AIDS epidemic.1, 3 The high success rate of TB infections has contributed to the emergence of multi-resistant strains of Mycobacterium tuberculosis.4 These multi-drug resistant tuberculosis strains (MDR-TB) are associated with high mortality (50-80%).5 Antibiotic resistance in M. tuberculosis primarily emerges because of inadequate treatments or patient noncompliance6, 7, which select for strains with decreased susceptibility to a specific drug.1, 8 Increased resistance to first line drugs9, such as isoniazid and rifampin, is creating a serious health crisis that requires the development of novel drugs to control future outbreaks of M. tuberculosis.10, 11
D-cycloserine (DCS) is a second line drug for tuberculosis that is primarily used with MDR-TB.12, 13 Second line drugs, like DCS, tend to have higher toxicity. Serious side-effects associated with DCS have included seizures and mental disorders (schizophrenia, depression, suicidal tendencies). DCS agonist activity against the N-methyl-D-aspartate (NMDA) receptors may contribute to these observed neural disorders.14 Nevertheless, the potent activity of DCS against MDR-TB makes this compound an attractive prototype for developing new antitubercle drugs.
DCS is a cyclic analogue of D-alanine, which is an essential metabolite for peptidoglycan synthesis.13, 15, 16 DCS has been shown to inhibit D-alanine racemase (Alr) and D-alanine-D-alanine ligase (Ddl) activity in vitro.17, 18 Recent studies have shown that mutations leading to the over production of Alr decrease the susceptibility of mycobacteria to DCS.19, 20 Similarly, an alr null mutant increases the susceptibility of mycobacteria to DCS, but surprisingly bacterial viability is not dependent on D-alanine.21 Thus, the lethal cellular target(s) of DCS is still undetermined, but since mycobacteria sensitivity to DCS depends on Alr activity, this suggests Alr is still a potential in vivo target contributing to the overall effect of DCS. Potentially, the therapeutic activity of DCS may require inhibiting multiple proteins. Identifying the in vivo target(s) and the associated metabolic pathways affected by DCS will provide better insight into the mechanism of DCS antitubercular activity.
The enzymes involved in peptidoglycan biosynthesis and D-alanine metabolism can serve as targets to monitor the in vivo activity of DCS through changes in the metabolome.22 Any change in the activity of these enzymes through mutations or drug activities will affect the network of metabolites associated with the corresponding pathways.23-25 Thus, monitoring changes in metabolite concentrations will reflect corresponding changes in protein activity caused by DCS or other drug activity. Nuclear magnetic resonance (NMR) is routinely used to monitor perturbations in the metabolome by collecting one dimensional (1D) 1H NMR spectra of cell extracts followed by multivariate data analysis.26-28 Principal component analysis (PCA) is routinely used to reduce the multivariate NMR data into a two dimensional (2D) scores plot. Each successive component captures the largest residual variance within the NMR spectral array, and is orthogonal to the previous components in the NMR spectra.29, 30 Each NMR spectrum is reduced to a single point in the 2D scores plot, where similar spectra will cluster close together and metabolome variants will form distinct and separate clusters.
The aim of this study is to apply NMR based metabolomics to understand the in vivo drug activity of DCS and to determine if D-alanine racemase is a target of DCS in nonpathogenic Mycobacterium smegmatis, a model system for tuberculosis for highly conserved pathways. The metabolome of wild type (mc2155)31, D-alanine racemase null mutant (TAM23)21, TAM23 complemented with wild-type alr gene (TAM23 pTAMU3), D-alanine racemase over producing and DCS resistant mutant (GPM14)19 and another DCS-resistant strain unrelated to alr mutations (GPM16)19 in the presence and absence of DCS was analyzed using 1H NMR and PCA.
Materials and methods
Analysis of colony morphology
M. smegmatis cultures were grown to saturation in MADC (OD600 of ∼1-2). Serial dilutions were plated on MADC agar supplemented with 50 mM D-alanine or 75ug/mL DCS as indicated and grown at 37°C. Digital images of representative colonies were taken with a Nikon CoolPix 8700 camera and processed for enhancement of brightness and contrast with GIMP v2.2.13 using the Retinex filter.
Preparation of NMR Metabolomic Samples
A total of 120 cultures of M. smegmatis samples were grown in the following 10 groups; (1) mc2155 (wild type), (2) TAM23 (D-alanine racemase null mutant), (3) TAM23 pTAMU3 (TAM23 complemented with wild-type alr gene) (4) GPM16 (DCS resistant mutant), (5) GPM14 (D-alanine racemase over producing mutant), (6) mc2155 with DCS, (7) TAM23 with DCS, (8) TAM23 pTAMU3 with DCS, (9) GPM16 with DCS and (10) GPM14 with DCS. Each culture is grown at 37°C with shaking at 200 rpm in 50mL of Middlebrook 7H9 broth (250mL flask) for 10-12 hours (OD600 = 0.6-1.0). Cultures were treated with 75 μg/ml DCS 2 hours before harvesting. Each culture was placed on ice for 5 minutes and then centrifuged for 10 minutes at 2,700 rpm. The cell pellets were washed twice with 30 ml of ice cold double distilled water. The cell pellets were resuspended with 10 ml of double distilled water and transferred to 30 ml Pyrex beakers. The cell pellets were then sonicated on a salt-ice water bath with a Vibra Cell Model VC600 for 5 minutes in the presence of 30% (vol/vol) type A-5 alumina. The cells were centrifuged again for 30 minutes at 15,000 rpm, and the supernatant was collected to remove any cell debris. The supernatant was transferred in 50 ml Corning orange cap tubes and frozen in an EtOH-dry ice bath to store at -80° C until ready to be analyzed. The supernatant was lyophilized and resuspended in 1.0 ml of 100% D2O containing 50 mM phosphate buffer (uncorrected pH = 7.2) with 50μM 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TMSP). The samples were stirred and centrifuged, where a 500 μL portion of the cell free extract was transferred to an NMR tube.
NMR Data Processing and Multivariate Data Analysis
The NMR spectra were collected on a Bruker 500 MHz Avance spectrometer equipped with a triple-resonance, Z-axis gradient cryoprobe. A BACS-120 sample changer with Bruker Icon software was used to automate the NMR data collection. 1H NMR spectra were collected with a standard Bruker pulse sequence (zgpr), solvent presaturation, a sweep width of 5482.5 Hz, 32K data points at 298K. A total of 4 dummy scans and 512 scans were used to obtain each of the NMR spectra. All peak positions were measured relative to the TMSP reference peak set to 0.0 ppm.
The 8 groups of 10 (GPM14, GPM16, TAM23, TAM23 pTAMU3 with/without DCS) and 2 groups of 20 (mc2155 with/without DCS) NMR spectra were processed automatically using a standard processing macro in the ACD/1D NMR manager version 8.0 (Advanced Chemistry Development, Inc., Toronto, Ontario). The residual H2O NMR resonance between 4.48 and 5.11 ppm was set to 0 and excluded from bucketing and PCA analysis. The baseline was corrected using spectrum averaging with a box half-width of 15 points and a noise factor of 2 to differentiate between real peaks and noise regions. Intelligent bucketing was used to integrate each region with a bin size of 0.025 ppm with a width looseness of 50%. The table of integrals was transferred to MS Excel, which was used to arrange all 120 samples based on the ten groups described above. An Excel macro was then used to combine the NMR spectra into a single file to normalize the binned intensities to a total integrated intensity of 1.0. The table was then imported into SIMCA (UMETRICS, Kinnelon, NJ) for PCA analysis using the program's standard parameters. Our previous analysis indicated that the presence of noise regions in the bucketing of NMR spectra can result in large and irrelevant variations in the PCA clustering32. Exclusion of the noise regions of the 1H NMR spectra was accomplished by an Excel macro that set the value of every bin below a certain intensity threshold to zero. Regions outside the spectral width of 0.0 ppm and 10.0 ppm and between 4.48 ppm to 5.11 ppm were removed to eliminate any undesired variations.
Results and Discussion
Mycobacterium smegmatis metabolome model system
Peptidoglycan biosynthesis is a conserved biological process present in most eubacteria including M. tuberculosis and M. smegmatis.33 Peptidoglycan forms the backbone of the lipid-rich mycobacterial cell wall and functions as a barrier against most drugs.34 It also withstands internal osmotic pressure and defines the cell shape.35 Peptidoglycan is formed via the peptidoglycan biosynthesis pathway, where in mycobacteria each chain is composed of alternating N-acetylglucosamine (GlcNAc) and, N-glycolylmuramic or N-acetylmuramic acid subunits.36 A pentapeptide is attached to each disaccharide unit at the muramic acid residue, where the final two D-alanine side chain amino acids are attached as a dipeptide.37, 38 This dipeptide participates in the cross-linking between peptidoglycan chains. Thus, the D-alanyl-D-alanine dipeptide is an essential building block of the peptidoglycan chain and is formed as part of the D-alanine pathway illustrated in Figure 1. D-alanine is a necessary precursor to the dipeptide and is converted from naturally occurring L-alanine by the enzyme alanine racemase (Alr).39, 40 The dipeptide is formed from the condensation of two D-alanine moieties by the enzyme D-alanine-D-alanine ligase (Ddl).35, 37
Figure 1.
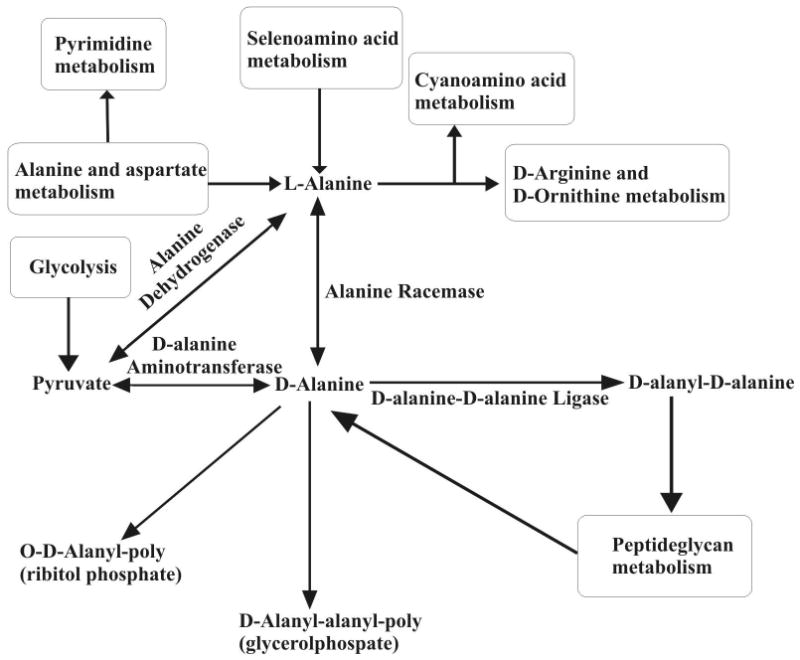
Alanine racemase converts L-alanine to D-alanine in the alanine pathway. D-alanyl-D-alanine ligase binds two molecules of D-alanine by peptide bonds, which is then used in the peptidoglycan biosynthesis pathway to form the cell wall.
The null mutant alr gene was used to generate the TAM23 M. smegmatis mutant strain where D-alanine racemase is inactivated, eliminating a pathway for the conversion of L-alanine into D-alanine.21 The alr gene has been found in many other bacterial species such as E. coli and streptococcus species.40 Two alr genes have been identified in E. coli41, 42, where alr encodes for biosynthesis activity and the dadX gene is involved in L-Ala catabolism.40, 42, 43 E. coli cell growth is completely dependent on supplemental D-alanine when both D-alanine racemase genes are inactivated.42, 43 Only one known alr gene is found in M. smegmatis.
DCS is effective against mycobacterium because it disrupts cell wall synthesis by inhibiting peptidoglycan biosynthesis. DCS is known to inhibit both Alr and Ddl in a concentration dependent manner.17, 18 However, over production of Alr contributes to DCS resistance suggesting that Alr may be an additional non-lethal binding target of DCS (Table 1). Nonetheless, Alr inhibition may contribute to the overall bactericidal effect as reflected by the hypersusceptibility of TAM23 to DCS.16, 19 The Alr over producing mutant (GPM14) strain and an unrelated DCS-resistant (GPM16) strain were identified from a group of M. smegmatis mutants that occurred spontaneously.
Table 1.
Summary of M. smegmatis cell strains and the minimal inhibitory concentration (MIC) used in the NMR metabolomics study.
| M. smegmatis Strain | Relevant Characteristics | MIC DCS
(μg/ml) |
Reference |
|---|---|---|---|
| mc2155 | Alr+; high efficiency plasmid transformation mutant of mc26 | 75.0 | Snapper et al. (1990)31 |
| GPM14 | First-step DCSr spontaneous mutant derived from mc2155; overproduces D-alanine racemase | 300 | Caceres et al. (1997)19 & Feng et al. (2003)16 |
| GPM16 | First-step DCSr spontaneous mutant derived from mc2155 | 150 | Caceres et al. (1997)19 |
| TAM23 | Alr- Kanr; M. smegmatis alr mutant derived from mc2155 | 2.56 | Chacon et al. (2002)21 |
| TAM23 pTAMU3 | Alr+ Hygr Kanr; M. smegmatis alr mutant complemented with wild-type gene integrated at the mycobacteriophage L5 attB site | 75.0 | Chacon et al. (2002)21 |
M. smegmatis dependence on D-alanine for growth
D-alanine is a major component in cell wall synthesis and is essential for cell survivability.38, 44 The production of D-alanine is a major component of the alanine pathway (Figure 1). The effect of the presence of D-alanine in the culture medium on the growth of wild type M. smegmatis (mc2155), the TAM23 null mutant and the TAM23 pTAMU3 alr complement mutant was explored by examining the colony morphology for the three strains in both the presence and absence of D-alanine (figure 2). Wild type M. smegmatis and the TAM23 and TAM23 pTAMU3 mutants were grown in MADC agar supplemented with 50 mM D-alanine, where a second culture of cells were grown without D-alanine. Similar cell cultures were obtained for the two DCS resistant (GPM14, GPM16) strains.
Figure 2.
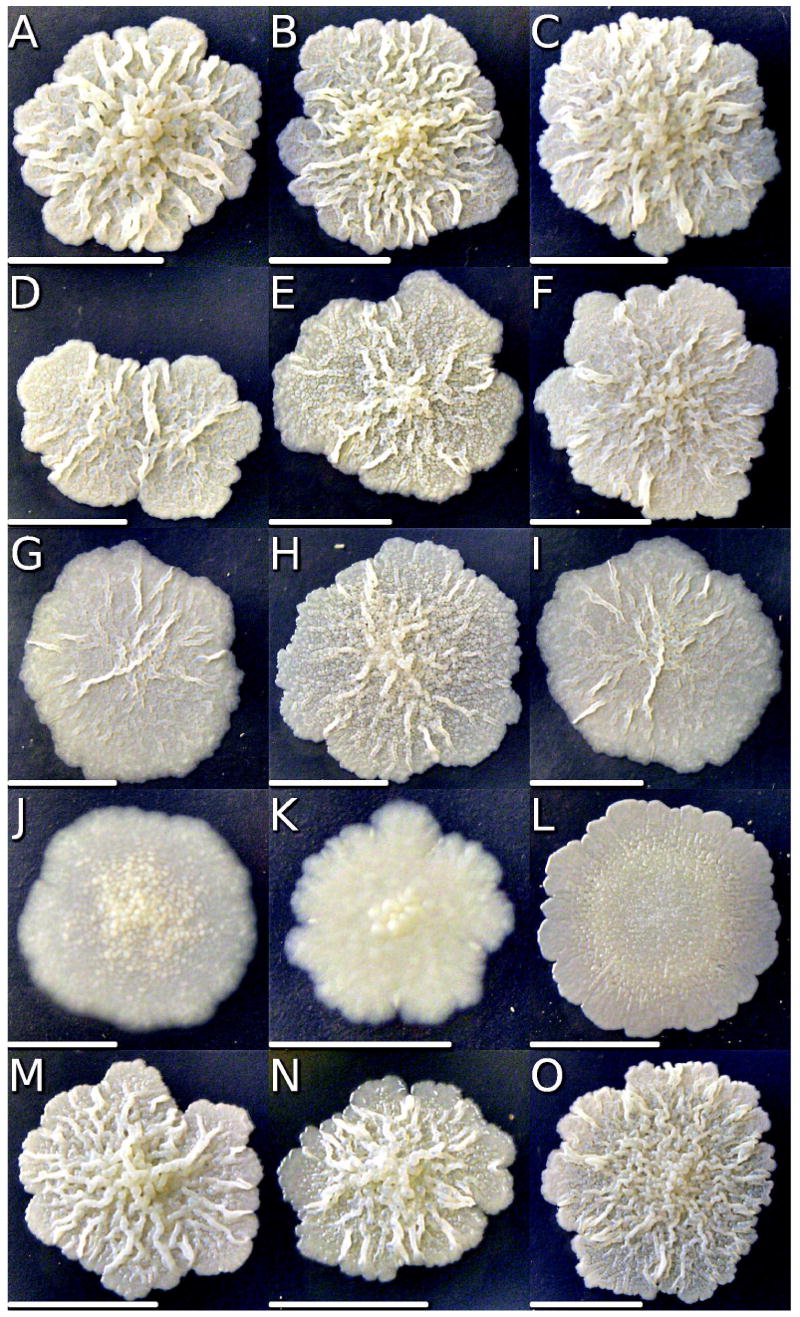
Wild type mc2155 (A-C) and mutants GPM 14 (D-F), GPM 16 (G-I), TAM 23 (J-L), and TAM23 pTAMU3 (M-O) cells were plated on MADC (A,D,G,J), MADC with 50 mM D-alanine (B,E,H,K), MADC with 50 μg/ml DCS (C,F,I), or MADC with 1.73 μg/ml DCS (L). Plates were incubated at 37°C until colony diameters were c.a. 1.0 cm and colony images were digitally captured, cropped, and scaled to similar sizes. Each picture has a scale bar (5.0 mm) to indicate the actual colony size.
If the only source of D-alanine for M. smegmatis is from the conversion of L-alanine by D-alanine racemase, then the viability of the TAM23 null mutant should be completely dependent on a supplemental source of D-alanine. Surprisingly, this is not the case.21 Wild-type M. smegmatis, TAM23 pTAMU3, DCS resistant strains (GPM14, GPM16) and TAM23 are able to grow both in the presence (Figure 2 B, E, H, K, N) and the absence of D-alanine (Figure 2 A, D, G, J, M). However, the growth is slower in the absence of D-alanine, where the cell wall of TAM23 appears to be weaker by its flaky appearance and irregular shaped borders. The ability for TAM23 to grow in the absence of D-alanine signifies that a secondary pathway for the production of D-alanine must be present, but the weaker appearance of the cell wall shows that the Alr is the primary enzyme for the production of D-alanine in M. smegmatis.
Effect of DCS on M. smegmatis colony morphology
As an indication of the DCS effect on the wild type and mutant strains, we determined colony morphologies of cells grown with DCS (Figure 2 C, F, I, L). As controls, cells were also grown in regular medium with and without D-alanine. Cells were grown in MADC broth to saturation, diluted and plated on MADC agar until colonies reached an appropriate size. M. smegmatis wild type, TAM23 pTAMU3, and DCS resistant strains had similar colony morphotypes regardless of the presence of D-alanine or DCS. The yellowish colonies displayed a rough and waxy appearance with irregular and lobate margins. However, the DCS hypersusceptible strain TAM23, on MADC medium lacking D-alanine, displayed colonies with a dryer and flatter appearance, and more smooth margins, as previously described.32 Addition of D-alanine partially restored the wild type morphotype, while DCS greatly exacerbated the colony morphology alterations yielding smooth flattened colonies. The effects of D-alanine and DCS on the TAM23 mutant directly correlate with the NMR metabolomic studies described below.
NMR spectral changes
The concentration of the metabolites in M. smegmatis cell extracts are directly related to peak intensities in the NMR spectra. Mutations or drugs that inhibit a particular enzyme will cause changes in NMR peak intensities pertaining to the affected metabolites. Detailed analysis of NMR spectral changes is possible because of recent efforts to compile metabolomic databases that incorporate detailed NMR assignment information.45-47 This includes the human metabolome database (http://www.hmdb.ca/scripts/Biofluid_browse.cgi), the Madison metabolomics consortium database (http://mmcd.nmrfam.wisc.edu/) and the NMR metabolomics database (http://www.liu.se/hu/mdl/main/). Characterization of the chemical composition of the metabolome and the availability of related NMR data is still incomplete, but the relative comparative difference between NMR spectra still provides some valuable insights. The NMR spectra from wild-type (mc2155) and TAM23 M. smegmatis mutant cells in the presence and absence of DCS are shown in Figure 3.
Figure 3.
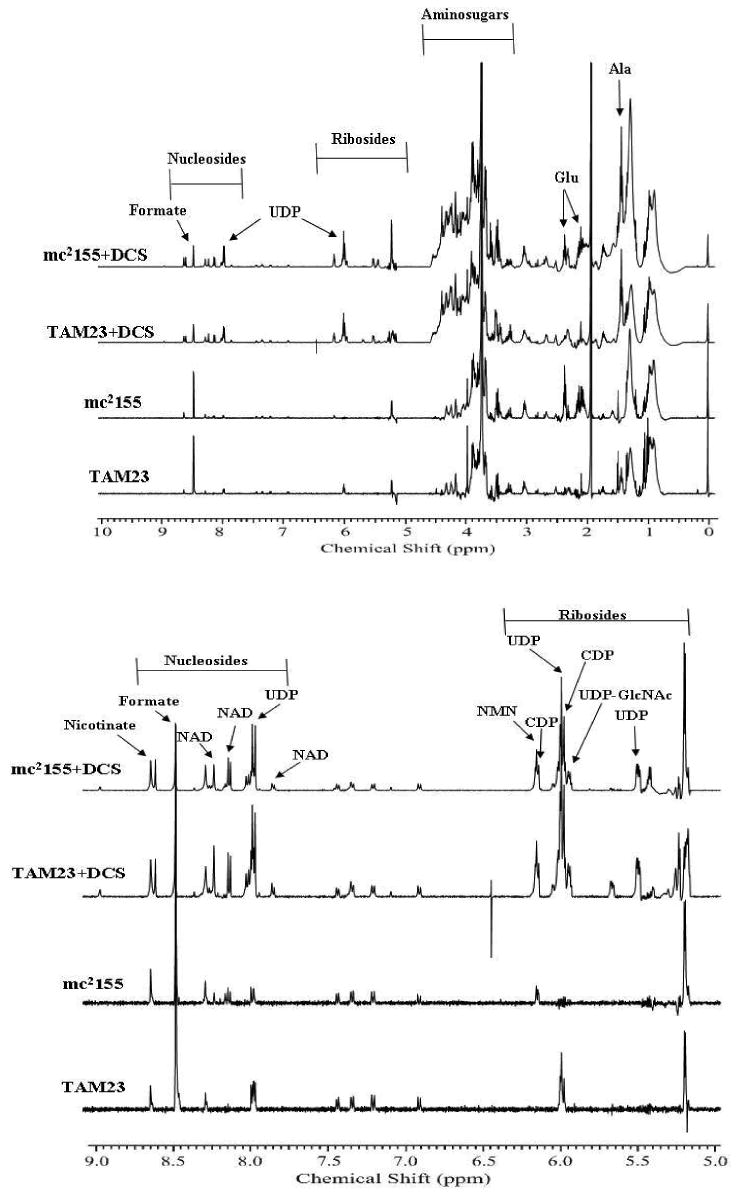
1D 1H NMR spectra in order from bottom to top are: TAM23, mc2155, TAM23 with DCS, and mc2155 with DCS.
The NMR spectrum of TAM23 cell extracts indicate that alanine (1.43 ppm) and UDP (5.50, 6.0 and 8.0 ppm) concentrations have increased, where these peak are absent in the wild-type spectrum. An increase in UDP would be expected from a stalled or disrupted peptidoglycan synthesis. Similarly, the concentration of D-alanine and L-alanine are expected to change when comparing the wild-type (mc2155) and TAM23 metabolome. Unfortunately, both these metabolites exhibit identical chemical shifts and are not differentiated in the NMR spectra of cell lysates.
Also, glutamate (2.10 and 2.33 ppm) concentration appears to be decreased by the inactivation of Alr in the TAM23 mutant. The region of the NMR spectrum that includes the glutamate peaks shows a distinct decrease in intensity between the wild type and TAM23 mutant cells. Since glutamate metabolism is an important precursor to peptidoglycan synthesis, this may suggest a second D-alanine synthesis pathway is active in TAM23 cells. The glutamate and alanine metabolism pathways are connected by the intervening pyrimidine metabolism pathway. Potentially, glutamate may become a source of D-alanine through a pyrimidine metabolism pathway that can produce D-alanine instead of L-alanine.
There also appears to be a dramatic decrease in the concentration of formate (8.46 ppm), a product of pyruvate metabolism in the formation of acetyl-CoA. Instead of producing acetyl-CoA, pyruvate may also be used with glutamate to produce D-alanine through a mechanism separate from the pyrimidine metabolism pathway. This may occur through the activity of D-alanine aminotransferase, which catalyzes the formation of D-alanine through a reaction that utilizes pyruvate and D-glutamate. It is important to note that while this enzyme has been identified in a number of bacterial genomes, it has not been identified in M. smegmatis. Glutamate racemase would convert L-glutamate to D-glutamate. An additional product of this reaction is 2-oxoglutarate, which is a metabolite in the tricarboxylic acid cycle (TCA). The TCA cycle connects with the nicotinate and nicotinamide metabolism pathways, which may also explain why these metabolites exhibit an increase in concentration with the addition of DCS.
These potential alternate D-alanine synthesis pathways implies that TAM23 does not simply contain a second D-alanine racemase, but that D-alanine must be synthesized through a second unique mechanism. This is consistent with the previous observations that alr null mutants do not exhibit D-alanine racemase activity.21 It is also consistent with the analysis of the cell morphology of TAM23 that indicates viable cells with weak cell walls in the absence of D-alanine.
The impact of DCS on the NMR spectra of TAM23 and wild-type M. smegmatis cells is even more dramatic than the difference between the TAM23 and wild-type cell strains. NMR peaks in the amino-sugar region (4.0-4.5 ppm), riboside region (5.0-6.5 ppm), and nucleoside region (7.75-8.75 ppm) exhibit the most significant changes in the drug treated culture. The amino-sugars identified are UDP-N-acetyl-D-glucosamine, UDP-N-acetylmuramate, N-acetyl-D-mannosamine, as well as other D-glucosamine and muramate groups. These amino-sugars are used as building blocks to help initiate the peptidoglycan synthesis. The amino-sugars concentrations are increased due to inadequate amounts of D-alanine required to form peptidoglycan chains. Thus, these metabolites result from failed peptidoglycan synthesis caused by enzymes inhibited by DCS.
Analysis of other NMR spectral changes due to the addition of DCS indicates an increase in nicotinate (8.61 ppm), NAD (7.85, 8.13, and 8.22 ppm), nicotinimide D-ribonucleotide (6.15 ppm). The concentration of UDP and CDP (5.95 and 6.14 ppm) are also increased in the presence of DCS, which accounts for the increased concentrations of UDP bound amino-sugars. These metabolites are associated with nicotinate and nicotinamide metabolism and pyrimidine metabolism. The increase in the concentrations of metabolites associated with the pyrimidine metabolism pathway is consistent with DCS activity because of the interconnection of the alanine metabolism and pyrimidine metabolism pathways. A similar relationship exists between the alanine metabolism and nicotinate and nicotinamide metabolism.
PCA of DCS activity in M. smegmatis
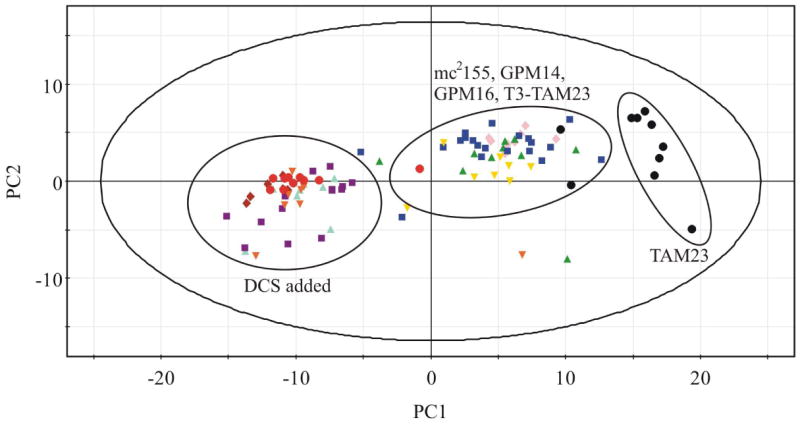
PCA analysis of the NMR metabolomic data provides an unbiased approach to determine an overall similarity between the various cell cultures. The complete set of 120 NMR spectra was analyzed using PCA, which reduces each individual spectrum to a single point in a 2D scores plot (Figure 4). The position of each point (spectrum) is based on the relative peak variations (metabolites) between each spectrum. NMR spectra of cell extracts with similar metabolite composition and concentrations will cluster close together in the 2D scores plot. Conversely, NMR spectra of cell extracts with variable metabolite composition and concentrations will form distinct clusters.
Figure 4.
PCA scores plot comparing mc2155 (
 ), TAM23 (●), GPM14 (
), TAM23 (●), GPM14 (
 ), GPM16 (
), GPM16 (
 ),TAM23 pTAMU3 (
),TAM23 pTAMU3 (
 ) mc2155 with DCS (
) mc2155 with DCS (
 ), and TAM23 with DCS (
), and TAM23 with DCS (
 ), GPM14 with DCS (
), GPM14 with DCS (
 ), GPM16 with DCS (
), GPM16 with DCS (
 ), and TAM23 pTAMU3 with DCS (
), and TAM23 pTAMU3 with DCS (
 ).
).
The 2D scores plot comparing each of the 120 NMR spectra using the two largest variations is shown in Figure 3. PC1 accounts for 37.5% of the variation between the spectra and PC2 accounts for an additional 9.7% of the variation. Three distinct clusters are readily apparent in the 2D scores plot. As expected, the wild-type M. smegmatis (mc2155) cells form a distinct and separate cluster from the alr null mutant (TAM23) cells. This is consistent with the inactivation of the D-alanine racemase, the corresponding impact on the metabolome for the TAM23 mutant and the detailed changes in the NMR spectra described above. Also as expected, the TAM23 pTAMU3 complement mutant clusters with wild-type (mc2155) cells since Alr activity has been restored. This clearly indicates that the distinct PCA clustering observed between wild-type and TAM23 cells is primarily caused by a difference in Alr activity. Surprisingly, the unrelated DCS resistant strain (GPM16) forms a broad cluster that overlaps with wild-type M. smegmatis. Thus, the mutation in GPM16 appears to have a minimal impact on the M. smegmatis metabolome relative to the D-alanine racemase null mutant.
Conversely, the close clustering between wild-type M. smegmatis and the Alr over producing mutant (GPM14) is not surprising since there was neither a loss nor gain of an enzymatic function. The increased level of Alr does not impact the metabolome probably because Ddl is a more efficient enzyme than Alr48, 49. Any increase in D-alanine would be transitory and rapidly converted to D-alanyl-D-alanine dipeptide.
The PCA of the M. smegmatis cell strains in the presence of DCS yielded interesting results. If the primary target of DCS is alanine racemase, then wild-type cells in the addition of DCS would have clustered with TAM23 in both the presence and absence of DCS. DCS would simply inhibit the activity of Alr and mimic TAM23, an alr null mutant. Moreover, if Alr is the sole lethal target, it would have required us to isolate a conditional rather than a null mutant to describe the metabolome associated with an Alr-deficient strain. Also, DCS would not induce any further changes in the TAM23 metabolome. This was not the case as evident by the large changes in the NMR spectra caused by the addition of DCS as seen in Figure 3. First, the 2D scores plot indicate that TAM23 with the addition of DCS forms a distinct cluster from TAM23 in the absence of DCS. This clearly indicates that DCS is inhibiting a second protein target besides Alr. Second, the wild-type cells with the addition of DCS also clustered closely with TAM23 in the presence of DCS. This establishes that Alr is also targeted by DCS. These results indicate that DCS is targeting multiple protein targets consistent with enzyme data demonstrating an inhibitory affect on both Alr and Ddl.17, 18
An unexpected result was the observation that the GPM14 and GPM16 DCS resistant mutants also clustered together with TAM23 and wild-type cells in the presence of DCS. This clearly indicates that the lethal target(s) for DCS is still inhibited in the resistant strains and that the over production of Alr in GPM14 or the unrelated mutation in GPM16 simply compensate for the inhibition of the DCS lethal target.
Is D-alanine-D-alanine ligase the primary lethal target of DCS? Our data clearly indicates that Alr is not the primary lethal target of DCS. Wild-type cells in the presence of DCS do not cluster with the alr null mutant cells (TAM23). Furthermore, all the cell cultures in the presence of DCS form a separate and distinct cluster from TAM23 in the absence of DCS. This would only occur if DCS is inhibiting a second protein besides Alr that results in further changes in the metabolome. This is clearly apparent when individual NMR spectra are compared with and without the addition of DCS. On a relative scale, smaller changes in the NMR spectra are observed when comparing wild-type and TAM23 cells. This is consistent with the simple induction of a secondary pathway in TAM23 to generate the essential D-alanine required for peptidoglycan synthesis, not a dramatic change that would be expected from a lethal inhibition of cell wall synthesis. These NMR results are also consistent with the observation that the alr null mutants is viable in the absence of D-alanine. Inactivating both D-alanine racemase genes in E. coli resulted in cell growth being completely dependent on supplemental D-alanine.42, 43
It is plausible that DCS is broadly active and inhibits multiple proteins involved in cell wall synthesis to result in cell death. Comparison of the NMR spectra for wild-type and TAM23 cells in the presence of DCS indicate that the difference in the metabolome caused by the alr null mutant is still present. Since this difference is attributed to the induction of a second D-alanine synthetic pathway, this pathway does not appear to be induced in wild-type cells from the addition of DCS. This also suggests that the inhibition of Alr in wild-type cells is not severe enough to induce this second pathway or that this second pathway is still active in TAM23 cells. Since the alr null mutation in TAM23 cells is not sufficient to induce cell death and the appearance that DCS is not inhibiting the secondary D-alanine synthetic pathway, DCS is probably acting through another primary lethal target besides Alr. These results are also consistent with the observation that TAM23 cells are more susceptible to DCS (Table 1). Cell morphology indicates that the TAM23 cell walls are weak with flaky and irregular shape. This implies that the second pathway for D-alanine synthesis is a poor substitute for the Alr pathway, and thus, cell death may occur before a complete inhibition of the primary lethal target of DCS occurs.
The NMR metabolomics data indicate that the DCS resistant mutant cells also cluster with wild-type and TAM23 cells in the presence of DCS. This indicates that the DCS resistant cells do not eliminate the impact of DCS on the metabolome, but simply alleviate the cell damage. DCS is still inhibiting enzymes in the cell wall synthetic pathway, but it is no longer capable of preventing cell wall synthesis. D-alanine-D-alanine ligase needs to bind two D-alanine molecules prior to forming the essential D-alanyl-D-alanine dipeptide. Presumably, DCS could compete with either D-alanine binding site to inhibit Ddl activity. Similarly, since DCS has been shown to efficiently inhibit Ddl, this enzyme appears a likely candidate for the primary lethal target of DCS. Increasing the cellular concentration of D-alanine, as it was shown to occur in GPM14,16 would enable it to compete with DCS and allow for the production of the D-alanyl-D-alanine dipeptide.
Why isn't Ddl simply overproduced to reduce the bacteria's susceptibility to DCS? The activity of D-alanine racemase is modulated by the induction of L-alanine dehydrogenase, which provides the cell with a mechanism to minimize any negative impact from overproducing Alr.50 A similar feed-back mechanism is not known to exist for Ddl. Similarly, D-alanine and L-alanine are ubiquitous metabolites with multiple uses in the cell. Conversely, the D-alanyl-D-alanine dipeptide is an essential end-product that may not be as well-tolerated if overproduced due to a mutation leading to high levels of Ddl over-productions. In this context, overproduction of Ddl in M. smegmatis from a recombinant plasmid leads to a lower level of resistance to ddl. Therefore, overproducing Ddl may not be the most efficient means to protect mycobacteria from DCS. Thus, the lethal action of DCS may act through the inhibition of Ddl while resistance to DCS is mediated by the overproduction of Alr.
Conclusion
A comparative metabolic study of the activity of DCS in M. smegmatis was conducted by Principal Component Analysis and NMR spectroscopy. The metabolome of wild type (mc2155), alr null mutant (TAM23), TAM23 complemented with wild-type alr gene (TAM23 pTAMU3) and DCS resistant mutants (GPM14, GPM16) were monitored to investigate the mechanism of action of DCS. Differences in the NMR spectra between wild-type and TAM23 cells suggest the presence of a second D-alanine synthetic pathway in the alr null mutant. This is consistent with the viability of the TAM23 cells in the absence of supplemental D-alanine. This secondary pathway appears to still be present in the TAM23 cells, but not induced in wild-type cells when both wild-type and TAM23 cells are exposed to DCS. This is consistent with the PCA results that indicate TAM23 and wild-type cells form separate and distinct clusters, but all five strains cluster together in the presence of DCS.
These results are consistent with multiple DCS targets that include Alr and Ddl in the peptidoglycan biosynthetic pathway. The clustering of the DCS resistant cells with wild-type and TAM23 cells indicate that DCS is still active and inhibiting cell wall pathway enzymes, but the detrimental impact has been minimized. Thus, D-alanine-D-alanine ligase appears to be the lethal target of DCS. This hypothesis is consistent with: (i) DCS resistant mutants that overproduce Alr, (ii) Alr is not a lethal target, (iii) secondary D-alanine pathway is unaffected by DCS, and (iv) DCS efficiently inhibits biosynthesis of D-alanine-D-alanine. It appears that the overproduction of Alr simply increases the efficiency by which D-alanine can compete with DCS to enable the production of the essential D-alanyl-D-alanine dipeptide.
Acknowledgments
This work was supported by grants from the Agricultural Research Division Interdisciplinary Research Program (Project NEB 39-142), Nebraska Tobacco Settlement Biomedical Research Development Funds and the Nebraska Research Council. Research was performed in facilities renovated with support from NIH (RR015468-01).
References
- 1.W.H.O. Global Tuberculosis Control: Surveillance, Planning and Financing. World Health Organization; Geneva, Switzerland: 2007. WHO/HTM/TB/2007.376. [Google Scholar]
- 2.Rattan A, Kalia A, Ahmad N. Multidrug-resistant Mycobacterium tuberculosis: molecular perspectives. Emerging Infectious Diseases. 1998;4(2):195–209. doi: 10.3201/eid0402.980207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams BG, Dye C. Antiretroviral drugs for tuberculosis control in the era of HIV/AIDS. Science (Washington, DC, United States) 2003;301(5639):1535–1537. doi: 10.1126/science.1086845. [DOI] [PubMed] [Google Scholar]
- 4.Telenti A. Genetics and pulmonary medicine. 5. Genetics of drug resistant tuberculosis. Thorax. 1998;53(9):793–7. doi: 10.1136/thx.53.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dooley SW, Jarvis WR, Martone WJ, Snyder DEJ. Multi-Drug resistant tuberculosis [editorial] Ann Intern Med. 1992;117:257–258. doi: 10.7326/0003-4819-117-3-257. [DOI] [PubMed] [Google Scholar]
- 6.Iseman MD. Treatment and implications of multidrug-resistant tuberculosis for the 21st century. Chemotherapy (Basel) 1999;45(Suppl 2):34–40. doi: 10.1159/000048480. [DOI] [PubMed] [Google Scholar]
- 7.Barry CE., III Modern drug development. Issues in Infectious Diseases. 2003;2:137–150. Mycobacteria and TB. [Google Scholar]
- 8.Gillespie SH. Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrobial Agents and Chemotherapy. 2002;46(2):267–274. doi: 10.1128/AAC.46.2.267-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyman CL. Tuberculosis: a survey and review of current literature. Curr Opin Pulm Med. 1995;1(3):234–42. [PubMed] [Google Scholar]
- 10.Udwadia ZF. India's multidrug-resistant tuberculosis crisis. Ann N Y Acad Sci. 2001;953:98–105. doi: 10.1111/j.1749-6632.2001.tb11365.x. [DOI] [PubMed] [Google Scholar]
- 11.Anonymous, Tuberculosis crisis. World Health Forum. 1994;15(3):292–3. [PubMed] [Google Scholar]
- 12.Joshi JM, Gothi D. Multi-drug resistant pulmonary tuberculosis. Current Respiratory Medicine Reviews. 2006;2(1):53–57. [Google Scholar]
- 13.Johnson R, Streicher EM, Louw GE, Warren RM, van Helden PD, Victor TC. Drug resistance in Mycobacterium tuberculosis. Current Issues in Molecular Biology. 2006;8(2):97–111. [PubMed] [Google Scholar]
- 14.Pitkanen M, Sirvio J, MacDonald E, Ekonsalo T, Riekkinen P., Sr The effects of d-cycloserine, a partial agonist at the glycine binding site, on spatial learning and working memory in scopolamine-treated rats. Journal of Neural Transmission: Parkinson's Disease and Dementia Section. 1995;9(23):133–44. doi: 10.1007/BF02259655. [DOI] [PubMed] [Google Scholar]
- 15.Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79(1):3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 16.Feng Z, Barletta RG. Roles of Mycobacterium smegmatis D-alanine:D-alanine ligase and D-alanine racemase in the mechanisms of action of and resistance to the peptidoglycan inhibitor D-cycloserine. Antimicrobial Agents and Chemotherapy. 2003;47(1):283–291. doi: 10.1128/AAC.47.1.283-291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert MP, Neuhaus FC. Mechanism of D-cycloserine action. Alanine racemase from Escherichia coli W. Journal of Bacteriology. 1972;110(3):978–87. doi: 10.1128/jb.110.3.978-987.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David HL, Takayama K, Goldman DS. Susceptibility of mycobacterial D-alanyl-D-alanine synthetase to D-cycloserine. American Review of Respiratory Disease. 1969;100(4):579–81. doi: 10.1164/arrd.1969.100.4.579. [DOI] [PubMed] [Google Scholar]
- 19.Caceres NE, Harris NB, Wellehan JF, Feng Z, Kapur V, Barletta RG. Overexpression of the D-alanine racemase gene confers resistance to D-cycloserine in Mycobacterium smegmatis. Journal of Bacteriology. 1997;179(16):5046–5055. doi: 10.1128/jb.179.16.5046-5055.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y. The magic bullets and tuberculosis drug targets. Annual Review of Pharmacology and Toxicology. 2005;45:529–564. doi: 10.1146/annurev.pharmtox.45.120403.100120. [DOI] [PubMed] [Google Scholar]
- 21.Chacon O, Feng Z, Harris NB, Caceres NE, Adams LG, Barletta RG. Mycobacterium smegmatis D-alanine racemase mutants are not dependent on D-alanine for growth. Antimicrobial Agents and Chemotherapy. 2002;46(1):47–54. doi: 10.1128/AAC.46.1.47-54.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forgue P, Halouska S, Werth M, Xu K, Harris S, Powers R. NMR Metabolic Profiling of Aspergillus nidulans to Monitor Drug and Protein Activity. Journal of Proteome Research. 2006;5(8):1916–1923. doi: 10.1021/pr060114v. [DOI] [PubMed] [Google Scholar]
- 23.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends in Biotechnology. 2004;22(5):245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Kell DB. Metabolomics and systems biology: making sense of the soup. Current Opinion in Microbiology. 2004;7(3):296–307. doi: 10.1016/j.mib.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Fiehn O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comparative and Functional Genomics. 2001;2(3):155–168. doi: 10.1002/cfg.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR in Biomedicine. 2005;18(3):143–162. doi: 10.1002/nbm.935. [DOI] [PubMed] [Google Scholar]
- 27.Ward JL, Beale MH. NMR spectroscopy in plant metabolomics. Biotechnology in Agriculture and Forestry. 2006;57:81–91. Plant Metabolomics. [Google Scholar]
- 28.Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Analytical and Bioanalytical Chemistry. 2007;387(2):525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 29.Griffin JL. The potential of metabonomics in drug safety and toxicology. Drug Discovery Today: Technologies. 2004;1(3):285–293. doi: 10.1016/j.ddtec.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Stoyanova R, Brown TR. NMR spectral quantitation by principal component analysis. NMR in biomedicine. 2001;14(4):271–7. doi: 10.1002/nbm.700. [DOI] [PubMed] [Google Scholar]
- 31.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Molecular Microbiology. 1990;4(11):1911–19. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 32.Halouska S, Powers R. Negative impact of noise on the principal component analysis of NMR data. Journal of Magnetic Resonance. 2006;178(1):88–95. doi: 10.1016/j.jmr.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 33.Van Heijenoort J. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001;11(3):25R–36R. doi: 10.1093/glycob/11.3.25r. [DOI] [PubMed] [Google Scholar]
- 34.Trias J, Benz R. Permeability of the cell wall of Mycobacterium smegmatis. Molecular Microbiology. 1994;14(2):283–90. doi: 10.1111/j.1365-2958.1994.tb01289.x. [DOI] [PubMed] [Google Scholar]
- 35.Nanninga N. Morphogenesis of Escherichia coli. Microbiology and Molecular Biology Reviews. 1998;62(1):110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahapatra S, Scherman H, Brennan PJ, Crick DC. N glycolylation of the nucleotide precursors of peptidoglycan biosynthesis of Mycobacterium spp. is altered by drug treatment. Journal of Bacteriology. 2005;187(7):2341–2347. doi: 10.1128/JB.187.7.2341-2347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Heijenoort J. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Natural Product Reports. 2001;18(5):503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 38.Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiology and Molecular Biology Reviews. 2005;69(4):585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denessiouk KA, Denesyuk AI, Lehtonen JV, Korpela T, Johnson MS. Common structural elements in the architecture of the cofactor-binding domains in unrelated families of pyridoxal phosphate-dependent enzymes. Proteins: Structure, Function, and Genetics. 1999;35(2):250–261. doi: 10.1002/(sici)1097-0134(19990501)35:2<250::aid-prot10>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 40.Noda M, Kawahara Y, Ichikawa A, Matoba Y, Matsuo H, Lee DG, Kumagai T, Sugiyama M. Self-protection Mechanism in D-Cycloserine-producing Streptomyces lavendulae. Gene cloning, characterization, and kinetics of its alanine racemase and D-alanyl-D-alanine ligase, which are target enzymes of D-cycloserine. Journal of Biological Chemistry. 2004;279(44):46143–46152. doi: 10.1074/jbc.M404603200. [DOI] [PubMed] [Google Scholar]
- 41.Lilley PE, Stamford NPJ, Vasudevan SG, Dixon NE. The 92-min region of the Escherichia coli chromosome: Location and cloning of the ubiA and alr genes. Gene. 1993;129(1):9–16. doi: 10.1016/0378-1119(93)90690-5. [DOI] [PubMed] [Google Scholar]
- 42.Wild J, Hennig J, Lobocka M, Walczak W, Klopotowski T. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Molecular and General Genetics. 1985;198(2):315–22. doi: 10.1007/BF00383013. [DOI] [PubMed] [Google Scholar]
- 43.Hols P, Defrenne C, Ferain T, Derzelle S, Delplace B, Delcour J. The alanine racemase gene is essential for growth of Lactobacillus plantarum. Journal of Bacteriology. 1997;179(11):3804–3807. doi: 10.1128/jb.179.11.3804-3807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Margolin W. Bacterial shape: Growing off this mortal coil. Current Biology. 2003;13(18):R705–R707. doi: 10.1016/j.cub.2003.08.043. [DOI] [PubMed] [Google Scholar]
- 45.Lundberg P, Vogel T, Malusek A, L PO, Cohen L, Dahlqvist O. MDL - The Magnetic Resonance Metabolomics Database. mdl.imv.liu.se. [Google Scholar]
- 46.Seavey BR, Farr EA, Westler WM, Markley JL. A Relational Database for Sequence-Specific Protein NMR Data. J Biomolecular NMR. 1991;1:217–236. doi: 10.1007/BF01875516. [DOI] [PubMed] [Google Scholar]
- 47.Wishart DS, et al. HMDB: The Human Metabolome Database. Nucleic Acids Research. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellsworth BA, Tom NJ, Bartlett PA. Synthesis and evaluation of inhibitors of bacterial D-alanine:D-alanine ligases. Chemistry & Biology. 1996;3(1):37–44. doi: 10.1016/s1074-5521(96)90082-4. [DOI] [PubMed] [Google Scholar]
- 49.Strych U, Penland RL, Jimenez M, Krause KL, Benedik MJ. Characterization of the alanine racemases from two Mycobacteria. FEMS Microbiology Letters. 2001;196(2):93–98. doi: 10.1111/j.1574-6968.2001.tb10547.x. [DOI] [PubMed] [Google Scholar]
- 50.Berberich R, Kaback M, Freese E. D-Amino acids as inducers of L-alanine dehydrogenase in Bacillus subtilis. Journal of Biological Chemistry. 1968;243(5):1008–13. [PubMed] [Google Scholar]


