Abstract
In eukaryotes, genes transcribed by RNA polymerase III (Pol III) carry their own internal promoters and as such, are transcribed as individual units. Indeed, a very few cases of dicistronic Pol III genes are yet known. In contrast to other hemiascomycetes, 5S rRNA genes of Yarrowia lipolytica are not embedded into the tandemly repeated rDNA units, but appear scattered throughout the genome. We report here an unprecedented genomic organization: 48 over the 108 copies of the 5S rRNA genes are located 3′ of tRNA genes. We show that these peculiar tRNA–5S rRNA dicistronic genes are expressed in vitro and in vivo as Pol III transcriptional fusions without the need of the 5S rRNA gene-specific factor TFIIIA, the deletion of which displays a viable phenotype. We also report the existence of a novel putative non-coding Pol III RNA of unknown function about 70 nucleotide-long (RUF70), the 13 genes of which are devoid of internal Pol III promoters and located 3′ of the 13 copies of the tDNA-Trp (CCA). All genes embedded in the various dicistronic genes, fused 5S rRNA genes, RUF70 genes and their leader tRNA genes appear to be efficiently transcribed and their products correctly processed in vivo.
INTRODUCTION
Co-transcription of two or more genes under the control of a single promoter is very scarce in the eukaryotic world (1). Very exceptionally several genes coding for proteins are cotranscribed as a single primary mRNA. For example, all genes from chromosome 3 of Leishmania major (with the exception of a single gene and one tRNA genes) are transcribed as two convergent polycistronic clusters (2). More numerous cases of polycistronic non-coding RNA (ncRNA) genes are known in plants. Besides the special case of snoRNAs encoded in introns of protein coding genes, non-intronic snoRNAs gene clusters are transcribed as polycistronic pre-snoRNAs and later processed. In Arabidopsis thaliana, 43 clusters representing 71 snoRNA genes were identified (3), while in Oryza sativa, 270 snoRNA genes are involved in 70 clusters (4). A single case of dicistronic snoRNAs is known in yeast Saccharomyces cerevisiae where snR190 and U14 are separated by only 67 nt and the primary transcript processed by the RNA endonuclease Rnt1 (5). In higher eukaryotes, many microRNAs are encoded in polycistronic transcripts (6,7). In S. cerevisiae also, seven box C/D snoRNAs are expressed as a 1.4 kb polycistronic RNA processed by Rnt1 and Rat1 nucleases (8). All the above examples concern genes transcribed by RNA polymerase II (Pol II). Pol I task is devoted to the transcription of a single polycistronic gene coding for the 35S rRNA, which is the precursor of the ribosomal 18S, 5.8S and 25S rRNAs.
In contrast with Pol I and Pol II genes, Pol III genes feature internal promoters (known as the A and B boxes in the cases of tRNA genes) and display at their 3′-end a transcription termination signal made of poly-T in the RNA-like strand (9,10). Therefore, clustering of Pol III genes appears at first glance unnecessary as well as unlikely. Nevertheless two types of such clustering are yet known, homologous and heterologous. First, in most hemiascomycetous yeast, tandemly repeated tRNA genes (tDNA) are quite common. For example, Debaryomyces hansenii displays 17 tandem tDNAs (11). In such Pol III tandems, the termination signal is located 3′ of the second gene and the primary transcript is probably processed into two functional tRNAs by the usual tRNA maturation machinery, as was demonstrated for tandem tRNA genes from Schizosaccharomyces pombe (12). The distance between the two genes is often very short, down to 3 nt; and the same types of tDNA pairs are encountered in neighbour genomes (13). Only two types of dicistronic heterologous Pol III transcripts are known yet: tRNA–snoRNA and tRNA–miRNA genes. In A. thaliana, 12 copies of the C/D snoRNA sno43 each located 3′ of copies of tDNA-Gly (GCC) were reported by Kruszka and coworkers (14). The same team also identified in rice two copies of a similar fusion between a novel C/D snoRNA and tDNA-eMet. tRNA–miRNA genes were discovered both in the C19MC cluster of the human genome (15) and in the mouse gammaherpes virus 68 (16). The present work reports the identification of two novel types of dicistronic heterologous Pol III transcripts in the genome of Yarrowia lipolytica. As a matter of fact, this hemiascomycete yeast contains multiple copies of dicistronic genes featuring in all cases a leading tRNA gene followed either by a 5S rRNA gene or a yet uncharacterized ncRNA gene.
In most hemiascomycetous yeasts, 5S rRNA genes are embedded in the highly repeated rDNA unit (∼10 kb, present in about 100 copies). In each active rDNA unit, Pol III and Pol I divergently transcribe 5S and 35S precursor RNA genes, respectively. Transcription of all Pol III genes (tRNA, 5S rRNA, RPR1 RNA, SCR1 RNA genes and a few others) first requires the binding onto the gene of the assembly transcription factor TFIIIC and, in a second step, the recruitment of TFIIIB which, in turn, recruits RNA polymerase III for multiple rounds of transcription (10). For all Pol III genes (except 5S rRNA genes), the primary recognition by TFIIIC relies on two short internal sequences known as the A- and B-boxes [TRGYnnAnnnG and GWTCRAnnC, respectively, see (13)]. Transcription of 5S rRNA genes (in which no B-box is present) is even more complex as it requires the binding of the additional factor TFIIIA, prior to the assembly of TFIIIC (17,18).
TFIIIA harbours nine zinc fingers forming three functional subdomains (19). In Xenopus, fingers 1–3 and 7–9 bind to the A and C promoter elements of the 5S rRNA gene (20) while the central fingers 4–6 interact with the intermediate promoter element (21,22). Besides its role in transcription, TFIIIA also interacts as a chaperone with the 5S rRNA transcript itself for nucleocytoplasmic transport and RNA storage (23–25). Different fingers form the minimal interaction domains with either type of nucleic acids: fingers 1–3 are necessary and sufficient for DNA binding (26) while fingers 4–6 form the minimal RNA binding domain (27–29). Although TFIIIA is an essential protein both in Xenopus and yeast, its only essential function, in S. cerevisiae, is the transcription of 5S rRNA genes (30). As a matter of fact, cells devoid of TFIIIA may survive if a single copy of the 5S rRNA gene is expressed from a RPR1 Pol III promoter, showing that the 5S rRNA transport and storage function of TFIIIA is dispensable in yeast. Consistent with this result is the survival of several mutants affecting the central fingers 4–6 shown to form the minimal 5S rRNA binding domain in Xenopus (27–29): disruption of yeast TFIIIA fingers 4, 5, 6, 4 + 5 or 4 + 6 yielded viable phenotypes (31).
In Y. lipolytica, the genomic organization of 5S rRNA genes is known to depart from that in other hemiascomycetes (32,33). As a matter of fact, none of the 5S rRNA genes are part of the rDNA unit but appear to lie dispersed in the whole genome (11). In this work, we noticed that nearly half (48 over 108) of the 5S rRNA gene copies of Y. lipolytica are closely located 3′ to tDNAs, suggesting that these peculiar tDNA-fused 5S rRNA genes might be expressed, without the need of TFIIIA, as Pol III transcriptional fusions, similarly to tandem tDNAs already known in hemiascomycetes (13). Again in Y. lipolytica, we also identified a novel 70 nt ncRNA present in 13 copies, each of them located 3′ to the 13 copies of the tRNA-Trp gene.
We first checked, by RT–PCR that all types of fusions are transcribed in vivo as a single RNA as well as in vitro by whole cell extracts (WCE) from either Y. lipolytica or S. cerevisiae. Through RNA analysis and northern experiments we then checked that both tRNA and 5S rRNA genes appear to be correctly matured. Finally, the deletion of the TFIIIA ortholog of Y. lipolytica yielded a viable phenotype suggesting that the transcription of only half of the 5S rRNA genes (those 3′-fused to tDNAs) can sustain viability. This very peculiar organization of 5S rRNA genes was searched for in other ascomycete genomes but, up to now, remains unique.
MATERIALS AND METHODS
Identification of 5S rRNA and Pol III composite genes
5S rRNA genes (5S rDNA) were searched with the Génolevures Blast facility (http://cbi.labri.fr/Genolevures/blast.php#) using the S. cerevisiae gene as seed. Of the 5S DNA, 117 copies were found of which eight are relics [deleted at either or both end(s)]. Out of the 109 remaining copies, 60 are regular isolated copies and 49 are located immediately downstream of five different types of tDNA. In one of this dicistronic tDNA–5S DNA, the 5S DNA is interrupted by a Ylt1 transposable element. Close examination of Pol III terminators (T-track) revealed a putative ncRNA [named RNA of unknown function about 70 nt-long (RUF70)] present at the 3′-end of all tDNA-Trp (CCA) genes. Detailed alignments of all 5S rRNA, tDNA–5S rDNA and tDNA-Trp (CCA)-RUF 70 genes are given in Supplementary Data 1, 2 and 3.
RT–PCR
Total RNA was extracted using the RNeasy Midi Kit (Qiagen, Courtaboeuf, France) and DNA contamination was removed with the Turbo DNA-free kit (Applied Biosystems/Ambion, Austin, Texas, USA). RT–PCR were performed with Ready-To-Go™ RT-PCR Beads (GE Healthcare Life Sciences, Orsay, France) and PCR control with PuReTaq Ready-To-Go™ PCR Beads (GE Healthcare Life Sciences). Primers used are listed in Supplementary Data 4.
Cloning of tRNA–5S rRNA and tRNA-Trp (CCA)–RUF70 genes
Genomic DNA from strain E150 (CLIB122, MatB lys11–23 ura3–302 leu2–270 xpr2–322) was prepared as described (34). PCR amplifications were carried out with the pfu DNA polymerase purified in the lab. Primers used are listed as Supplementary Data 4. PCR products were then introduced into an EcoRV-digested pBluescript SK and competent cells of strain DH10B were transformed by electroporation (1.7 kV/mm, 25 μF, 100 Ω). Transformants were selected on LB media with ampicillin (100 μg/ml) and Xgal (40 μg/ml), verified by both PCR amplification with primers used for DNA amplification and plasmid sequencing.
In vitro transcriptions
Transcription in yeast (Y. lipolytica or S. cerevisiae) WCE [50 μg of S100 (35)] was carried out with the same buffer conditions as for the reconstituted system (see below). 50 μg of S100 extract were incubated with 60 ng of the indicated templates, 1 h at 25°C. After the addition of NTP mix [0.6 mM (ATP, GTP, CTP), 0.03 mM UTP and 1 μl of 32P UTP at 10 μCi/μl], a further 20 min incubation was allowed before stopping the reaction. The transcripts were analysed by electrophoresis on 6% acrylamide–urea gel.
Standard in vitro transcriptions were performed as previously described (36,37) using affinity purified whole recombinant rTFIIIC, partially purified B″ fraction (38,39), pure rBdp1, rTBP, rBrf1, highly purified RNA polymerase III and the respective plasmid DNA template [see (40) for details].
Deletion of ylTFIIIA
The complete deletion of Y. lipolytica TFIIIA gene (ylTFIIIA, YALI0F05104g) was performed as follows (41). Promoter (P = 990 bp) and terminator (T = 588 bp) regions of haploid strain PO1d (CLIB139, MatA ura3–302 leu2–270 xpr2–322) were amplified by PCR with primers Tf3A-P1 (caccagaatcttcatcatcagcctactg), Tf3A-P2 (CATTACCCTGTTATCCCTACaaacatgctcttagagctgactctcgag) including an I-SceI restriction site in bold, Tf3A-T1 (CTAGGGATAACAGGGTAATCtatggagtactcgtccaatgaagaggtc) also with an I-SceI restriction site and Tf3A-T2 (gtagacagttcatcagcgcataacgtac). A ∼1.9 kb I-SceI_LEU2_I-SceI fragment [ML cassette (41)] was introduced into the PT cassette at the I-SceI restriction site. Cells of PO1d were transformed by the lithium acetate method (42) with about 400 ng of purified disruption cassette. Transformants were selected on YNB medium with NH4Cl (5 g/l), glucose (10 g/l), sodium potassium phosphate buffer, pH 6.8 (50 mM), agar (2%) and uracyl (100 μg/ml). Verification of the correct ylTFIIIA deletion was realized by PCR with primers external to the disruption cassette, i.e. upstream of P (V1 gttgcatctaagtgctatccacg) and downstream of T (V2 atgcagcatgcagccaggtattg). As the PCR products of the wild copy and of the disrupted copy are the same size, PCR amplification was followed by an EcoRI restriction that generates three restriction fragments for the wild copy and four for the disrupted one. Three independent clones were retained (ΔylTFIIIA C6, C7 and C8).
Northern blot analyses
For northern blot experiments, total RNA extractions were performed using the hot phenol method as described (43) and not using RNeasy Midi Kit to obtain large amount of small RNA products. Total RNA (20 μg) was analysed by northern blot using the following oligonucleotides according to the previously described procedure (44,45)
Trp: TGGAGTCGAAAGCTCTACCATTG
RUF 70: The various oligonucleotides were mixed during the hybridization step.
WX01:GCTATGGGACTTAAACCCACAAT; WX06:GCTAGGGGAATTAAACCCCATA; WX09:GCGATGGGACTCTAACCCATATT; WX10:GCTAGGGGAGTCAAACCCCTTAT; WX11:GCTACCGGAATCAAACCGGCTTA; WX12:GCTACCGGAATTTAACCGGCTAT; WX13:AACAGAGGAATTAAACCTCCTTG.
RESULTS
Variety of 5S rRNA genes organization throughout eukaryotic genomes
In most hemiascomycetes 5S rRNA genes are embedded in the rDNA unit (Table 1). In S. cerevisiae, the 35S rRNA which is the precursor of 18S, 5.8S and 25S ribosomal RNAs is synthesized by RNA polymerase I machinery while a single copy of 5S rRNA gene is synthesized by RNA polymerase III with the help of the TFIIIA, -C and -B transcription factors. The tandemly repeated rDNA units form a single cluster internal to one chromosome (as in S. cerevisiae) or multiple clusters often located in subtelomeric regions (as in Candida glabrata). Yarrowia lipolytica is presently the only fully sequenced hemiascomycete in which 5S rRNA genes are not included in the rDNA units, similarly to S. pombe and most other eukaryotes.
Table 1.
Variability of the genomic organization of 5S rRNA and other rRNA genes in yeasts
| Number of rDNA cluster | Internal or Subtelomeric | rDNA units number | rDNA unit length (bp) | 5S rRNA genes status included (in rDNA unit) or gene number if external | References | |
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | Unique | I | ∼100 | 9137 | Included | SGD |
| Candida glabrata | 2 | S | ? | – | Included two copies/repeat | (11) |
| Zygosaccharomyces rouxii | ? | I | ? | 9868 | Included | (46) |
| Lachancea kluyveri | Unique | I | ? | 8656 | Included | (47) |
| Lachancea thermotolerans | Unique | I | ∼200 | 8506 | Included | (48) (46) |
| Kluyveromyces lactis | Unique | I | ? | 8711 | Included | (49) (46) |
| Eremothecium gossypii | Unique | I | ∼50 | 8187 | Included | (50) |
| Debaryomyces hansenii | 3 | I | ? | 7705 | Included two copies/repeat | (51) |
| Yarrowia lipolytica | 6 | S | ∼100 | – | 109 | (32) (11) |
| Schizosaccharomyces pombe | 2 | S | ∼100 | – | 30 | (32, 52) |
The rDNA unit comprises 18S, 5.8S and 25S ribosomal RNA genes, which are transcribed as a single 35S precursor rRNA by RNA polymerase I. The 5S rRNA genes are transcribed by RNA polymerase III. In all yeasts but Y. lipolytica and S. pombe, one copy of 5S gene is embedded in the rDNA (two copies for C. glabrata and D. hansenii). S. cerevisiae data were obtained from the Saccharomyces Genome Database (SGD, http://www.yeastgenome.org/).
In Y. lipolytica half of the 5S rRNA gene copies are located 3′ of tRNA genes
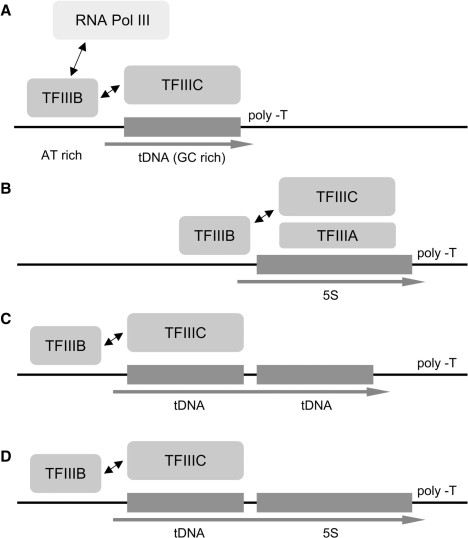
In Y. lipolytica, the 108 full length 5S rRNA genes are dispersed throughout the whole genome. The genome also contains eight 5S rRNA gene relics (with various 5′ or 3′ or 5′ and 3′ deletions, see alignment in Supplementary Data 1). Very remarkably, 48 of the 5S rRNA genes are located immediately 3′ of tDNAs. Some examples of dicistronic Pol III genes are already known: dual tDNAs in S. pombe (12,53) or hemiascomycetes (13) and tRNA–snoRNA genes (14); but tRNA–5S rRNA composite genes are an unprecedented case. Figure 1 summarizes the different elements of the Pol III transcription machinery requested for transcription of tRNA or 5S rRNA genes. The in vivo transcription of a duplex tRNA gene is known to rely solely on the promoter elements of the upstream gene (12). In the present case of a dicistronic tRNA–5S rRNA gene, we hypothesized that, for steric reasons, TFIIIA may not bind the downstream 5S rRNA gene and therefore that this composite gene could be expressed without the need of TFIIIA.
Figure 1.
Overview of the transcription mechanisms of tRNA, 5S rRNA and tRNA–5S rRNA dicistronic genes. (A) In S. cerevisiae, transcription of a tRNA gene by Pol III requires the assembly of transcription factor TFIIIC onto the tDNA (GC-rich) followed by that of TFIIIB (onto a AT-rich region) that finally recruits Pol III for multiple cycles of transcription. (B) A regular 5S rRNA gene is first recognized by its specific factor TFIIIA; then the 5S rRNA gene–TFIIIA complex is bound by TFIIIC and the next steps of transcription are identical to that of tRNA genes. In both cases, transcription stops and efficiently recycles when Pol III reaches the terminal T-track in the RNA-like strand. (C) Transcription of a dicistronic tRNA–tRNA gene by a unique TFIIIC molecule binding on the upstream gene. Assembly of TFIIIB onto the upstream gene (GC-rich) triggered by TFIIIC bound on the downstream gene to transcribe this only gene is penalized in vivo. (D) Hypothetical transcription of a dicistronic tRNA–5S rRNA gene may proceed similarly through the recognition of the promoter elements of the upstream gene by TFIIIC. In this case a single primary RNA (arrow) is produced and later matured into two functional products (tRNA and 5S rRNA) without the need of TFIIIA (see text for details).
The type of the tDNAs to which 5S rRNA genes are linked appears to be non-random (Table 2). Of the 30 tDNA-Gly (GCC), 29 are followed by 5S rRNA genes but none of the 11 tDNA-Gly (TCC). In one of these 29 dual genes, the 5S rRNA gene is interrupted by an Ylt1 element. Of the 28 tDNA-Asp (GTC) and 27 tDNA-Glu, 8 and 9 are followed by 5S rRNA genes, respectively. Two single tDNA-Thr (AGT) and tDNA-Gln (CTG) are also followed by 5S rRNA genes. The distance separating nt 73 of the tDNAs and nt 1 of the mature 5S rRNA is most of time 4 or 5 nt and only once, 7 nt. We also observed the presence of a single tricistronic gene tRNA-Lys (CTT)-tRNA-Glu (CTC)-5S rRNA gene with intergenic distances of 17 and 4 nt. From the examination of their sequence, all 3′ fused 5S rRNA genes appear functional (with the exception of a single one 3′ deleted by the insertion of an Ylt1 element). The sequences of the 5S rRNA fused genes do not depart from that of isolated genes. Coordinates of these 47 dicistronic and of the single tricistronic genes are given in Supplementary Data 2.
Table 2.
Genomic organization of the 47 tRNA–5S RNA dicistronic genes and of a single tRNA–tRNA–5S RNA tricistronic gene in Y. lipolytica
| tRNA (type) | Number of tRNA gene copies | Number of 5S rRNA gene-fused copies (full length) | Intergenic distance (nt) |
|---|---|---|---|
| Gly (GCC) | 30 | 28 | 4 or 5 |
| Asp (GTC) | 28 | 8 | 4 or 5 |
| Glu (CTC) | 27 | 9 | 4 |
| Gln (CTG) | 13 | 1 | 4 |
| Thr (AGT) | 23 | 1 | 7 |
| Lys (CTT)-Glu (CTC) | 3 | 1 | 17 and 4 |
A novel putative ncRNA, ‘RUF70’, is located 3′ of the 13 tRNA-Trp genes of Y. lipolytica
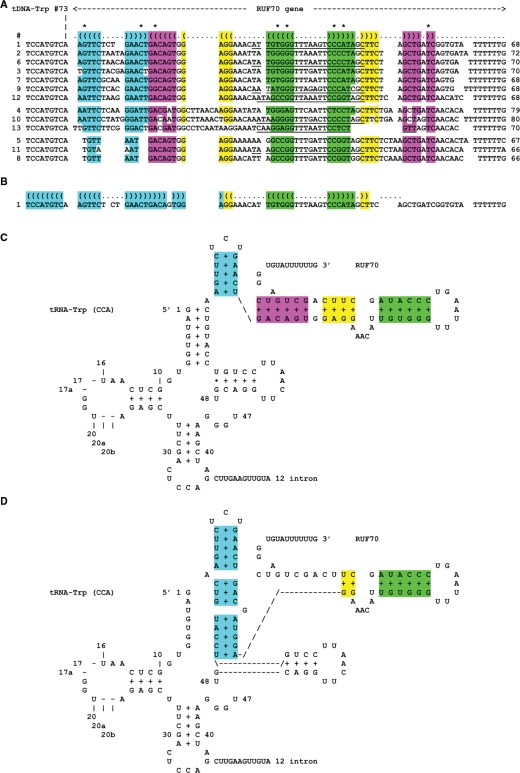
Following the discovery of tRNA genes located immediately upstream of about half of the 5S rRNA genes, we thoroughly checked for the presence of a Pol III transcriptional terminator (T-track) in the 50 nt at the 3′-end of all other tRNA genes. With the exception of a few tandem tRNA genes already reported (11), a feasible terminator was always identified except in all 13 copies of the essential tRNA-Trp (CAA) genes in which the Pol III terminator was always found about 70 nt downstream of the tDNA 3′-end. According to previously used nomenclature (54), we named this putative ncRNA ‘RUF70’. As shown in Figure 2A, a common tertiary structure is possible for all 13 copies of Y. lipolytica RUF70. Noteworthy, base changes leading to compensatory base pairing (shown with stars in Figure 2A) support conservation of secondary structure and argue positively for function. An alternative structure is shown in Figure 2B and the corresponding 2D structures in Figure 2C and D. Detailed coordinates of the 13 tDNA-Trp (CCA)–RUF70 dicistronic genes are given in Supplementary Data 3.
Figure 2.
Structural alignment of the 13 RUF70 genes located beyond the 13 copies of tDNA-Trp (CCA). (A) For sake of clarity, the DNA sequence, instead of RNA sequence, is used. Nucleotides 67–73 of the tDNA-Trp (CCA) are shown at left followed by the sequence of the RUF70 genes (sorted according to similarity). Coloured background in stems indicate correct base pairing (including GT pairs, GU in RNA). Stars above base pairing line (shown as opening and closing brackets) denote compensatory base pairing (e.g. TA or GC or CG pairs exchange) in stems. The length indicated at right is computed from the nucleotide following nt 73 of tDNA-Trp to the transcription termination signal (T-track, not included). The oligonucleotides used in northern experiments (see Figure 3D) hybridize with the underlined sequences. Detailed coordinates are given in Supplementary Data 3. (B) Alignment showing an alternative structure expanding the first stem at the expense of the tRNA acceptor stem. (C) 2D RNA structure corresponding to alignment shown in (A) for the first sequence. (D) Same for the alignment shown in (B).
Characterization of the tRNA–5S rRNA and tRNA-Trp– RUF70 genes dicistronic transcripts
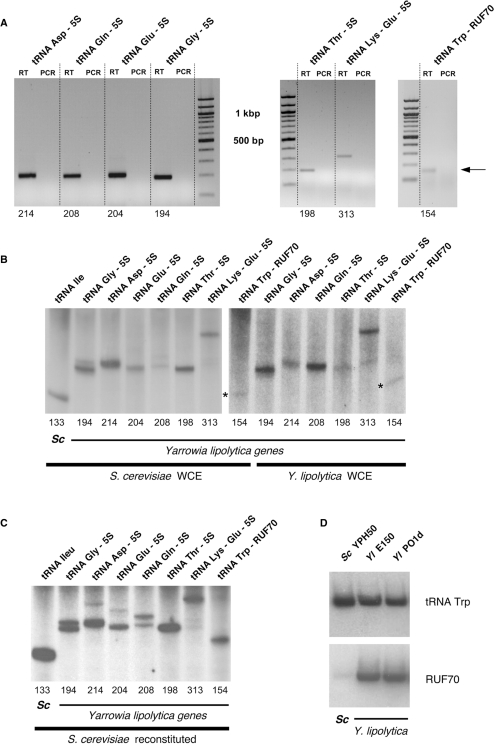
Performing RT–PCR, we demonstrated that tRNA–5S rRNA and tRNA-Trp–RUF70 genes are transcribed into a single dicistronic pre-RNA. Oligonucleotides were designed to amplify, after reverse transcription, a PCR double stranded DNA extending from nt 1 of the mature tRNA to nt 118 of the 5S rRNA (or nt 68 in the case of RUF70). Figure 3A shows that, in all cases, PCR bands of the expected length were obtained.
Figure 3.
Expression detected by RT–PCR, in vitro trancriptions and northern analyses. (A) Expression of the five different tRNA–5S rRNA, one tRNA–tRNA–5S rRNA and tRNA-Trp–RUF70 composite genes detected by RT–PCR. ‘RT’ refers to regular RT–PCR experiment, the reverse transcriptase was omitted in the lanes labelled ‘PCR’. The predicted length of the RT–PCR products is indicated at bottom. Only a faint band was obtained when testing the expression of the tRNA-Trp–RUF70 gene (arrow). (B) In vitro transcription of cloned copies of the same composite genes with WCE from either S. cerevisiae or Y. lipolytica as indicated. A longer exposure was used for the rightmost seven lanes. A tRNA-Ile (TAT) gene from S. cerevisiae (labelled ‘Sc’) is used as a control in the left lane. Expected lengths are indicated at bottom. Asterisks denote the faint bands obtained in the transcription experiments of tRNA-Trp–RUF70 gene. (C) In vitro transcription of the same genes using a fully recombinant TFIIIC reconstituted system from S. cerevisiae. (D) Northern blot analysis of the mature transcription products of the tRNA-Trp (CCA)–RUF70 composite genes. Total RNAs were extracted from the indicated strains growing in exponential phase. RUF70 genes were probed with a mixture of oligos targeted at the main stem-loop (sequences underlined in Figure 2A). The positive response of S. cerevisiae RNAs is due to the nearly perfect sequence conservation of the tRNA-Trp (CCA) genes between S. cerevisiae and Y. lipolytica.
One copy of each type of fusions was then selected and cloned into an EcoRV-digested pBluescriptSK vector for further transcriptional studies. Using WCE from both S. cerevisiae and Y. lipolytica strains, we observed one major RNA species for the various DNA templates that, according to their size, may represent the primary transcripts (Figure 3B). As previously described (12,53,55), our data confirmed that the tRNA and the 5S rRNA or the tRNA-Trp and RUF70 genes are transcribed coordinately, starting from the promoter of the leader tRNA gene. The same results are obtained using an in vitro reconstituted Pol III transcription system. In fact, in this case some minor bands with slower mobility can be detected, suggesting that some processing events may have occurred (Fig. 3C), as previously described (40). Yeast S. cerevisiae cell free extracts were previously shown to be able to process precursor tRNA but also tRNA–tRNA fusion (55,56). However, under our experimental conditions, none of our tRNA fusions were completely cleaved (Figure 3C), suggesting that these fusions are not correctly processed in our in vitro experiments.
Characterization of the matured RNA products
To assess the correct maturation of the tRNA-Trp and RUF70 molecules, we perform northern experiments with RNA extracted from Y. lipolytica and S. cerevisiae growing in exponential phase (Figure 3D). Only one strong signal with the same size was observed with tRNA-Trp specific oligonucleotides in total RNA prepared from both species (Figure 3D, top pannel). As a matter of fact, tDNA-Trp from both species are nearly identical [only 3 nt are different]. On the other hand, oligonucleotides targeted to RUF70 RNAs revealed a signal only in RNA extracted from Y. lipolytica as expected (bottom panel). It corresponds to a band that migrates slightly more rapidly that the tRNA-Trp (∼70 versus 78 nt, data not shown). We hardly detected the dicistronic transcript tRNA-Trp–RUF70 (Figure 3A), however, its presence in vivo as a mature ∼70 base product is unambiguously evidenced by northern experiment (Figure 3D). We, therefore, favour the hypothesis of Pol III dicistronic transcripts for the expression of the 13 tRNA-Trp–RUF70 genes.
Although two types of 5S rRNA are present in Y. lipolytica (expressed from the isolated genes and potentially from the tDNA-fused genes), only a single species of 5S rRNA is revealed when total RNA was analysed in acrylamide–urea gel (Figure 4A, lanes 2 and 3). Altogether these data strongly suggest that all kinds of discistronic genes are both expressed as a primary RNA product that is next correctly processed into tRNA, 5S rRNA or RUF70 functional components.
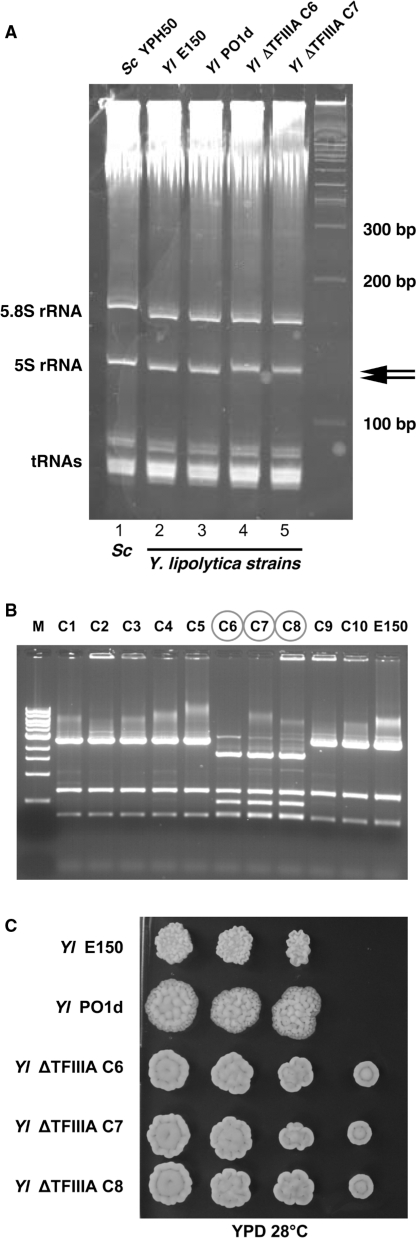
Figure 4.
Phenotypic and 5S rRNA analyses of Y. lipolytica ΔylTFIIIA mutants. (A) Total RNA content of wild-type S. cerevisiae strain (YPH500), wild-type Y. lipolytica strains (E150 and PO1d) and two ΔylTFIIIA Y. lipolytica deleted strains (ΔylTFIIIA C6 and ΔylTFIIIA C7, similar results were obtained for ΔylTFIIIA C8, not shown). Cells were grown in exponential phase, total RNA was extracted and analysed in 6% acrylamide–urea gel and stained with ethidium bromide. The migration of tRNA, 5.8S and the two distinct 5S rRNA species (arrows at right) are indicated. (B) Restriction pattern of the ylTFIIIA amplified locus of Y. lipolytica transformants. Only three out 10 transformants lost accurately the ylTFIIIA gene (C6, C7 and C8). Strain E150 was used as a control for the wild restriction pattern. M, molecular marker, 1 kb DNA ladder (New England Biolabs). (C) Effects of the disruption of the ylTFIIIA gene (YALI0F05104g) on growth ability. Serial dilutions (from 3 × 103 to 3 cells) of overnight cultures of the wild-type strains (E150 and PO1d) and of three independent (ΔylTFIIIA C6, C7 and C8) mutant strains were inoculated on YPD plates (or YNB supplemented, not shown) and incubated at 28°C (or 18°C, not shown) for 3 days.
Inactivation of ylTFIIIA yields a viable phenotype in Y. lipolytica and suggests that ylTFIIIA is possibly implicated in 5S rRNA maturation
Since 5S rRNA expressed by the transcription of dicistronic tRNA–5S rRNA genes appear to be correctly matured, we wondered whether TFIIIA was really, in Y. lipolytica, an essential protein. In S. cerevisiae, TFIIIA is encoded by YPR186c (Tfc2, also name Pzf1); this protein is essential for the transcription of 5S rRNA genes and the deletion of its gene is lethal for the cells. (57). Using BlastP, we unambiguously identified orthologs in C. glabrata, Kluyveromyces lactis, D. hansenii and Y. lipolytica (with E values e-118, e-118, e-76 and e-29, respectively). Sequences and alignment of TFIIIA orthologs in hemiascomycetes are given in Supplementary Data 5. We performed the deletion of the identified ortholog (YALI0F05104g gene, see Materials and Methods) in the Y. lipolytica haploid strain PO1d. Among the numerous resulting clones, 10 independent clones were analysed by PCR and EcoRI restriction to verify the deletion. Three of them (ΔylTFIIIA C6, C7, C8, circled in Figure 4B) showed only the expected restriction pattern of the disrupted cassette and not the pattern of the wild copy. This implies that ylTFIIIA gene was successfully deleted in these transformants, with no remaining wild gene copy. These transformants displayed viable phenotype with normal growth at 18°C and 28°C and only a slight morphological phenotype: colonies are smooth instead of rough (Figure 4C). We then examined the total RNA extracted from both WT and ΔylTFIIIA mutant strains (Figure 4A). In both types of cells, we observed a single band for matured 5S rRNA; however in the mutant strains, the 5S rRNA appear to be slightly longer suggesting that ylTFIIIA could be, in some way, implicated in the maturation of 5S rRNA [compare lanes 4 or 5 (wild-type) with lanes 2 or 3 (mutant) in Figure 4A]. This difference in length appears to be comparable to the difference between 5S rRNA from S. cerevisiae (lane 1) and Y. lipolytica (lane 2) which is only 1 nt.
Evolution of TFIIIA factor throughout eukaryotes: specificity of the Y. lipolytica ortholog
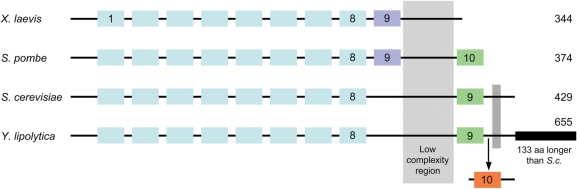
The first TFIIIA sequence was obtained from a Xenopus laevis cDNA (58). Proteolytic digestion at 3-K interval and the presence of 9–11 atoms of zinc per 7S particle (5S rRNA + TFIIIA) together with the repetitive presence of numerous Cys and His residues suggested the 9 zinc-finger model (19). Further sequencing of the X. laevis TFIIIA gene itself revealed that exon–intron boundaries also correspond closely to the repetitive unit, supporting the idea that TFIIIA evolved by successive duplication of a primordial 30-residue unit (59). The second TFIIIA sequence, that of S. cerevisiae was shown to diverge extensively from the Xenopus sequence, especially fingers 8 and 9 were separated by an essential 81-residue intervening sequence (60,61). The gene is unique in S. cerevisiae and TFIIIA is essential. Next came the sequence of S. pombe (52) and a careful comparison of the three sequences revealed that the S. pombe ortholog has in fact 10 zinc-finger, with its finger 9 corresponding to finger 9 of the Xenopus sequence and finger 10 corresponding to finger 9 of the yeast sequence (62); this is schematized in Figure 5. Curiously, the S. pombe ortholog terminates immediately after finger 10. Examination of several hemiascomycete TFIIIA sequences shows that they are all similar to the S. cerevisiae model: the Cys and His residues of the last but one finger (numbered 8 in X. laevis) were lost (see multiple alignment in Supplementary Data 5). Interestingly, the Y. lipolytica ortholog (ylTFIIIA) is the longest among TFIIIA proteins (655 residues) and it displays two specific extensions with respect to the sequences of other hemiascomycetes. A first 37-residue intervening sequence is located between fingers 9 and the last homology block common to all hemiascomycetes, a 10th zinc finger is present in this extension; it has no equivalent in other TFIIIA's. A second long N-terminal extension extends the sequence by 133 residues with respect to S. cerevisiae ortholog (Figure 5).
Figure 5.
Evolution of TFIIIA sequence throughout eukaryotes. The nine filled rectangles in the top sequence (X. laevis) symbolize the nine zinc fingers. The last zinc finger in S. cerevisiae TFIIIA is also traditionally numbered nine because this sequence was obtained prior to that of S. pombe that exhibits 10 zinc fingers. Multiple sequences alignment (not shown) confirm that finger 9 of S. pombe sequence corresponds to finger 9 of X. laevis sequence and that fingers 9 of S. cerevisiae and Y. lipolytica correspond to finger 10 of S. pombe. In TFIIIA from Y. lipolytica, an extra 10th zinc is present between finger 9 and the last block of homology common to all hemiascomytes (small dark vertical rectangle). See also alignment in Supplementary Data 5.
DISCUSSION
In this work we report the identification and first characterization of two novel types of dicistronic Pol III genes in Y. lipolytica. In both types, similarly to other cases already known, the leader gene encodes a tRNA and the second gene encodes here either a 5S rRNA (48 cases) or a novel RNA (13 cases), RUF70, of unknown function. These two types enlarge the present collection of dicistronic Pol III genes: tandem tRNA genes in S. pombe and hemiascomycetes (11–13); tRNA–snoRNA genes in A. thaliana and O. sativa (14); tRNA–miRNA genes in human (15) and mouse gammaherpes virus 68 (16). Nearly half of the 5S rRNA gene copies present in Y. lipolytica are implicated in Pol III fusions (48 over 108 full length genes), splitting the 5S rRNA genes into two families: the isolated genes (60 copies) and the fused genes (48 copies). The existence of such dicistronic genes linking a tRNA gene and a 5S rRNA gene is an unprecedented and still unique case. Interestingly, ylTFIIIA becomes no more essential in normal growth conditions and its absence revealed a possible role of ylTFIIIA in 5S rRNA processing.
The various composite genes appear to be active in normal growth conditions. Pol III promoter elements, defined as containing the sequences TRGYnnAnnnG and GWTCRAnnC as the A- and B-block, respectively (13), are present and functional as evidenced by in vitro experiments performed with a reconstituted Pol III transcription system. Furthermore, these Pol III promoter elements are correctly cross recognized through species (Figure 3B and C) since transcription can be detected with the Pol III transcription machinery derived from S. cerevisiae. Transcription of these genes occurs in vivo and starts with the synthesis of a primary RNA product expressing both genes, as demonstrated by RT–PCR (Figure 3A). Further experiments demonstrate that these RNA are processed in vivo. All copies of the tDNA-Gly(GCC) but two (28 out of 30) are followed by a full length 5S rRNA gene, RUF70 is only present in dicistronic genes in fusion with tDNA-Trp. However, neither uncleaved tRNA-Gly (GCC)-5S rRNA, nor tRNA-Trp–RUF70 primary product could be detected by northern blot analysis contrary to the matured tRNA-Gly, tRNA-Trp, 5S rRNA and RUF70 (Figure 3D and data not shown). As shown experimentally by Kruszka and coworkers, the accurate processing of a snoRNA adjacent to tRNA-Gly (GCC) in A. thaliana implicates the regular tRNA 3′-processing activity performed by tRNase Z (14). In S. cerevisiae, the 3′-tRNase activity is supported by Trz1 (encoded by YKR079c) (63,64) and this protein appears to be well conserved among hemiascomycetes (E-value e-32 for the ortholog of Y. lipolytica, YALI0A13057g, with respect to that of S. cerevisiae). Yeast cell free extracts were generally found to allow the synthesis of processed transcripts, including internal cleavage when dicistronic genes are used (55,56), contrary to what we observed using both S. cerevisiae and Y. lipolytica extracts (Figure 3B), suggesting that the reaction conditions are not optimal for the cleavage. For instance, metal ion were found to be required for in vitro pre-tRNA processing reactions catalyzed by TRZ1, the tRNase Z from A. thaliana (65). tRNAse Z also appears to request in vitro finely tuned pH and salt concentration values from one species to another (66) and we concluded that the cleavage by tRNase Z is not observed in our transcription experiments due to unsuitable pH and salt conditions.
Tandem tRNA gene transcription was first investigated two decades ago with a tDNA-Ser–tDNA-Met from S. pombe. Both genes were shown to be transcribed as a dimeric tRNA precursor followed by processing of the two tRNAs (12). In vivo transcription was drastically reduced by mutations in either the A-box (A at position 19) or the B-Box (A at 53) of the leader tRNA-Ser gene. This provided a direct evidence that the trailer tRNA-Met gene is derived through processing of the dimeric precursor and not initiated independently from the internal tRNA-Met gene promoters (12). The novel case of tRNA–5S rRNA genes in Y. lipolytica raises the question of a possible conflict between the Pol III transcription factors requested for both genes. We reasoned similarly in the case of the composite tRNA–5S rRNA genes of Y. lipolytica and assumed that ylTFIIIA might be dispensable in this special case. We also considered a possible steric conflict between ylTFIIIA bound on the 5S rRNA gene and TFIIIC bound on the leader tRNA gene. Furthermore, if ylTFIIIA and TFIIIC bind on the 5S rRNA gene, TFIIIB would have to bind DNA inside the leader tRNA gene, the high GC content of which is known to prevent efficient TFIIIB binding (67). For these reasons, we attempted the deletion of the Y. lipolytica ortholog of S. cerevisiae TFIIIA.
Deletion of ylTFIIIA turned out to be viable and leads only to a slight morphological phenotype. More important, 5S rRNA appears slightly longer (a few nucleotides) in the deletant strains and we noticed that a single 5S rRNA species, homogenous in length is observed in both the wild-type strains and in the ylTFIIIA-deleted strains. This may appear illogical, at first glance, if one considers that, in wild-type strains, two types of 5S rRNAs coexist: those expressed from the isolated 5S rRNA gene and those resulting from the maturation of the dicistronic primary transcript of the tRNA–5S rRNA genes (both types being present in nearly equal gene amounts). In the ylTFIIIA-deleted strains, in which the transcription of isolated genes is no more possible, only the latter type of 5S rRNA is present. The defect in 5S RNA processing we observed in our ΔylTFIIIA strains (a few nucleotides longer) suggests that ylTFIIIA itself could participate, in some way, in this processing possibly through its role of 5S RNA-chaperone (23–25) or in functional interaction with the YL3 protein (68). However, it also indicates that the final processing event that occurs in the presence of ylTFIIIA is not absolutely required for 5S rRNA to be functional, at least under normal growth conditions. As a matter of fact, 3′-extended 5S RNA was also observed in a strain deleted for the 3′ exoribonuclease Rex1p exhibiting no growth defect (69). With respect to TFIIIA proteins of other hemiascomycetes, ylTFIIIA harbours a 10th zinc finger and a long C-terminal extension. These specific features might be also implicated in novel roles for TFIIIA in Y. lipolytica.
Our results nicely parallel those obtained earlier on the artificial Pol III fusion RPR1 promoter-5S rRNA studied by Camier and coworkers (30). A chimeric gene was constructed in which the first 84 nt of the RPR1 gene containing a tRNA-like promoter sequence was fused to a 5S rRNA gene. This gene was transcribed in an in vitro Pol III transcription assay and a single RNA transcript was observed at the expected length for initiation at the RPR1 promoter, however no cleavage of the two products took place. No transcript initiating at the 5S promoter was observed, even after addition of TFIIIA, suggesting that the RPR1 promoter is functional and dominant over the downstream 5S promoter. In vivo, cells devoid of TFIIIA could survive if they carry such a construction on a multicopy plasmid. Similarly with our present observations, 5S rRNA expressed from RPR1-5S rRNA hybrid genes was slightly longer than that produced from endogenous 5S rRNA gene copies.
A number of tandem tDNAs are already known in hemiascomycetes (11). We first checked that the Pol III gene fusions we observed in the sequenced strain of Y. lipolytica (strain E150) were also present in the strain investigated during earlier studies [strain PO1d, (70)]. We then checked the set of nine hemiascomycetes + S. pombe previously studied (13) for the presence of a Pol III terminator 3′ to all tDNAs (within 40 nt 3′). No new cases of Pol III fusions other than tandem tDNA genes already reported were discovered. We then extended our search to other ascomycetes: Pichia stipitis, Neurospora crassa, Magnaporthe grisea, Coprinus cinereus, Aspergillus nidulans, Fusarium graminearum, Ustilago maydis and Podospora anserina. No other type of Pol III fusions than tandem tDNAs was discovered (with the exception of a triple tDNA-Lys (CTT) in Pichia stipitis). We also further investigated two recently published plant genomes: Populus trichocarpa and Vitis vinifera. In P. trichocarpa, we disclosed 12 copies of the tDNA-Gly (GCC)–snoRNA (Figure 6) identical to those of A. thaliana (14) and only two copies in V. vinifera. We finally noticed that, curiously, the very same type of tDNA gene, namely tDNA-Gly (GCC) is implicated in Pol III gene fusions in several organisms: tRNA-Gly (GCC)–snoRNA genes in A. thaliana (14) and also in P. trichocarpa (this work), tDNA-Gly (GCC)-5S rRNA genes in Y. lipolytica (this work). We do not know whether this might indicate for tDNA-Gly (GCC) a propensity to generate fusions more easily than other tRNAs. Remarkably, the other tDNA-Gly, with anticodon (TGC), is never implicated in fusions.
Figure 6.
Structural alignment of the 10 C/D box snoRNA genes located beyond the tDNA-Gly (GCC) genes of P. trichocarpa. Ten dicistronic tRNA-Gly (GCC)–snoRNA genes were identified in the genome of P. trichocarpa. The 10 copies of tDNA are identical and are shown under the usual cloverleaf structure (top). The 10 C/D snoRNA gene sequences shown (bottom) immediately follows nt 73 of the tDNAs. Dashes indicate the antisense element and the asterisk indicates the nucleotide targeting the methylation. Most conserved residues (in bold) are displayed in the last line.
Remarkably, the Y. lipolytica genome contains another type of Pol III fusions: each of the 13 copies of the tRNA-Trp (CCA) gene is followed by an extension of about 70 nt. This extension is cotranscribed and appears to be stable enough as northern blot can detect it. We noticed that between the 13 copies, the sequence is only loosely conserved, suggesting some degeneration. We first thought about a RNA implicated in the maturation of the tRNA-Trp or in the splicing of its intron. However, this tRNA is highly conserved throughout hemiascomycetes (only 3 nt are changed with respect to the S. cerevisiae tRNA-Trp) and the intron is not present in some species such as D. hansenii. We noticed some structural similarity (stems/loops) with the ncRNA CeN76 identified in the non-coding transcriptome of Caenorhabditis elegans [Figure 2C in (71)]. In the model we propose (Figure 2A and C), a stem is immediately adjacent to the 3′ end of the tRNA, similarly with the models proposed for CeN76 and the snoRNA following the tRNA-Gly (GCC) in A. thaliana [Figure 6 in (14)]. Attempt to identify a similar ncRNA in other ascomycetes and S. pombe remained unsuccessful. The role of RUF70 in Y. lipolytica remains unknown as are unknown the role of many ncRNAs recently discovered in higher eukaryotes (54,71).
In each yeast species, all tRNA genes encoding a given tRNA type (same anticodon) remain identical or nearly so. This intraspecific sequence homogeneity, combined with the rapid variation of the number of paralogous gene copies, is puzzling. It contrast with the rapid sequence variation between paralogous copies of protein coding genes, suggesting the existence of a mechanism for a quick loss and proliferation for tRNA genes in genome. The existence of a proliferation mechanism is further suggested by the frequent occurrence, in yeast genomes, of a variety of Pol III gene fusion involving a leader tRNA gene. Such fusions might appear unnecessary since every Pol III gene carries its own internal promoter elements. This apparently unlikely gene fusion event is now known to have occurred many more times in hemiascomycetes where a variety of pairs of tRNA genes separated by a very short distance are observed often in multiple copies in genomes (11). Multiple paralogous copies of dicistronic tRNA–snoRNA are also present in plants (14). In Y. lipolytica, fusions event including a 5S rRNA gene have occurred at least six times (as six different tDNA types are implicated), some of the fusions being amplified up to 29 times in the case of the tRNA-Gly (GCC)-5S rRNA gene. Similarly, the tRNA-Trp–RUF70 gene was amplified into 13 dispersed copies. We still do not know the possible mechanisms of Pol III gene fusion and proliferation in hemiascomycetes.
A strikingly similar case of 5′ maturation by tRNase Z was very recently reported for the 5S rRNA gene in the euryarchaeota H. volcanii (72). The 5S gene is expressed from the ribosomal operon that also includes the 16S and 23S rRNA genes and a distal tRNA gene. The 5S rRNA gene is not preceded by a tRNA gene but the sequences immediately upstream fold into a tRNA-like structure and therefore the 5′ splicing of the 5S rRNA (from the primary transcript) can be performed by tRNase Z. Beside that of H. volcanii, tRNase Z from A. thaliana or S. cerevisiae were also able to perform this maturation in vitro. With this information in mind, we examined the sequences upstream the mature snR52, RPR1 and Zod1 RNA genes of S. cerevisiae (and related species) which contains regular A- and B-promoter elements (13,73). However, we could not arrange any of these sequences in a way similar to the tRNA-like structure discovered in H. volcanii. The quite similar case of tRNA-helped 5S rRNA 5′ processing we report here in Y. lipolytica is therefore the only one yet known in eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We wish to thank Bernard Dujon (Institut Pasteur), Jean-Luc Souciet (Université Louis Pasteur, Strasbourg) and Henri Grosjean (Université Paris-Sud, Orsay) for many stimulating discussions. R.K.-L. acknowledges a fellowship from the French Ministère de l’Enseignement Supérieur et de la Recherche.
FUNDING
Agence Nationale pour la Recherche (Project # ANR-05-BLAN-0331); GDR CNRS 2354 (Génolevures-3). Funding to open access charge: iBiTec-S.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blumenthal T. Operons in eukaryotes. Brief. Funct. Genomic. Proteomic. 2004;3:199–211. doi: 10.1093/bfgp/3.3.199. [DOI] [PubMed] [Google Scholar]
- 2.Worthey EA, Martinez-Calvillo S, Schnaufer A, Aggarwal G, Cawthra J, Fazelinia G, Fong C, Fu G, Hassebrock M, Hixson G, et al. Leishmania major chromosome 3 contains two long convergent polycistronic gene clusters separated by a tRNA gene. Nucleic Acids Res. 2003;31:4201–4210. doi: 10.1093/nar/gkg469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown JW, Clark GP, Leader DJ, Simpson CG, Lowe T. Multiple snoRNA gene clusters from Arabidopsis. RNA. 2001;7:1817–1832. [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CL, Liang D, Zhou H, Zhuo M, Chen YQ, Qu LH. The high diversity of snoRNAs in plants: identification and comparative study of 120 snoRNA genes from Oryza sativa. Nucleic Acids Res. 2003;31:2601–2613. doi: 10.1093/nar/gkg373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 7.Thatcher EJ, Bond J, Paydar I, Patton JG. Genomic organization of zebrafish microRNAs. BMC Genomics. 2008;9:253. doi: 10.1186/1471-2164-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu LH, Henras A, Lu YJ, Zhou H, Zhou WX, Zhu YQ, Zhao J, Henry Y, Caizergues-Ferrer M, Bachellerie JP. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 1999;19:1144–1158. doi: 10.1128/mcb.19.2.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 10.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 12.Willis I, Hottinger H, Pearson D, Chisholm V, Leupold U, Söll D. Mutations affecting excision of the intron from eukaryotic dimeric tRNA precursor. EMBO J. 1984;3:1573–1580. doi: 10.1002/j.1460-2075.1984.tb02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marck C, Kachouri-Lafond R, Lafontaine I, Westhof E, Dujon B, Grosjean H. The RNA polymerase III-dependent family of genes in hemiascomycetes: comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res. 2006;34:1816–1835. doi: 10.1093/nar/gkl085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruszka K, Barneche F, Guyot R, Ailhas J, Meneau I, Schiffer S, Marchfelder A, Echeverria M. Plant dicistronic tRNA-snoRNA genes: a new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J. 2003;22:621–632. doi: 10.1093/emboj/cdg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 16.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 17.Wang CK, Weil PA. Purification and characterization of Saccharomyces cerevisiae transcription factor IIIA. J. Biol. Chem. 1989;264:1092–1099. [PubMed] [Google Scholar]
- 18.Gottesfeld J, Bloomer LS. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982;28:781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- 19.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pieler T, Hamm J, Roeder RG. The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell. 1987;48:91–100. doi: 10.1016/0092-8674(87)90359-x. [DOI] [PubMed] [Google Scholar]
- 21.Clemens KR, Liao XL, Wolf V, Wright PE, Gottesfeld JM. Definition of the binding sites of individual zinc fingers in the transcription factor IIIA-5S RNA gene complex. Proc. Natl Acad. Sci. USA. 1992;89:10822–10826. doi: 10.1073/pnas.89.22.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes JJ, Tullius TD. Structure of the TFIIIA-5 S DNA complex. J. Mol. Biol. 1992;227:407–417. doi: 10.1016/0022-2836(92)90897-s. [DOI] [PubMed] [Google Scholar]
- 23.Pelham HRB, Brown DD. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc. Natl Acad. Sci. USA. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guddat U, Bakken AH, Pieler T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in xenopus oocytes. Cell. 1990;60:619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- 25.Honda BM, Roeder RG. Association of a 5S gene transcription factor with 5S RNA and altered levels of the factor during cell differentiation. Cell. 1980;22:119–126. doi: 10.1016/0092-8674(80)90160-9. [DOI] [PubMed] [Google Scholar]
- 26.Liao XL, Clemens KR, Tennan L, Wright PE, Gottesfeld JM. Specific interaction of the first three zinc fingers of TFIIIA with the internal control region of the Xenopus 5S RNA gene. J. Mol. Biol. 1992;223:857–871. doi: 10.1016/0022-2836(92)90248-i. [DOI] [PubMed] [Google Scholar]
- 27.Theunissen O, Rudt F, Guddat U, Mentzel H, Pieler T. RNA and DNA binding zinc fingers in Xenopus TFIIIA. Cell. 1992;71:679–690. doi: 10.1016/0092-8674(92)90601-8. [DOI] [PubMed] [Google Scholar]
- 28.Clemens KR, Wolf V, McBryant SJ, Zhang P, Liao X, Wright PE, Gottesfeld JM. Molecular basis for specific recognition of both RNA and DNA by a zinc finger protein. Science. 1993;260:530–533. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- 29.Lee BM, Xu J, Clarkson BK, Martinez-Yamout MA, Dyson HJ, Case DA, Gottesfeld JM, Wright PE. Induced fit and “lock and key” recognition of 5S RNA by zinc fingers of transcription factor IIIA. J. Mol. Biol. 2006;357:275–291. doi: 10.1016/j.jmb.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Camier S, Dechampesme AM, Sentenac A. The only essential function of TFIIIA in yeast is the transcription of 5S rRNA genes. Proc. Natl Acad. Sci. USA. 1995;92:9338–9342. doi: 10.1073/pnas.92.20.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothfels K, Rowland O, Segall J. Zinc fingers 1 and 7 of yeast TFIIIA are essential for assembly of a functional transcription complex on the 5 S RNA gene. Nucleic Acids Res. 2007;35:4869–4881. doi: 10.1093/nar/gkm517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Heerikhuizen H, Ykema A, Klootwijk J, Gaillardin C, Ballas C, Fournier P. Heterogeneity in the ribosomal RNA genes of the yeast Yarrowia lipolytica; cloning and analysis of two size classes of repeats. Gene. 1985;39:213–222. doi: 10.1016/0378-1119(85)90315-4. [DOI] [PubMed] [Google Scholar]
- 33.Clare JJ, Davidow LS, Gardner DC, Oliver SG. Cloning and characterisation of the ribosomal RNA genes of the dimorphic yeast, Yarrowia lipolytica. Curr. Genet. 1986;10:449–452. doi: 10.1007/BF00419872. [DOI] [PubMed] [Google Scholar]
- 34.Querol A, Barrio E, Huerta T, Ramon D. Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl. Environ. Microbiol. 1992;58:2948–2953. doi: 10.1128/aem.58.9.2948-2953.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huet J, Manaud N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 1996;273:249–267. doi: 10.1016/s0076-6879(96)73024-0. [DOI] [PubMed] [Google Scholar]
- 36.Deprez E, Arrebola R, Conesa C, Sentenac A. A subunit of yeast TFIIIC participates in the recruitment of TATA- binding protein. Mol. Cell. Biol. 1999;19:8042–8051. doi: 10.1128/mcb.19.12.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huet J, Conesa C, Carles C, Sentenac A. A cryptic DNA binding domain at the COOH terminus of TFIIIB70 affects formation, stability and function of preinitiation complexes. J. Biol. Chem. 1997;272:18341–18349. doi: 10.1074/jbc.272.29.18341. [DOI] [PubMed] [Google Scholar]
- 38.Kassavetis GA, Joazeiro CA, Pisano M, Geiduschek EP, Colbert T, Hahn S, Blanco JA. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 39.Ferrari R, Rivetti C, Acker J, Dieci G. Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation. Proc. Natl Acad. Sci. USA. 2004;3:13442–13447. doi: 10.1073/pnas.0403851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ducrot C, Lefebvre O, Landrieux E, Guirouilh-Barbat J, Sentenac A, Acker J. Reconstitution of the yeast RNA polymerase III transcription system with all recombinant factors. J. Biol. Chem. 2006;281:11685–11692. doi: 10.1074/jbc.M600101200. [DOI] [PubMed] [Google Scholar]
- 41.Fickers P, Le Dall MT, Gaillardin C, Thonart P, Nicaud JM. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J. Microbiol. Methods. 2003;55:727–737. doi: 10.1016/j.mimet.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Le Dall MT, Nicaud JM, Gaillardin C. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr. Genet. 1994;26:38–44. doi: 10.1007/BF00326302. [DOI] [PubMed] [Google Scholar]
- 43.Oficjalska-Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, Sentenac A, Lefebvre O. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol. Cell. 2006;22:623–632. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Sethy-Coraci I, Moir RD, Lopez-de-Leon A, Willis IM. A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res. 1998;26:2344–2352. doi: 10.1093/nar/26.10.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Moir RD, Sethy-Coraci IK, Warner JR, Willis IM, Huang Y, McGillicuddy E, Weindel M, Dong S, Maraia RJ. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 2000;20:3843–3851. doi: 10.1128/mcb.20.11.3843-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The Génolevures-3 Consortium. Comparative genomics of five hemiascomycetous species identifies the basic yeast genome repertoire. Genome Research. 2008 in preparation. [Google Scholar]
- 47.Neuvéglise C, Bon E, Lépingle A, Wincker P, Artiguenave F, Gaillardin C, Casaregola S. Genomic exploration of the hemiascomycetous yeasts: 9. Saccharomyces kluyveri. FEBS Lett. 2000;487:56–60. doi: 10.1016/s0014-5793(00)02280-8. [DOI] [PubMed] [Google Scholar]
- 48.Malpertuy A, Llorente B, Blandin G, Artiguenave F, Wincker P, Dujon B. Genomic exploration of the hemiascomycetous yeasts: 10. Kluyveromyces thermotolerans. FEBS Lett. 2000;487:61–65. doi: 10.1016/s0014-5793(00)02281-x. [DOI] [PubMed] [Google Scholar]
- 49.Bolotin-Fukuhara M, Toffano-Nioche C, Artiguenave F, Duchateau-Nguyen G, Lemaire M, Marmeisse R, Montrocher R, Robert C, Termier M, Wincker P, et al. Genomic exploration of the hemiascomycetous yeasts: 11. Kluyveromyces lactis. FEBS Lett. 2000;487:66–70. doi: 10.1016/s0014-5793(00)02282-1. [DOI] [PubMed] [Google Scholar]
- 50.Wendland J, Pohlmann R, Dietrich F, Steiner S, Mohr C, Philippsen P. Compact organization of rRNA genes in the filamentous fungus Ashbya gossypii. Curr. Genet. 1999;35:618–625. doi: 10.1007/s002940050460. [DOI] [PubMed] [Google Scholar]
- 51.Lépingle A, Casaregola S, Neuvéglise C, Bon E, Nguyen H, Artiguenave F, Wincker P, Gaillardin C. Genomic exploration of the hemiascomycetous yeasts: 14. Debaryomyces hansenii var. hansenii. FEBS Lett. 2000;487:82–86. doi: 10.1016/s0014-5793(00)02285-7. [DOI] [PubMed] [Google Scholar]
- 52.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 53.Reyes VM, Newman A, Abelson J. Mutational analysis of the coordinate expression of the yeast tRNAArg-tRNAAsp gene tandem. Mol. Cell. Biol. 1986;6:2436–2442. doi: 10.1128/mcb.6.7.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chakrabarti K, Pearson M, Grate L, Sterne-Weiler T, Deans J, Donohue JP, Ares M., Jr Structural RNAs of known and unknown function identified in malaria parasites by comparative genomics and RNA analysis. RNA. 2007;13:1923–1939. doi: 10.1261/rna.751807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engelke DR, Gegenheimer P, Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J. Biol. Chem. 1985;260:1271–1279. [PubMed] [Google Scholar]
- 56.Klekamp MS, Weil PA. Specific transcription of homologous class III genes in yeast-soluble cell-free extract. J. Biol. Chem. 1982;257:8432–8441. [PubMed] [Google Scholar]
- 57.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 58.Ginsberg AM, King BO, Roeder RG. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984;39:479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- 59.Tso JY, Van Den Berg DJ, Korn LJ. Structure of the gene for Xenopus transcription factor TFIIIA. Nucleic Acids Res. 1986;14:2187–2200. doi: 10.1093/nar/14.5.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Archambault J, Milne CA, Schappert KT, Baum B, Friesen JD, Segall J. The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J. Biol. Chem. 1992;267:3282–3288. [PubMed] [Google Scholar]
- 61.Milne CA, Segall J. Mapping regions of yeast transcription factor IIIA required for DNA binding, interaction with transcription factor IIIC and transcription activity. J. Biol. Chem. 1993;268:11364–11371. [PubMed] [Google Scholar]
- 62.Schulman DB, Setzer DR. Identification and characterization of transcription factor IIIA from Schizosaccharomyces pombe. Nucleic Acids Res. 2002;30:2772–2781. doi: 10.1093/nar/gkf385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takaku H, Minagawa A, Takagi M, Nashimoto M. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 2003;31:2272–2278. doi: 10.1093/nar/gkg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y, Beck A, Davenport C, Chen Y, Shattuck D, Tavtigian SV. Characterization of TRZ1, a yeast homolog of the human candidate prostate cancer susceptibility gene ELAC2 encoding tRNase Z. BMC Mol. Biol. 2005;6:12. doi: 10.1186/1471-2199-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Späth B, Settele F, Schilling O, D’Angelo I, Vogel A, Feldmann I, Meyer-Klaucke W, Marchfelder A. Metal requirements and phosphodiesterase activity of tRNase Z enzymes. Biochemistry. 2007;46:14742–14750. doi: 10.1021/bi7010459. [DOI] [PubMed] [Google Scholar]
- 66.Späth B, Schubert S, Lieberoth A, Settele F, Schutz S, Fischer S, Marchfelder A. Two archaeal tRNase Z enzymes: similar but different. Arch Microbiol. 2008 doi: 10.1007/s00203-008-0368-4. ePub. [DOI] [PubMed] [Google Scholar]
- 67.Joazeiro CA, Kassavetis GA, Geiduschek EP. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 68.Lee Y, Nazar RN. Terminal structure mediates 5 S rRNA stability and integration during ribosome biogenesis. J. Biol. Chem. 2003;278:6635–6641. doi: 10.1074/jbc.M212220200. [DOI] [PubMed] [Google Scholar]
- 69.van Hoof A, Lennertz P, Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Souciet J, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, Casaregola S, de Montigny J, Dujon B, et al. Genomic exploration of the hemiascomycetous yeasts: 1. A set of yeast species for molecular evolution studies. FEBS Lett. 2000;487:3–12. doi: 10.1016/s0014-5793(00)02272-9. [DOI] [PubMed] [Google Scholar]
- 71.Deng W, Zhu X, Skogerbo G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, Liu C, et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis and expression. Genome Res. 2006;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hölzle A, Fischer S, Heyer R, Schütz S, Zacharias M, Walther P, Allers T, Marchfelder A. Maturation of the 5S rRNA 5′ end is catalyzed in vitro by the endonuclease tRNase Z in the archaeon H. volcanii. RNA. 2008;14:928–937. doi: 10.1261/rna.933208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guffanti E, Percudani R, Harismendy O, Soutourina J, Werner M, Iacovella MG, Negri R, Dieci G. Nucleosome depletion activates poised RNA polymerase III at unconventional transcription sites in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:29155–29164. doi: 10.1074/jbc.M600387200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.