Abstract
The ubiGmccBA operon of Clostridium acetobutylicum is involved in methionine to cysteine conversion. We showed that its expression is controlled by a complex regulatory system combining several RNA-based mechanisms. Two functional convergent promoters associated with transcriptional antitermination systems, a cysteine-specific T-box and an S-box riboswitch, are located upstream of and downstream from the ubiG operon, respectively. Several antisense RNAs were synthesized from the downstream S-box-dependent promoter, resulting in modulation of the level of ubiG transcript and of MccB activity. In contrast, the upstream T-box system did not appear to play a major role in regulation, leaving antisense transcription as the major regulatory mechanism for the ubiG operon. The abundance of sense and antisense transcripts was inversely correlated with the sulfur source availability. Deletion of the downstream promoter region completely abolished the sulfur-dependent control of the ubiG operon, and the expression of antisense transcripts in trans did not restore the regulation of the operon. Our data revealed important insights into the molecular mechanism of cis-antisense-mediated regulation, a control system only rarely observed in prokaryotes. We proposed a regulatory model in which the antisense RNA controlled the expression of the ubiG operon in cis via transcriptional interference at the ubiG locus.
INTRODUCTION
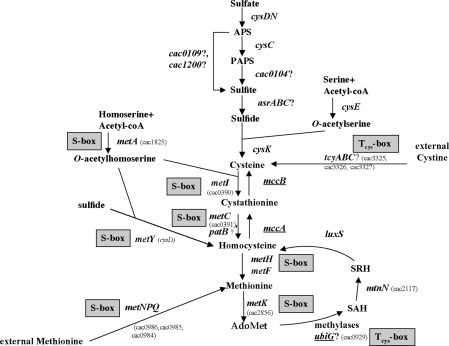
Micro-organisms can frequently use sulfate for the synthesis of organic sulfur metabolites, mostly cysteine, methionine and S-adenosyl-methionine (AdoMet). The sulfate assimilation pathway involves uptake and activation of inorganic sulfate followed by a stepwise reduction to sulfide. An O-acetylserine-thiol-lyase further catalyzes the reaction of sulfide and O-acetylserine (OAS) to give cysteine (1–3). Cysteine is then converted into methionine by the transsulfuration pathway followed by a methylation due to methionine synthases (4,5). Methionine is also a sulfur source for several bacteria indicating an efficient conversion of methionine into cysteine usually through the AdoMet recycling pathway and then the reverse transsulfuration pathway (Figure 1). This pathway, which is present in mammals, Saccharomyces cerevisiae, Pseudomonas aeruginosa and Bacillus subtilis, requires the sequential action of cystathionine β-synthases and cystathionine γ-lyases (6–8). In B. subtilis, the mccB and mccA genes encoding a cystathionine γ-lyase and a cystathionine β-synthase form an operon together with yrrT, mtnN and yrhC. The mtnN gene encodes the AdoHcy nucleosidase, involved in AdoMet recycling and yrrT, a putative AdoMet-dependent methyl-transferase. The master regulator of cysteine metabolism, CymR, represses the transcription of this operon in the presence of cysteine (6,9,10). Both CymR and CysK, the OAS-thiol-lyase, are required for DNA binding (6). The probable formation of a CysK–CymR complex may induce conformational changes to help the CymR-dependent binding to its targets. Physiological data indicate that OAS, the direct precursor of cysteine is important in the signaling pathway of CymR (6).
Figure 1.
Sulfur metabolism in C. acetobutylicum. The genes controlled by antisense RNAs are underlined. ‘S-box’ indicates the genes controlled by an S-box riboswitch. And ‘Tcys-box’ indicates the genes controlled by a cysteine-specific T-box system. A question mark indicates the genes probably involved in this pathway. AdoMet, S-adenosyl-methionine; Serine O-acetyltransferase, cysE; OAS-thiol-lyase, cysK; methionine adenosyltransferase, metK; adenosylhomocysteine nucleosidase, mtnN; S-ribosylhomocysteine lyase, luxS; cystathionine β-synthase, mccA; cystathionine γ-lyase, mccB; homoserine acetyl-transferase, metA; cystathionine γ-synthase, metI; cystathionine β-lyase, metC and patB; methionine synthase, metH; ATP sulfurylase, cysDN; APS kinase, cysC; anaerobic sulfite reductase, asrABC (11,12). The cac numbers for C. acetobutylicum genes correspond to those of AcetoList (http://bioinfo.hku.hk/GenoList/index.pl?database=acetolist) (38).
Clostridium acetobutylicum is one of the best-studied solventogenic clostridia and the genome of strain ATCC824 has been sequenced (11). In C. acetobutylicum, cysteine and homocysteine can be synthesized either directly from reduced sulfur (thiolation pathway) or by their interconversion involving the transsulfuration and the reverse transulfuration pathways (Figure 1) (12). An ubiGmccBA operon encodes putative AdoMet-dependent methyl-transferase (sharing low similarity with YrrT), cystathionine γ-lyase and cystathionine β-synthase. A cysteine-specific T-box is located upstream of this operon (12). In Firmicutes, the T-box riboswitch regulates the expression of various genes encoding aminoacyl-tRNA synthetases or is involved in amino acid biosynthesis by premature termination of transcription (13–16). Gene expression is controlled by competition between two mutually exclusive structures, a terminator and a less stable antiterminator. The choice between alternative leader RNA structures is determined by interactions of the nascent transcript with an uncharged tRNA, specific for the amino acid class of the regulated gene. The specificity of the response is dependent on pairing of the anticodon of the tRNA with a single codon (the ‘specifier sequence’) in the leader sequence (13,17). The acceptor end of the uncharged tRNA also pairs with residues in the antiterminator element, preventing the formation of the competing terminator helix (13). Each gene regulated by T-box is specifically induced in response to a decrease in aminoacylation of the cognate tRNA, and therefore signals the deficiency in the corresponding amino acid or aminoacyl-tRNA synthetase.
Interestingly, Rodionov et al. (12) have proposed that the ubiGmccBA operon is followed by a convergent S-box regulatory element. The S-box riboswitch controls the expression of genes involved in methionine transport, biosynthesis and recycling, in response to methionine availability (18). The leader transcript of S-box-controlled genes may fold in mutually exclusive antiterminator or terminator structures. S-box RNA directly senses the level of AdoMet in vivo and in vitro, and functions as an AdoMet-dependent riboswitch. In the presence of methionine, the concentration of AdoMet increases. The binding of this compound to the leader mRNA stabilizes the terminator structure leading to premature termination of transcription (19–21). Clostridia mainly use S-box-dependent riboswitches to control their methionine metabolism as observed for Bacillales (Figure 1) (12).
The ubiGmccBA operon is unique in the sense that two convergent promoters and two different systems of regulation by transcription antitermination are present upstream of and downstream from this operon. This suggested a mechanism of control involving an antisense RNA (12). In the past few years, the importance of regulatory mechanisms based on the action of RNA molecules became evident. Natural antisense transcripts have been found in both prokaryotes and eukaryotes (22–27). Antisense RNAs can be divided into cis-encoded RNAs, which are transcribed from the opposing DNA strand at the same locus and are thus fully complementary to their target RNAs; and trans-encoded RNAs, which are transcribed from separate loci and are only partially complementary to their target RNAs (26). While in eukaryotes, cis-encoded antisense RNAs are widespread (28), in prokaryotes, most of the antisense RNAs discovered until recently are trans-encoded (29–32). Cis-antisense RNAs are mainly found in plasmids, phages and transposons (27,33) and little is known about cis-antisense RNAs encoded by bacterial chromosome (34).
Since only few genetic tools are available in clostridia, we decided to study the regulation of the expression of the C. acetobutylicum ubiG operon mainly in B. subtilis. We then confirmed the sulfur-dependent control of the ubiG operon by experiments in the original host, C. acetobutylicum. We identified cis-encoded antisense RNAs complementary to the 3′-part of the C. acetobutylicum ubiGmccBA operon. This brings important insights into a novel regulatory mechanism, which involves antisense transcription repressed in the presence of methionine as a control of the methionine to cysteine conversion pathway.
MATERIALS AND METHODS
Bacterial strains and culture conditions
The B. subtilis strains used in this work are listed in Table 1. Escherichia coli strains were grown in LB broth. Bacillus subtilis was grown in SP medium or in minimal medium (10) supplemented with a sulfur source as stated: 1 mM K2SO4, 1 mM l-methionine, 1 mM l-cystine, 1 mM dl-homocysteine or 5 mM glutathione. For strain BFS2063 or strains derived from SSB353, 1 mM isopropyl β-d-thiogalactoside (IPTG) was added allowing the expression of genes under the control of the Pspac promoter. When required, antibiotics were added to the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 5 μg ml−1; spectinomycin, 100 μg ml−1; kanamycin, 5 μg ml−1; erytromycin 1 μg ml−1; lincomycin 25 μg ml−1. Standard procedures were used to transform E. coli and B. subtilis (35,36). Clostridium acetobutylicum ATCC824 strain was grown under anaerobic conditions (10% H2, 5% CO2, 85% N2), in a synthetic medium MS supplemented with 1 mM methionine or 1 mM cysteine (37).
Table 1.
Strains and vectors used in this study
| Genotype | Source | |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| BFS2063a | trpC2 ΔmccA::lacZ-ermb | Schumann |
| BSIP1165 | trpC2 ΔmccB::aphA3 | (6) |
| BSIP1303 | trpC2 ΔmccAB::aphA3 | (6) |
| BSIP1751 | trpC2 amyE::(cat Pc-S1box(−236, +349)Cacd-lacZ) | pDIA5715→168e |
| BSIP1761 | trpC2 amyE::(cat P-Tbox (−302, +392)Cac-lacZ) | pDIA5720→168 |
| BSIP1802 | trpC2 ΔmccAB::aphA3 amyE::(cat P-Tbox-ubiGmccBA-Sbox-PCac) | Materials and methods |
| BSIP1832 | trpC2 ΔmccA::lacZ-erm amyE::(cat P-Tbox-ubiGmccBA-Sbox-PCac) | BSIP1802→ BFS2063 |
| BSIP1833 | trpC2 ΔmccB::aphA3 amyE::(cat P-Tbox-ubiGmccBA-Sbox-PCac) | BSIP1802→ BSIP1165 |
| BSIP1845 | trpC2 amyE::(cat P-S2box(−236, +1137)Cac-lacZ) | pDIA5759→168 |
| BSIP1858 | trpC2 ΔmccAB::aphA3 amyE::(cat P-Tbox-ubiGmccBACac) | Materials and methods |
| BSIP1863 | trpC2 ΔmccB::aphA3 amyE::(cat P-Tbox-ubiGmccBACac) | BSIP1858 →BSIP1165 |
| BSIP1864 | trpC2 ΔmccA::lacZ-erm amyE::(cat P-Tbox-ubiGmccBACac) | BSIP1858 →BFS2063 |
| BSIP1894 | trpC2 Δrnc::spc | 168 →BG322 |
| BSIP1895 | trpC2 ΔmccB::aphA3 amyE::(cat P-Tbox-ubiGmccBA-Sbox-PCac) ΔrnjB::spc | SSB353→ BSIP1833 |
| BSIP1896 | trpC2 ΔmccB::aphA3 amyE::(cat P-Tbox-ubiGmccBA-Sbox-PCac) Pspac rnjA::erm | SSB353→ BSIP1833 |
| BSIP1898 | trpC2 ΔmccB::aphA3 Δrnc::spc | BSIP1165→ BSIP1894 |
| BSIP1899 | trpC2 ΔmccB::aphA3 amyE::(cat P-Tbox-ubiGmccBA-Sbox-PCac) ΔrnjB::spcPspac rnjA::erm | SSB353→ BSIP1895 |
| BSIP1900 | trpC2 ΔmccB::aphA3 amyE::(cat P-Tbox-ubiGmccBA-Sbox-PCac) Δrnc::spc | BSIP1802→ BSIP1898 |
| BSIP1919 | trpC2 lacA::erm Sbox (−227, +232)Cac | pDIA5786→ 1A785 |
| BSIP1920 | trpC2 lacA::erm AS (−227, +1137)Cac | pDIA5787→ 1A785 |
| BSIP1946 | trpC2 ΔmccAB::aphA3 amyE::(cat P-Tbox-ubiGmccBACac) lacA::erm Sbox (−227, +232)Cac | BSIP1919→ BSIP1858 |
| BSIP1947 | trpC2 ΔmccAB::aphA3 amyE::(cat P-Tbox-ubiGmccBACac) lacA::erm AS (−227, +1137)Cac | BSIP1920→ BSIP1858 |
| SSB353 | trpC2 amyE::(cat thrS'-lacZ) Pspac rnjA::erm ΔrnjB::spc | H. Putzer |
| BG322 | trpC2 thr-5 Δrnc::spc | (75) |
| 1A785 | trpC2 lacA::spc | (76) |
| E. coli | ||
| DH5α | F-φ80 lacZΔM15 ΔlacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk−, mk−) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| C. acetobutylicum | ||
| ATCC824 | Wild type | ATCCg |
| Vectors | markersf reference | |
| pAC6 | plasmid allowing the construction of transcriptional fusions with the lacZ gene integrated at the B. subtilis amyE locus | Ap, Cm (39) |
| pHM2 | plasmid allowing the integration of DNA fragments at the B. subtilis amyE locus | Ap, Cm H. Putzer |
| pAXO1 | plasmid allowing the integration of DNA fragments at the B. subtilis lacA locus. The inserted fragments are placed under the control of a promoter inducible by xylose | Ap, EL (76) |
aThe mccA mutant is nonpolar when we add IPTG in the culture medium. The genes located downstream are expressed under the control of the Pspac promoter inducible by IPTG.
bcat, erm, aphA3 and spc genes encode proteins leading to chloramphenicol, erythromycin, kanamycin and spectinomycin resistance.
cP, promoter.
dCac, C. acetobutylicum.
eThe arrow indicates a construction by transformation.
fAp, Cm and EL correspond to the ampicillin, the chloramphenicol and the erythromycin plus lincomycin resistance markers.
gAmerican Type Culture Collection Manassas, VA, USA.
AS, antisense region; S1box(−236, +349) and S2box(−236, +1137), short and long S-box containing regulatory region.
Plasmid and strain construction
Plasmids from E. coli and chromosomal DNA from B. subtilis were prepared according to standard procedures. Restriction enzymes, DNA polymerase and phage T4 DNA ligase were used as recommended by the manufacturers. DNA fragments were purified from agarose gels with the Nucleospin Extract II kit (Macherey-Nagel, Germany). DNA sequences were determined using the dideoxy-chain termination method with plasmid DNA as template and the Big Dye Terminator v.3.1 kit (Applied Biosystems, USA).
The metB and cac0931 genes (http://bioinfo.hku.hk/GenoList/index.pl?database=acetolist) (38) were renamed mccB and mccA according to their sequence similarities with the genes of B. subtilis, respectively. All primers used in this study are listed in Supplementary Table S1. The S-box region of the ubiGmccBA operon from C. acetobutylicum (−236 to +349 relative to the transcriptional start site of the antisense RNA) and the S-box region with the 800 last bases of the mccA gene (−236 to +1137) were amplified by PCR. We used primers containing a 5′-BamHI or a 3′-EcoRI site (Table S1). These fragments were inserted between the BamHI and EcoRI sites of pAC6 (39) resulting in plasmids pDIA5715 and pDIA5759. The T-box promoter region (−302 to +392 relative to the transcriptional start point of the ubiG operon) was cloned similarly into pAC6 to give plasmid pDIA5720. The plasmids pDIA5715, pDIA5720 and pDIA5759 were linearized by ScaI and used to transform B. subtilis 168 to give strains BSIP1751, BSIP1761 and BSIP1845, respectively (Table 1).
We failed to clone the ubiGmccBA operon from C. acetobutylicum in E. coli. This could be due to a possible toxicity of the MccB protein. The ubiG operon was therefore cloned directly in B. subtilis. Two DNA fragments containing the ubiGmccBA operon with the S-box promoter region (−294 to +3672 relative to the transcriptional start site of the operon), or deleted of the S-box (−294 to +3195) were amplified by PCR using primers containing a 5′-ClaI or a 3′-BamHI site (Supplementary Table S1). These fragments were inserted between the ClaI and BamHI sites of pHM2. The ligation mixtures were then linearized by XhoI and SalI and used to transform strain BSIP1303 to give strains BSIP1802 and BSIP1858, respectively (Table 1). To construct strains expressing the antisense RNA in trans, we cloned the DNA corresponding to the downstream part of the ubiG operon in the vector pAXO1 (Table 1). The sequences corresponding to the S-box (−227 to +232 relative to the antisense transcriptional start site), and the S-box region with the 800 last bases of the mccA gene (−227 to +1137) were amplified by PCR with primers containing a 5′-BamHI or a 3′-SacII site (Supplementary Table S1). These fragments were inserted into pAXO1 resulting in plasmids pDIA5786 and pDIA5787. The inserted fragments of pDIA5786 and pDIA5787 were integrated into the chromosome of 1A785 at the lacA locus by a double crossing-over event to give strain BSIP1919 or BSIP1920. Strain BSIP1858 deleted in the S-box promoter region was subsequently transformed with chromosomal DNA of strain BSIP1919 or BSIP1920 to give strain BSIP1946 or BSIP1947 (Table 1).
Enzyme assays
β-Galactosidase specific activity was measured as described by Miller (40) with crude extracts obtained by lysozyme treatment. Concentration of proteins was determined by the method of Bradford. One unit of β-galactosidase is defined as the amount of enzyme that produces 1 nmol min−1 of O-nitrophenol (ONP) at 28°C. The mean value of at least three independent experiments is presented.
Zymogram was performed to detect homocysteine γ-lyase activity of MccB. Cells were harvested during the exponential growth phase and collected by centrifugation. A 40 μg of unboiled crude extracts were loaded onto a native polyacrylamide gel (12% polyacrylamide gel in Tris–glycine buffer). After electrophoresis, the gel was washed twice for 10 min in 50 ml of water and twice for 10 min in 50 ml of Tris–HCl (50 mM, pH 7.4). The gel was then incubated at 37°C for 2 h in the following solution: 50 mM Tris–HCl (pH 7.4), 10 mM MgCl2, 10 mM homocysteine, 0.5 mM Pb(Ac)2, 5 mM dithiothreitol and 0.4 mM pyridoxal phosphate. H2S formed during the enzymatic reaction precipitated as insoluble PbS allowing the detection of homocysteine γ-lyase activity. The signal was then quantified with the ‘Quantity One’ software (Bio-Rad, USA). We have verified that the signal increase was linear depending on the protein quantity using the purified B. subtilis MccB protein (6).
RNA isolation and northern blot analysis
We extracted total RNA from various exponentially growing B. subtilis strains and the C. acetobutylicum ATCC824 strain as previously described (41). For northern blot analysis, 10 μg or 50 μg of total RNA was separated in a 1.5% denaturing agarose gel containing 2% formaldehyde, and transferred to Hybond-N+ membrane (Amersham Biosciences, UK) in 20× SSC buffer (3 M NaCl, 0.3 M sodium citrate pH 7). Prehybridization was carried out for 2 h at 68°C in 10 ml of prehybridization buffer ULTRAHyb (Ambion, USA). Hybridization was performed overnight at 68°C in the same buffer in the presence of a single strand RNA [α-32P]-labeled probe. The probe was synthesized from a PCR product containing a T7 phage promoter sequence on one of its extremities (see Supplementary Table S1 for the primers). One microgram of this PCR product was used as a matrix for in vitro transcription reaction with phage T7 RNA polymerase, 0.5 mM each ATP, GTP, CTP and 50 μCi of [α-32] UTP using Maxiscript kit (Ambion). The probe was then treated with TURBO DNAse I and purified on ‘Nucaway spin column’ (Ambion). After hybridization, membranes were washed twice for 5 min in 50 ml 2 × SSC 0.1% SDS buffer and twice for 15 min in 50 ml 0.1 × SSC 0.1% SDS buffer. Radioactive signal was detected with a Typhoon (Amersham) and the RNA quantity was estimated with the ‘Image Quant’ software (Amersham Biosciences). Verification of equal loading was achieved by measurement of RNA concentration by absorbance at 260 nm, by direct comparison of rRNA band intensities after staining by ethidium bromide (‘Quantity One’ software quantification) and by hybridization with ptsH and/or gyrA RNA probes as independent controls. The size of the transcripts was estimated by comparison with RNA molecular weight standards (Ambion).
Quantitative RT–PCR
We heated at 65°C for 5 min 5 μg or 2.5 μg of total RNA, extracted from various B. subtilis strains and the C. acetobutylicum ATCC824 strain. After a slow cooling, cDNAs were synthesized for 1 h at 55°C with Superscript III Reverse Transcriptase (Invitrogen, UK), and 1 pmol of strand-specific 3′ oligonucleotide primers: O20 and O26 are complementary to the B. subtilis or C. acetobutylicum gyrA, respectively (Supplementary Table S1). O22 and O23 are complementary to the mccA gene of the ubiGmccBA mRNA or antisense RNA. The reverse transcriptase was inactivated by incubation at 70°C for 15 min. Real-time quantitative PCR was performed twice in a 20 μl reaction volume containing 100 ng or 1 μg of cDNA, 12.75 μl of the SYBR PCR master mix (Applied Biosystems), and 400 nM of gene-specific primers. Primers used are listed in ‘Quantitative RT–PCR’ section in Supplementary Table S1. Amplification and detection were performed as previously described (42). In each sample, the quantity of cDNA for each gene was normalized to the quantity of the gyrA cDNA, which is considered as a stably expressed gene. The relative change in gene expression was recorded as the ratio of normalized target concentrations (ΔΔct) (43).
5′ and 3′ RACE
5′RACE (Rapid amplification of cDNA ends) assays were performed on total RNA extracted from strains BSIP1802 and BSIP1899 using a 5′RACE System kit (Invitrogen). Reverse transcription was performed with the Superscript II enzyme at 42°C. The cDNAs were purified on SNAP column and a polyC tail was added with Terminal deoxynucleotidyl transferase. PCR was then performed with the Triple Master polymerase (Qiagen, Germany) using a primer hybridizing with the polyC tail and either a T-box-specific primer (O5), or an S-box-specific primer (O27) (see Supplementary Table S1). Cycling conditions were as follows: 94°C/2 min; 35 cycles of 94°C/30 s, 55°C/30 s, 72°C/1 min; 72°C 7 min. PCR amplification products purified with Nucleospin extract II PCR Purification kit (Macherey-Nagel) were sequenced.
To identify the 3′-end of the ubiGmccBA transcript, we used RNAs isolated from strain BSIP1802 grown with methionine as sulfur source. For the antisense RNAs 3′-end identification, the 3′RACE experiments were performed using RNAs isolated from strains BSIP1802, BSIP1900 and BSIP1899 (Table 1) grown with glutathione as sulfur source. 3′RACE assays were carried out as previously described (44) with some modifications. A total of 15 μg of total RNA was dephosphorylated 1 h at 50°C with calf intestine alkaline phosphatase (Roche, Germany). Reactions were stopped by phenol/chloroform extraction, followed by ethanol precipitation. RNA was then mixed with 500 pmol of 3′ RNA adapter (5′P-UUCACUGUUCUUAGC-GGCCGCAUGCUC-idT-3′) (Dharmacon Research, USA), heat-denaturated at 95°C for 5 min and then quick-chilled on ice. The adapter was ligated overnight at 17°C with 5 U of T4 RNA ligase (New England Biolabs, USA). After phenol/chloroform extraction and ethanol precipitation, RNA (5 μg) was reverse-transcribed using the Thermoscript Reverse Transcriptase kit (Invitrogen) with 100 pmol of a primer complementary to the RNA adapter (O30) in three subsequent steps of 20 min at 55, 60 and 65°C. Treatment with RNase H was then performed for 20 min at 37°C. One microliter of the product of reverse transcription was used for amplification in the presence of 250 μM of each dNTP, 1 unit of Triple Master polymerase, 25 pmol of primers and 1 × Triple Master polymerase buffer containing 2.5 mM MgCl2. For the ubiGmccBA transcript, an adapter-specific primer (O30) and O28, which hybridizes with mccA were used. For the antisense RNA, the PCR was performed with the adapter-specific primer and O16 (Supplementary Table S1). Cycling conditions were as follows: 95°C/10 min; 35 cycles of 95°C/30 s, 55°C/40 s, 72°C/2 min; 72°C 10 min. Nested PCR was performed in the same conditions on the first PCR products to increase the amplification specificity. We used either an internal mRNA primer (O29) or an internal antisense RNA primer (O3). The PCR products were then purified with the MinElute Reaction Cleanup Kit (Qiagen), cloned using the TOPO TA Cloning Kit (Invitrogen) and sequenced.
RESULTS
Sulfur-dependent regulation of the expression of the T-box and S-box promoter regions of the C. acetobutylicum ubiGmccBA operon in B. subtilis
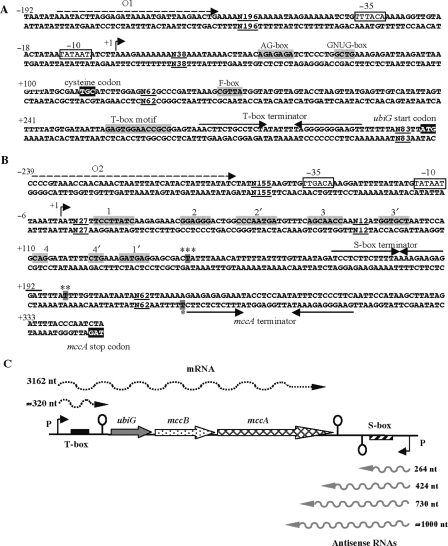
We set out to investigate the regulatory elements involved in the control of the C. acetobutylicum ubiGmccBA operon by first determining its transcriptional start point by 5′RACE analysis. Transcription is initiated 397-bp upstream of the ubiG translational initiation codon. Hexamers −10 (TATAAT) and −35 (TTTACA) showing similarity to a σA consensus sequence and separated by an 18-bp spacer are found upstream of the transcriptional start site (Figure 2A). A T-box antitermination system containing a cysteine specifier codon and all conserved elements characteristic for T-box genes is located downstream from this promoter suggesting a mechanism of premature termination of transcription in response to cysteine availability (Figure 2A). To study the regulation of the ubiG operon, we constructed a transcriptional fusion containing the T-box regulatory region fused to the E. coli lacZ gene. The P-T-box(−302, + 392)-lacZ fusion was introduced at the amyE locus of the B. subtilis strain 168. The resulting strain was grown in minimal medium with various sulfur sources. The β-galactosidase activities obtained are presented in Table 2. The P-T-box-lacZ fusion was slightly regulated in response to sulfur availability. The expression of the fusion was only 1.7-fold lower in the presence of 1 mM cystine than in the presence of 5 mM glutathione or 20 μM cystine (Table 2). However, a small transcript corresponding to premature termination of transcription at the T-box terminator was observed by northern blot using a T-box-specific probe (Supplementary Figure S1). To test more drastic conditions of cysteine starvation, the fusion was introduced into a mutant inactivated for the two major cystine transporters, TcyP and TcyJKLMN (45). The growth of a ΔtcyPΔtcyJKLMN was reduced in the presence of cystine as sulfur source. The expression of the P-T-box-lacZ fusion was not derepressed in a ΔtcyPΔtcyJKLMN mutant even in the presence of 20 μM cystine (Supplementary Figure S1). A cysteine auxotrophic B. subtilis cysE strain inactivated for the serine-acetyltransferase, an enzyme involved in cysteine biosynthesis (46), was used to try to deplete the intracellular pool of cysteine. No derepression of the transcriptional P-T-box-lacZ fusion was observed in this strain, as well as in double and triple mutant strains inactivated in addition for one or two cystine transporters (tcyP and tcyJKLMN) (Supplementary Figure S1). Thus, the expression of the ubiGmccBA operon under the control of the cysteine-specific T-box was hardly responding to sulfur availability. This suggested the existence of additional more efficient control mechanism. We therefore turned our attention to the convergent S-box system immediately downstream from the operon (12).
Figure 2.
The T-box and the S-box regulatory region of the ubiG operon of C. acetobutylicum. (A) T-box regulatory region. (B) S-box regulatory region. The transcriptional start sites ‘+1’ for sense and antisense transcripts are identified by 5′RACE and are indicated by broken arrows. The −10 and −35 regions are boxed. The transcriptional terminators are indicated by arrows. The numbers indicate positions relative to the transcriptional start sites. The regions indicated in gray correspond to the conserved motives of the S-box (1, 1′, 2, 2′, 3, 3′, 4, 4′) and the T-box (T-box motif, AG, GNUG, F-boxes). Oligonucleotides (O1) and (O2), which are indicated by dashed arrows, are used for amplification of the entire ubiG operon region with regulatory sequences. The ubiG start codon, the mccA stop codon and the cysteine-specific codon (TGC) of the T-box system are indicated in black. The 3′-end of the ubiGmccBA transcript indicated by a star was identified by 3′RACE. The 3′-end of the short transcript terminated in the S-box region is indicated by two stars when identified by 3′RACE with RNA extracted from a ▵rnjBpspac-rnjA mutant (BSIP1899) and by three stars when identified by 3′RACE with RNA extracted from the wild-type or a rnc mutant (BSIP1900). (C) Scheme of the ubiG region showing different detected sense (above) and antisense (below) transcripts. The antisense RNAs are indicated by gray arrows, and the mRNA is indicated by black dashed arrows. Their estimated lengths are indicated at right. Broken arrows with ‘P’ symbol indicate the position of the upstream and downstream promoters; loops indicate putative transcriptional terminators for the S-box, T-box and for the 3′-end of the ubiG operon.
Table 2.
Effect of the sulfur source on the expression of P-T-box-lacZ and P-S-box-lacZ transcriptional fusions
 |
ND: non-determined,  :T-box;
:T-box;  :S-box;
:S-box;  :mccA gene;
:mccA gene;  : transcription terminator;
: transcription terminator;  : lacZ gene: P: promoter. Sulfur sources were added to culture medium to the following concentrations: glutathione, 5 mM; methionine, 1 mM; sulfate, 1 mM; homocysteine.
: lacZ gene: P: promoter. Sulfur sources were added to culture medium to the following concentrations: glutathione, 5 mM; methionine, 1 mM; sulfate, 1 mM; homocysteine.
a1mM Cystine.
b20 μM Cystine. β-galactosidase activities are presented in nmol ONP/ min−1/ mg of protein−1.
To demonstrate the presence of a functional promoter linked to the S-box system, we performed a 5′RACE analysis. A single start site was identified 3426-bp downstream from the ubiGmccBA mRNA transcriptional start site, on the complementary strand. This downstream promoter perfectly matches to the σA consensus sequences with the −10 (TATAAT) and −35 (TTGACA) boxes separated by 16 bp (Figure 2B). Between this promoter and the end of the mccA gene, a S-box conserved motif followed by a terminator is present. This prompted us to test whether this downstream element was regulated by methionine. Two lacZ fusions corresponding either to the S-box and its promoter [PS1box(−236, + 349)-lacZ], or the S-box, its promoter, and the 800 last bp of the mccA gene [PS2box(−236, + 1137)-lacZ] were constructed and introduced into B. subtilis as described above. The PS1 box(−236, + 349)-lacZ fusion was highly expressed in the presence of cystine, sulfate or glutathione but 9-fold repressed when the strain was grown with methionine (Table 2). In the presence of homocysteine, the expression was 2-fold reduced when compared to that obtained with glutathione. These results indicated that the S-box from C. acetobutylicum is functional in B. subtilis. However, the expression of the second fusion PS2box (−236, + 1137)-lacZ containing sequences complementary to the 3′-end of mccA (800 last bases) was low and not regulated by the sulfur source (Table 2). This suggested that the transcription from the S-box-dependent promoter could stop before the fusion point to lacZ (i.e. before the mccA translational start site). Alternatively, a long untranslated region upstream of lacZ could destabilize the whole RNA molecule in this construction.
Complementation of the B. subtilis mccA and mccB mutants by the C. acetobutylicum mccBA genes
Since only few genetic tools are available in clostridia, we first studied the regulation of the expression of the C. acetobutylicum ubiG operon in B. subtilis. We constructed B. subtilis strains containing the complete C. acetobutylicum ubiGmccBA operon (BSIP1802), or the operon deleted of the S-box promoter region (BSIP1858). The operon was directly cloned in the B. subtilis strain BSIP1303 that contains a deletion of the mccAB genes (see Materials and methods section and Table 1). This ΔmccAB mutant cannot grow in the presence of methionine as sole sulfur source, due to the absence of the cystathionine β-synthase, MccA (6). The MccA and MccB proteins of C. acetobutylicum shared 52 and 60% identity with MccA and MccB from B. subtilis, respectively. Assuming that the mccBA genes of C. acetobutylicum can complement a mccAB mutant of B. subtilis, we selected transformants that restored the growth of strain BSIP1303 in the presence of methionine as sole sulfur source.
To confirm the function of MccA and MccB from C. acetobutylicum, we complemented either the mccA or the mccB mutant from B. subtilis. The DNA of BSIP1802 or BSIP1858 was therefore introduced by transformation into a B. subtilis mccA mutant (BFS2063). The resulting strains, BSIP1832 and BSIP1864, were grown in minimal medium in the presence of methionine and IPTG (this allows the expression of mccB). The growth of the B. subtilis mccA mutant was restored in the strains containing the C. acetobutylicum mccBA genes integrated at the amyE locus (data not shown). This strongly suggested that the C. acetobutylicum MccA protein was a cystathionine β-synthase that was functional in B. subtilis.
A B. subtilis ΔmccB mutant can grow in the presence of methionine, and previous results indicated that there was a second uncharacterized cystathionine γ-lyase in this organism (6). As a consequence, we could not use this enzymatic activity to specifically detect MccB. We therefore measured the MccB-specific homocysteine γ-lyase activity. Crude extracts of strains 168, BSIP1165, BSIP1833 and BSIP1863 grown in the presence of methionine were prepared. The release of sulfide from homocysteine due to homocysteine γ-lyase activity was directly detected by the precipitation of insoluble PbS in native gels. As expected, a single band was detected in the wild-type strain. This signal disappeared in the ΔmccB mutant (BSIP1165) (data not shown). The integration of the C. acetobutylicum ubiGmccBA operon at the amyE locus of the B. subtilis ΔmccB mutant restored the detection of homocysteine γ-lyase activity. This indicated that MccB from C. acetobutylicum had homocysteine γ-lyase activity in vitro.
Modulation of MccB synthesis in response to sulfur availability
The regulation of the C. acetobutylicum MccB synthesis in response to sulfur availability was first studied in the B. subtilis strain BSIP1802 containing the C. acetobutylicum ubiGmccBA operon with the two convergent promoters. This strain was grown in minimal medium in the presence of glutathione, methionine, cystine, or sulfate as sole sulfur sources. The homocysteine γ-lyase activity was tested by zymogram. The detected activity was high when the strain BSIP1802 was grown with methionine, and 15-, 10- and 50-fold reduced in the presence of glutathione, sulfate or cystine, respectively (Figure 3A). The effect of the deletion of the downstream promoter and the S-box motif (BSIP1858) on MccB synthesis was further tested. The homocysteine γ-lyase activity detected in strain BSIP1858 was the same in the presence of all these sulfur sources (Figure 3B). These results indicated that the MccB synthesis is repressed by glutathione, sulfate or cystine only when the downstream promoter region is present. The modulation of MccB activity was not correlated with the variation of expression of the P-T-box (−302, +392)-lacZ fusion indicating the existence of another mechanism of regulation. The requirement of a downstream convergent promoter suggested that an antisense RNA could participate in the regulation of the ubiG operon.
Figure 3.
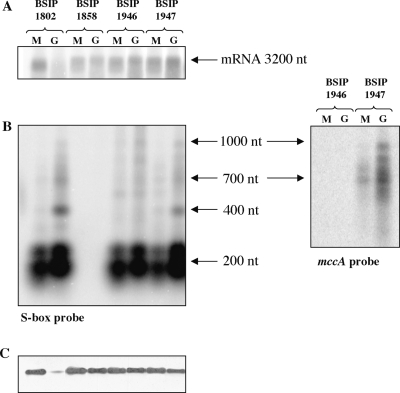
Modulation of MccB synthesis in response to sulfur availability. The homocysteine γ-lyase activity of MccB is detected on zymogram. A total of 40 μg of crude extracts were charged on a native polyacrylamide gel (12%). The release of sulfide from homocysteine due to homocysteine γ-lyase activity was directly detected by the precipitation of insoluble PbS (see Materials and methods section). The proteins were extracted from B. subtilis strains BSIP1802 (P-Tbox-ubiGmccBA-Sbox-P) (A), BSIP1858 (P-Tbox-ubiGmccBA) (B) or C. acetobutylicum ATCC824 strain (C); strains were grown in the presence of 5 mM glutathione (G), 1 mM methionine (M), 1 mM K2SO4 (S) or 1 mM cystine or cysteine (C). These results are representative of at least two independent experiments.
Detection of antisense RNAs and characterization of their 3′-end
To determine whether a stable antisense RNA could be synthesized from the S-box promoter, we performed northern blot experiments using strand-specific RNA probes. One probe hybridized with the 130 first bases of the RNA transcribed from the downstream promoter (+1 to +130 relative to the transcriptional start point), and the second corresponded to the mccA gene (+662 to +446 relative to the mccA translation start point) (Figures 2 and 4; Supplementary Table S1). The B. subtilis strains BSIP1802 and BSIP1858 were grown in minimal medium in the presence of methionine, conditions where the PS1box-lacZ fusion is highly repressed, or glutathione, conditions where the expression of the P-T-box-lacZ fusion seems to be slightly increased. After extraction, RNAs were analyzed by Northern blot. We detected transcripts of about 200 (double band), 400, 700 and 1000 nt with the probe hybridizing with the S-box region (Figures 2, 4C and Supplementary Figure S2). Longer exposure time was required for better detection of the 400, 700 and 1000 nt transcripts. The smallest transcript detected (approximately 200 nt) might correspond to premature termination of transcription at the S-box terminator. For the mccA probe, only the 700 and 1000 nt transcripts were detected according to the probe position (Figure 4B). In contrast, no transcript was detected with the same probes in strain BSIP1858 (Figure 4E). These results showed that several antisense transcripts were synthesized from the downstream promoter and that the deletion of this promoter abolished their synthesis.
Figure 4.
Northern blot analysis of sense and antisense RNAs. The RNAs were extracted from the strains BSIP1802 (P-Tbox-ubiGmccBA-Sbox-P) (A–C) or BSIP1858 (P-Tbox-ubiGmccBA) (D and E) grown with 1 mM methionine (M) or 5 mM glutathione (G). The hybridizations were performed with single strand RNA probes. The positions of the probes are indicated by arrows on the corresponding genomic region scheme. Broken arrows with ‘P’ symbol indicate the position of the upstream and downstream promoters; loops indicate putative transcriptional terminators for the S-box, T-box and for the 3′-end of the ubiG operon. (A and D) The ubiG probe; (B and E) mccA probe; (C) S-box probe. The main transcripts observed and their sizes are indicated by arrows at the right of the northern blot figure. Longer exposure time was required for better detection of the 400, 700 and 1000 nt transcripts. The RNA size is determined with single strand RNA size markers ‘RNA millennium’ and ‘RNA century-Plus’ (Ambion). These results are representative of at least three independent experiments.
To further characterize these transcripts and to determine the possible overlap between the ubiGmccBA mRNA and the antisense RNAs, their 3′-ends were determined. 3′RACE analysis was performed by the ligation of an adapter to the 3′-hydroxyl group of RNAs, followed by adapter- and sense- or antisense-specific amplifications. The size of the ubiGmccBA mRNA was determined to be 3162 bases. The stop of transcription occurred at a T-nucleotide at the end of an inverted repeat, which corresponds to a predicted terminator located downstream from mccA (Figure 2B). For the antisense RNAs, the sequencing of about 40 independent clones identified several 3′-ends. The most abundant 3′-RACE fragments placed the 3′-ends at position +264, +424 and +730 from the transcriptional start site of the downstream promoter (Supplementary Figure S2). Additional minor sites were located at position +324, +347, +618, +678 and +808. Interestingly, the same sites were identified for antisense RNAs isolated from a host strain mutated in the rnc gene encoding the double strand-specific RNase III (see below Results section for the effects of the rnc gene mutation). The length of antisense RNAs corresponding to the major termination stop sites is in agreement with the approximate sizes of these RNAs estimated by northern blots (Figure 4C). No stem and loop structures are predicted at position +264, +424 and +730 suggesting that transcription could stop independently from classical terminators. For the small transcripts corresponding to the S-box region, the major 3′-end of the transcript corresponded to position +140 from the transcriptional start site both in the wild-type strain and in a mutant inactivated for the RNase III. Surprisingly, this 3′-end is located upstream of the S-box terminator (Figure 2B) suggesting a cleavage of the transcript. In a strain inactivated for the RNases J1 and J2, we identified a 3′ extremity located at position +198 downstream from the S-box terminator (Figure 2B). These RNases are known to cleave the RNA of the thrS leader region between the T-box motif and the terminator (47). Our results suggested that a cleavage between the S-box motif and its terminator could also be possible.
Inverse regulation of the expression of the ubiGmccBA mRNA and antisense RNAs
We then compared the level of antisense RNAs detected in the presence of methionine or glutathione. The short transcript of about 200 nt probably terminated at the S-box terminator was present in both growth conditions as observed for the B. subtilis metIC operon (48). The expression of the longer read-through transcripts was high in the presence of glutathione and reduced in the presence of methionine. This showed that the repression of antisense RNA synthesis in the presence of methionine was mediated by premature termination of transcription at the S-box motif. To further quantify the variation of the antisense RNA abundance as a function of the sulfur source, we performed quantitative RT–PCR analysis on RNA extracted from the B. subtilis strains BSIP1802 (containing the complete C. acetobutylicum ubiG operon) and BSIP1858 (containing the operon deleted of the S-box promoter region). We used two oligonucleotides located at the end of mccA. The expression of antisense RNAs in strain BSIP1802 was high in the presence of glutathione or cystine and 26- or 21-fold reduced in the presence of methionine. In contrast, the antisense RNAs were not detectable in strain BSIP1858 (Table 3). This confirmed the results obtained by northern blots (Figure 4B, C and E).
Table 3.
Quantification of the mRNA and the antisense RNAs by quantitative RT–PCR
|
mRNA |
Antisense RNA |
|||
|---|---|---|---|---|
| Met/GR | Met/Cys | Met/GR | Met/Cys | |
| BSIP1802 | 4.173 ± 0.049 | 6.078 ± 2.15 | 0.038 ± 0.004 | 0.047 ± 0.007 |
| BSIP1858 | 0.901 ± 0.08 | 1.365 ± 0.07 | ND | ND |
| BSIP1946 | 0.805 ± 0.13 | 1.525 ± 0.233 | ND | ND |
| BSIP1947 | 0.69 ± 0.08 | 1.125 ± 0.346 | 0.039 ± 0.01 | 0.034 ± 0.001 |
| ATCC824 | – | 5.169 ± 0.178 | – | 0.166 ± 0.008 |
The quantification was carried out by quantitative RT–PCR on total RNA extracted from B. subtilis strains BSIP1802 (P-Tbox-ubiGmccBA-Sbox-P), BSIP1858 (P-Tbox-ubiGmccBA), BSIP1946 (P-Tbox-ubiGmccBA lacA::erm Sbox(−227,+232)), BSIP1947 [P-Tbox-ubiGmccBA lacA::erm AS(−227,+1137)] and C. acetobutylicum ATCC824 strain (wild type) after culture with methionine (Met), glutathione (GR) or cystine (Cys). Values presented are the ratio obtained from the quantity detected with methionine relative to the quantity detected with glutathione (Met/GR) or cystine (Met/Cys) and normalized to gyrA expression. ‘ND’, non-detectable; the limit of detection is estimated at < 10−15g. ‘−’, not determined.
To study the impact of the presence of antisense RNAs on the quantity of the ubiG mRNA, we performed northern blot experiments on RNA extracted from strains BSIP1802 and BSIP1858 grown in minimal medium with methionine or glutathione. To specifically detect the ubiG mRNA, we used a single strand RNA probe that hybridized with ubiG (+388 to +648 relative to the ubiG transcriptional start point) (Supplementary Table S1, Figure 4). As expected for the ubiG operon, a transcript of about 3200 nt was detected in both strains (Figure 4A and D) in accordance with the size determined by 5′ and 3′RACE experiments (3162 nt). The quantity of the ubiG transcript detected in strain BSIP1802 in northern blot was markedly higher in the presence of methionine than in the presence of glutathione (Figure 4A). Using quantitative RT–PCR, we showed that the level of the ubiGmccBA mRNA in strain BSIP1802 was 4-fold reduced in the presence of glutathione and 6-fold reduced in the presence of cystine as compared to that obtained after growth with methionine (Table 3). In contrast, no variation of the ubiGmccBA mRNA quantity was observed in strain BSIP1858 under the same conditions (Figure 4D and Table 3). This indicated that the expression of the C. acetobutylicum ubiG operon becomes independent of the sulfur source when the S-box promoter region is deleted in agreement with the measurements of MccB activity obtained by zymogram (Figure 3A and B). This demonstrated the existence of an inverse correlation between the sense and antisense transcript expression patterns.
Effect of antisense RNA synthesis in trans on the control of ubiGmccBA expression
To investigate the molecular mechanisms implicated in antisense-mediated control of the ubiG operon, we first tested the possible role of specific RNases in this regulatory system. Our 3′RACE results showed a likely overlap of up to 700 nt between the transcript and the antisense RNAs (Figure 2C). Indeed, if a sense/antisense duplex formation promotes degradation of these RNAs, the involvement of some double strand-specific RNases may be proposed. RNase III, or RNases shown to cleave RNA at a position adjacent to double strand regions, i.e. B. subtilis RNaseJ1 and J2, are good candidates for this degradation. However, by northern blot as well as by zymogram analysis, we observed no major effects of inactivation of these RNases on the regulation of the ubiG operon (data not shown).
In most chromosomal regulation systems involving antisense RNAs, the target and regulatory RNA-encoding genes are located at different loci in the chromosome. In contrast, only cis-located elements control the ubiG operon of C. acetobutylicum. To address the question whether the antisense RNA interferes with transcription of the ubiG operon in cis or in trans, the effect of the expression of the antisense RNA in trans on the ubiGmccBA mRNA abundance was tested in B. subtilis. We first cloned the sequences corresponding to the S-box region (−227 to +232) and the longest detected antisense RNA of about 1000 nt (−227 to +1137) in the vector pAXO1. These constructions were then inserted into the lacA locus of strain BSIP1858 (deleted of the downstream promoter and the S-box motif) giving strains BSIP1946 and BSIP1947 (Table 1). We subsequently performed northern blot and quantitative RT–PCR experiments using RNA extracted from strains BSIP1802, BSIP1858, BSIP1946 and BSIP1947 after growth in the presence of methionine, glutathione or cystine. When expressed in cis (BSIP1802) or in trans (BSIP1947), the same antisense RNA molecules in similar amounts were detected and the expression pattern in response to sulfur availability was conserved (Figure 5B, Table 3). Indeed, the expression of antisense RNAs was high in the presence of glutathione or cystine and 25- or 29-fold reduced in the presence of methionine. As expected, no antisense RNAs were detected for strain BSIP1858 while only short transcripts of about 200 nt were observed for strain BSIP1946 (Figure 5B, Table 3). Furthermore, the ubiGmccBA transcript was synthesized at similar levels in the presence of methionine or glutathione in strain BSIP1858, in strain BSIP1946 expressing the short S-box transcript and in strain BSIP1947 expressing the antisense RNA in trans (Figure 5A, Table 3). This indicated that the expression of the antisense RNA in trans is unable to restore the sulfur-dependent regulation observed in BSIP1802. Accordingly, the MccB homocysteine γ-lyase activity observed in zymogram was reduced in the presence of glutathione only in strain BSIP1802 (Figure 5C). Altogether these results strongly suggested a mechanism of action for the antisense RNAs in cis.
Figure 5.
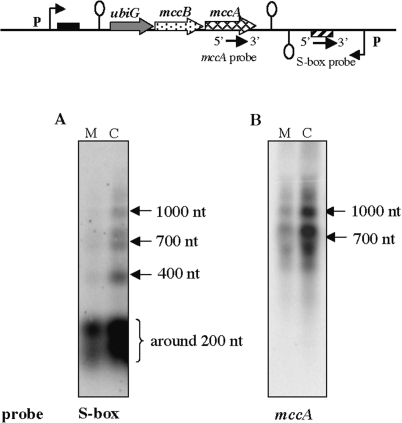
Effect of the expression of the antisense RNA in trans on the ubiGmccBA regulation. Northern blot analysis was carried out on total RNA extracted from BSIP1802 (P-Tbox-ubiGmccBA-Sbox-P), BSIP1858 (P-Tbox-ubiGmccBA), BSIP1946 [P-Tbox-ubiGmccBA lacA::erm Sbox(−227, + 232)] or BSIP1947 [P-Tbox-ubiGmccBA lacA::erm AS(−227, + 1137)], after growth with 1 mM methionine (M) or 5 mM glutathione (G). (A) The mRNA expression was detected with the probe hybridizing with the ubiG gene. (B) The antisense RNAs were detected with the probe complementary to the S-box region. On the right of this figure, detection with a mccA probe is also shown for BSIP1946 and BSIP1947. The arrows show the different antisense RNAs detected with their estimated size. (C) The MccB homocysteine γ-lyase activity was detected on zymogram. These results are representative of at least two independent experiments.
Sulfur-dependent regulation of the ubiG operon in C. acetobutylicum
To confirm the expression of antisense RNA species in C. acetobutylicum, we performed northern blot and quantitative RT–PCR on RNAs extracted from the original host. The C. acetobutylicum ATCC824 strain was grown in synthetic medium in the presence of methionine or cysteine. Using the same probes as for B. subtilis experiments, antisense RNAs of about 200, 400, 700 and 1000-nt long were detected by northern blot (Figure 6A). This corresponds to the estimated length of the antisense RNAs observed in B. subtilis (Figure 4C). We further compared the sulfur-dependent regulation of the ubiG and antisense RNAs in C. acetobutylicum. The quantity of the antisense RNAs detected by Northern blot was higher in the presence of cysteine than in the presence of methionine (Figure 6A and B). Using quantitative RT–PCR, we showed that the level of the antisense RNAs was 6-fold reduced in ATCC824 strain grown in the presence of methionine, as compared with cysteine (Table 3). We also showed by quantitative RT–PCR that the ubiG operon expression was 5-fold reduced in the presence of cysteine as compared to methionine (Table 3). Thus, as in B. subtilis, we observed inverse correlation between the expression patterns of sense and antisense transcripts depending on sulfur source availability. Moreover, the homocysteine γ-lyase activity of MccB on zymogram was 71-fold reduced in the presence of cysteine (Figure 3C). All these data are in agreement with the results obtained in B. subtilis, and strongly suggest that the sulfur-dependent control of the ubiG operon in C. acetobutylicum is similar to that observed in B. subtilis.
Figure 6.
Expression of the antisense RNAs in C. acetobutylicum. Northern blot analysis was carried out on total RNA extracted from strain ATCC824 after growth with 1 mM methionine (M) or 1 mM cysteine (C). The antisense RNAs were detected with the probe hybridizing with the S-box motif (A) or the mccA gene (B). The positions of the probes are indicated by arrows on the corresponding genomic region scheme. Broken arrows with ‘P’ symbol indicate the position of the upstream and downstream promoters; loops indicate putative transcriptional terminators. The arrows show the different antisense RNAs detected with their estimated size. The RNA size is determined with single strand RNA size markers ‘RNA millennium’ (Ambion). These results are representative of at least two independent experiments.
DISCUSSION
In this study, we have analyzed a complex RNA-mediated regulatory system involving an S-box riboswitch and antisense RNAs that governs the expression of the C. acetobutylicum ubiG operon. Even though most of the studies were carried out in the heterologous host B. subtilis the major aspects of this control could be confirmed in the original host, C. acetobutylicum validating the experimental approach used here. By transcriptional fusion analysis, we observed a weak regulatory response of the cysteine-specific T-box antitermination system located downstream from the promoter of this operon even in drastic conditions of cysteine starvation (Table 2). However, we demonstrated premature termination of transcription at the T-box terminator by northern blot analysis strongly suggesting the functional capability of the T-box (Figure S1). We cannot exclude that the T-box system may poorly function in the heterologous host B. subtilis but may have been conserved in C. acetobutylicum to ensure a secondary regulatory function under particular conditions. Another possibility could be that the T-box of ubiG in C. acetobutylicum has partially lost its responsiveness to cysteine due to a recent recruitment of a more efficient control system involving an antisense RNA. Indeed, transcription of antisense RNAs from a downstream promoter attenuated by an S-box riboswitch plays a major role in the sulfur-dependent regulation of the ubiG operon. The deletion of the downstream promoter region completely abolished this regulation (Figure 4D). In the presence of methionine, glutathione or cystine, the abundance of the ubiG transcript was inversely correlated to that of antisense transcripts as shown by northern blot experiments and quantitative RT–PCR analyses in B. subtilis and C. acetobutylicum (Figure 4A–C, Table 3). In the presence of methionine, premature termination of transcription at the S-box riboswitch (Table 2, Figures 4C and 6A) leads to a low level of antisense RNAs and concomitant increase in ubiG mRNA encoding the enzymes required for methionine to cysteine conversion (Figure 4A, Table 3). Accordingly, the homocysteine γ-lyase activity of MccB was high in the zymogram assay under this condition (Figure 3). In the presence of cystine or glutathione, which probably trigger methionine starvation, transcription from the downstream promoter proceeds through the S-box terminator due to the increased formation of the antiterminator structure in the absence of AdoMet (Table 2, Figures 4B, C and 6). Under these conditions, antisense RNAs overlapping the ubiG mRNA were synthesized maintaining the quantity of the ubiGmccBA mRNA and the MccB activity at a low level (Figure 3 and 4A, Table 3). In the presence of cystine, a weak additional T-box-dependent premature termination of transcription could occur, as observed by northern blot analysis with a T-box-specific probe in both B. subtilis and C. acetobutylicum (Figure S1). This might be responsible for the slightly increased factor of regulation observed when we compared methionine to cystine instead of methionine to glutathione conditions (Table 3). This complex mechanism allows the expression of the ubiG operon in the presence of methionine and its downregulation in the presence of cystine or glutathione. This perfectly fits to the physiological function of the ubiGmccBA operon that is involved in methionine to cysteine conversion as shown by its ability to complement the B. subtilis mccA and mccB mutants.
Several antisense RNA species were detected in the presence of glutathione or cystine. By northern blot analysis with strand-specific riboprobes and by subsequent 5′/3′ RACE experiments, we have identified major antisense RNAs of about 260, 400, 700 and 1000 nt in length (Figures 4, 6 and S2). We did not find Rho-independent terminators in these regions in antisense orientation. Interestingly, multiple RNA species have been frequently observed for antisense RNAs both in bacteria and eukaryotes. They correspond either to alternative transcription initiation or termination sites, to alternative splicing species, to processed forms or may result from transcriptional interference (23,26,28,34,44,49–53). For the ubiG system, the pattern of antisense molecules remained the same in mutants inactivated for RNase III or RNases J1/J2 (data not shown), suggesting that the shorter antisense RNAs do not represent cleavage products by these cellular RNAses. The various antisense RNAs could correspond to several preferential sites of transcription stops or pauses in the elongation of the transcriptional machinery.
The identification of 3′-ends for the ubiGmccBA mRNA and the antisense RNAs shows the existence of a large region of overlapping between these sense and antisense transcripts (up to 700 nt) (Figure 2C), leading to the possible formation of an RNA–RNA duplex. In both eukaryotes and prokaryotes, it is proposed that the antisense RNA action might be mainly mediated through the activation of double-stranded RNA-dependent pathways (24,29,31,54). To date, nearly all known cis-encoded antisense RNAs on bacterial chromosomes seem to affect either translation or mRNA stability, although the detailed mechanisms remain to be elucidated (34). In our case, an antisense-mediated inhibition of translation of the genes of the ubiG operon seems unlikely. By northern blot and 3′RACE analysis, we identified only antisense RNA species terminated within the mccA coding sequence (Figures 2C, 4B, C and 6). In addition, no significant effect was observed when antisense transcripts are synthesized in trans (Figure 5). Finally, we showed that neither RNase III [an endoribonuclease specific for double-stranded RNA (34,53,55–57)] nor RNases J1/J2 sharing functional analogies with E. coli RNAse E (47,58,59) have major roles in the antisense-dependent regulation of the ubiG operon (data not shown). All our results are rather in favor of another mechanism like transcriptional interference allowing antisense RNA action in cis.
A recent report has proposed that there is a significant correlation between the regulatory mechanism of antisense function and the relative expression pattern of antisense transcripts and their sense targets (28). For example, double-stranded RNA-dependent mechanisms require coexpression with their target, whereas transcriptional interference rather implicates mutual exclusion of sense and antisense transcripts (24). We observed an inverse correlation in the abundance of sense and antisense transcripts for the C. acetobutylicum ubiG system (Figure 4A–C, Table 3). Moreover, the antisense RNAs were able to control their target only in cis, since they have no important effect when expressed in equal amounts in trans (Figure 5, Table 3). Altogether, these data suggest the implication of a transcriptional interference mechanism, which refers to a direct negative influence in cis of one transcriptional process on a second (60,61). In this case, the presence of overlapping transcriptional units may lead to collisions of the two RNA polymerase complexes on opposite strands. Depending on the strength of convergent promoters, this might result in anti-correlated expression levels of sense and antisense or in repression of both transcripts (61). We compared the strength of the ubiG promoter and of the antisense promoter in both C. acetobutylicum and B. subtilis. We first quantified the transcripts initiated from the sense and antisense promoters by northern blot analysis with probes complementary to T-box region (for sense) and S-box region (for antisense). By this approach, we estimated that the antisense promoter is about 3-fold stronger that the sense one. In addition, these results were confirmed by quantitative RT–PCR analysis using oligonucleotides within T-box or S-box regions (data not shown). However, examples of proved transcriptional collision are still rare (24,52,62). One recent study in Saccharomyces cerevisiae demonstrated the cis-antisense-mediated regulation of IME4, a gene required for initiation of meiosis, by a transcriptional interference mechanism with stronger antisense and weaker sense promoters (63). This mechanism has been also proposed for inversely expressed genes at imprinted loci in mouse (64). Another possibility of transcriptional interference could involve the accumulation of positive supercoils ahead of the transcription machinery from efficient S-box riboswitch-dependent promoter, changing the conformation of the ubiG promoter into an inactive form. In bacterial and yeast chromosomes, transcription-induced positive supercoils are able to diffuse and affect the transcriptional process several kilobases away from the site of origin (65–67). Further studies will be required to determine the exact nature of the molecular mechanism involved in the cis-acting antisense control of the C. acetobutylicum ubiG operon.
Previous searches of regulatory RNAs in bacteria have focused on trans-encoded chromosomal small RNAs from intergenic regions (30,68,69). The number and importance of cis-encoded antisense RNA regulators are probably underestimated. In Gram-positive bacteria, few studies on regulatory RNAs have been performed (69–72). Only one example of cis-antisense RNA has been described in B. subtilis (53) while an antisense RNA has also been involved in the control of glutamate metabolism in C. acetobutylicum (73,74). The ubiG system, which provides an example of a new unexpected mechanism for the control by cis-antisense RNAs in bacteria (34), may serve as a model for antisense-mediated regulation of gene expression in cis both in prokaryotes and in eukaryotes. The complex regulatory system identified here combines several RNA-based mechanisms: two transcriptional antitermination systems, the S-box riboswitch and cysteine-specific T-box, and an antisense RNA transcription. To our knowledge, it is the first report of large cis-encoded chromosomal antisense RNAs (up to 1000 nt in length) participating in the control of important metabolic processes in bacteria. Clostridia representing an evolutionary ancient group of bacteria may have developed a complex RNA-based regulatory mechanism to tightly control methionine to cysteine conversion instead of a transcriptional repressor as found in B. subtilis (6).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
‘Centre National de la Recherche Scientifique’ (URA2171); the ‘Institut Pasteur’; ‘Ministère de l’Education Nationale de la Recherche et de la Technologie’ PhD fellowship (to G.A.). Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
Supplementary Material
ACKOWLEDGEMENTS
We are grateful to Bruno Dupuy and Stéphanie Raffestin for helpful discussions. We thank M. Gelfand and D. Rodionov for pointing out this original system and for helpful discussions at the beginning of this work. I.M.-V. and O.S. are associate and assistant professors at the ‘Université Paris 7’, respectively.
REFERENCES
- 1.Guédon E, Martin-Verstraete I. In: Amino Acid Biosynthesis-pathways, Regulation and Metabolic Engineering. Wendisch VF, editor. Germany: Springer; 2007. pp. 195–218. [Google Scholar]
- 2.Schelle MW, Bertozzi CR. Sulfate metabolism in mycobacteria. Chembiochem. 2006;7:1516–1524. doi: 10.1002/cbic.200600224. [DOI] [PubMed] [Google Scholar]
- 3.Sekowska A, Kung HF, Danchin A. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2000;2:145–177. [PubMed] [Google Scholar]
- 4.Greene RC. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. Neidhardt FC, editor. Washington, DC: ASM Press; 1996. pp. 542–560. [Google Scholar]
- 5.Ludwig ML, Matthews RG. Structure-based perspectives on B12-dependent enzymes. Annu. Rev. Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 6.Hullo MF, Auger S, Soutourina O, Barzu O, Yvon M, Danchin A, Martin-Verstraete I. The conversion of methionine to cysteine in Bacillus subtilis and its regulation. J. Bacteriol. 2007;189:187–197. doi: 10.1128/JB.01273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vermeij P, Kertesz MA. Pathways of assimilative sulfur metabolism in Pseudomonas putida. J. Bacteriol. 1999;181:5833–5837. doi: 10.1128/jb.181.18.5833-5837.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi SY, Reyes D, Leelakriangsak M, Zuber P. The global regulator Spx functions in the control of organosulfur metabolism in Bacillus subtilis. J. Bacteriol. 2006;188:5741–5751. doi: 10.1128/JB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Even S, Burguière P, Auger S, Soutourina O, Danchin A, Martin-Verstraete I. Global control of cysteine metabolism by CymR in Bacillus subtilis. J. Bacteriol. 2006;188:2184–2197. doi: 10.1128/JB.188.6.2184-2197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolling J, Breton G, Omelchenko MV, Makarova KS, Zeng QD, Gibson R, Lee HM, Dubois J, Qiu DY, Hitti J, et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 2001;183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004;32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundy FJ, Henkin TM. The T box and S box transcription termination control systems. Front Biosci. 2003;8:20–31. doi: 10.2741/908. [DOI] [PubMed] [Google Scholar]
- 14.Grundy FJ, Moir TR, Haldeman MT, Henkin TM. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res. 2002;30:1646–1655. doi: 10.1093/nar/30.7.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Putzer H, Gendron N, Grunberg-Manago M. Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 1992;11:3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in Bacillus subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 18.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol. Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 19.Epshtein V, Mironov AS, Nudler E. The riboswitch-medited control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl Acad. Sci. USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 24.Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G. In search of antisense. Trends Biochem. Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Silby MW, Levy SB. Overlapping protein-encoding genes in Pseudomonas fluorescens Pf0-1. PLoS Genet. 2008;4:e1000094. doi: 10.1371/journal.pgen.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 27.Wagner EG, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 28.Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 2007;10:257–261. doi: 10.1016/j.mib.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Weaver KE. Emerging plasmid-encoded antisense RNA regulated systems. Curr. Opin. Microbiol. 2007;10:110–116. doi: 10.1016/j.mib.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Vasconcelos I, Girbal L, Soucaille P. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J. Bacteriol. 1994;176:1443–1450. doi: 10.1128/jb.176.5.1443-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang G, Ho C, Qiu Y, Cubas V, Yu Z, Cabau C, Cheung F, Moszer I, Danchin A. Specialized microbial databases for inductive exploration of microbial genome sequences. BMC Genomics. 2005;6:14. doi: 10.1186/1471-2164-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stülke J, Martin-Verstraete I, Zagorec M, Rose M, Klier A, Rapoport G. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 1997;25:65–78. doi: 10.1046/j.1365-2958.1997.4351797.x. [DOI] [PubMed] [Google Scholar]
- 40.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 41.Guillouard I, Auger S, Hullo MF, Chetouani F, Danchin A, Martin-Verstraete I. Identification of Bacillus subtilis CysL, a regulator of the cysJI operon, which encodes sulfite reductase. J. Bacteriol. 2002;184:4681–4689. doi: 10.1128/JB.184.17.4681-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milohanic E, Glaser P, Coppee JY, Frangeul L, Vega Y, Vazquez-Boland JA, Kunst F, Cossart P, Buchrieser C. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 2003;47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EGH, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 45.Burguiere P, Auger S, Hullo MF, Danchin A, Martin-Verstraete I. Three different systems participate in L-cystine uptake in Bacillus subtilis. J. Bacteriol. 2004;186:4875–4884. doi: 10.1128/JB.186.15.4875-4884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dedonder RA, Lepesant JA, Lepesant-Kejzlarova J, Billault A, Steinmetz M, Kunst F. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl. Environ. Microbiol. 1977;33:989–993. doi: 10.1128/aem.33.4.989-993.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auger S, Huen WH, Danchin A, Martin-Verstraete I. The metIC operon involved in methionine biosynthesis in Bacillus subtilis is controlled by transcription antitermination. Microbiology. 2002;148:507–518. doi: 10.1099/00221287-148-2-507. [DOI] [PubMed] [Google Scholar]
- 49.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 50.Enerly E, Sheng Z, Li KB. Natural antisense as potential regulator of alternative initiation, splicing and termination. In Silico Biol. 2005;5:367–377. [PubMed] [Google Scholar]
- 51.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osato N, Suzuki Y, Ikeo K, Gojobori T. Transcriptional interferences in cis natural antisense transcripts of humans and mice. Genetics. 2007;176:1299–1306. doi: 10.1534/genetics.106.069484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silvaggi JM, Perkins JB, Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Storz G, Opdyke JA, Wassarman KM. Regulating bacterial transcription with small RNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:269–273. doi: 10.1101/sqb.2006.71.033. [DOI] [PubMed] [Google Scholar]
- 55.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darfeuille F, Unoson C, Vogel J, Wagner EG. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Condon C. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 59.de la Sierra-Gallay IL, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nat. Struct. Mol. Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- 60.Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW. Transcriptional interference: an unexpected layer of complexity in gene regulation. J. Cell Sci. 2007;120:2755–2761. doi: 10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- 61.Shearwin KE, Callen BP, Egan JB. Transcriptional interference - a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crampton N, Bonass WA, Kirkham J, Rivetti C, Thomson NH. Collision events between RNA polymerases in convergent transcription studied by atomic force microscopy. Nucleic Acids Res. 2006;34:5416–5425. doi: 10.1093/nar/gkl668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Joh K, Masuko S, Yatsuki H, Soejima H, Nabetani A, Beechey CV, Okinami S, Mukai T. The mouse Murr1 gene is imprinted in the adult brain, presumably due to transcriptional interference by the antisense-oriented U2af1-rs1 gene. Mol. Cell. Biol. 2004;24:270–279. doi: 10.1128/MCB.24.1.270-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.El Hanafi D, Bossi L. Activation and silencing of leu-500 promoter by transcription-induced DNA supercoiling in the Salmonella chromosome. Mol. Microbiol. 2000;37:583–594. doi: 10.1046/j.1365-2958.2000.02015.x. [DOI] [PubMed] [Google Scholar]
- 66.Gartenberg MR, Wang JC. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc. Natl Acad. Sci. USA. 1992;89:11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moulin L, Rahmouni AR, Boccard F. Topological insulators inhibit diffusion of transcription-induced positive supercoils in the chromosome of Escherichia coli. Mol. Microbiol. 2005;55:601–610. doi: 10.1111/j.1365-2958.2004.04411.x. [DOI] [PubMed] [Google Scholar]
- 68.Huttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heidrich N, Chinali A, Gerth U, Brantl S. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 2006;62:520–536. doi: 10.1111/j.1365-2958.2006.05384.x. [DOI] [PubMed] [Google Scholar]
- 71.Licht A, Preis S, Brantl S. Implication of CcpN in the regulation of a novel untranslated RNA (SR1) in Bacillus subtilis. Mol. Microbiol. 2005;58:189–206. doi: 10.1111/j.1365-2958.2005.04810.x. [DOI] [PubMed] [Google Scholar]
- 72.Zemanova M, Kaderabkova P, Patek M, Knoppova M, Silar R, Nesvera J. Chromosomally encoded small antisense RNA in Corynebacterium glutamicum. FEMS Microbiol. Lett. 2008;279:195–201. doi: 10.1111/j.1574-6968.2007.01024.x. [DOI] [PubMed] [Google Scholar]
- 73.Fierro-Monti IP, Reid SJ, Woods DR. Differential expression of a Clostridium acetobutylicum antisense RNA: implications for regulation of glutamine synthetase. J. Bacteriol. 1992;174:7642–7647. doi: 10.1128/jb.174.23.7642-7647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Janssen PJ, Jones DT, Woods DR. Studies on Clostridium acetobutylicum glnA promoters and antisense RNA. Mol. Microbiol. 1990;4:1575–1583. [PubMed] [Google Scholar]
- 75.Herskovitz MA, Bechhofer DH. Endoribonuclease RNase III is essential in Bacillus subtilis. Mol. Microbiol. 2000;38:1027–1033. doi: 10.1046/j.1365-2958.2000.02185.x. [DOI] [PubMed] [Google Scholar]
- 76.Hartl B, Wehrl W, Wiegert T, Homuth G, Schumann W. Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J. Bacteriol. 2001;183:2696–2699. doi: 10.1128/JB.183.8.2696-2699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.