Abstract
We evaluate here the use of real-time quantitative PCR (q-PCR) as a method for screening for homologous recombinants generated in mammalian cells from either conventional gene-targeting constructs or whole BAC-based constructs. Using gene-targeted events at different loci, we show that q-PCR is a highly sensitive and accurate method for screening for conventional gene targeting that can reduce the number of clones requiring follow-up screening by Southern blotting. We further compared q-PCR to fluorescent in situ hybridization (FISH) for the detection of gene-targeting events using full-length BAC-based constructs designed to introduce mutations either into one gene or simultaneously into two adjacent genes. We find that although BAC-based constructs appeared to have high rates of homologous recombination when evaluated by FISH, screening by FISH was prone to false positives that were detected by q-PCR. Our results demonstrate the utility of q-PCR as a screening tool for gene targeting and further highlight potential problems with the use of whole BAC-based constructs for homologous recombination.
INTRODUCTION
Homologous recombination in mouse embryonic stem cells has provided a powerful tool for the generation of gene-targeted mice carrying mutations of defined loci (1,2). However, since homologous recombination can be a relatively infrequent event in mammalian cells (3,4), detection of homologous recombination events can require the screening of large numbers of clones. The gold standard for screening is Southern blot analyses, which both identifies correctly targeted clones and permits analysis of the structure of the gene-targeted event, including verification of single copy insertion of the construct. However, Southern analyses can be time-consuming, particularly in cases of loci with low-frequency targeting where hundreds of cell lines may need to be screened.
The use of PCR-based strategies, most of which utilize one primer outside of the targeting constructs in conjunction with a primer present on the selectable marker, has permitted more rapid screening for homologous recombinants (5–8). Other PCR strategies, including screening for loss of plasmid sequences from the targeting construct, have also been used (9). However, conventional PCR provides limited information on the number and structure of inserts. Moreover, conventional PCR-based strategies are prone to false negatives resulting from problems with long-range PCR or low amounts of DNA. Conversely, false positives can arise due to annealing of products initiated by the different primers, giving rise to products consistent with appropriate targeting but that actually derive from clones containing random insertions. Thus, PCR-based screens cannot replace the information gained through Southern analyses, although this technique can reduce the numbers of clones to be evaluated in greater detail.
Recent data have suggested that whole BAC-based gene-targeting vectors can provide an alternative method allowing for more rapid generation of homologous recombinants (10–12). However, screening for homologous recombination after introduction of such vectors can be complicated by the long length of homologous arms used, which most often preclude screening by either Southern analyses or PCR strategies. To bypass these assay problems, several alternatives have been utilized. These include: (i) the use of one short-length arm on the targeting construct to permit screening by Southern analyses using an outside probe (10); (ii) the use of fluorescent in situ hybridization (FISH) to detect the number of BAC sequences integrated as well as their chromosomal locations (12); and (iii) the use of real-time quantitative PCR (q-PCR) to screen for the loss of sequences deleted in the targeting construct (11). The latter two techniques permit use of long targeting arms that are predicted to increase recombination frequencies. However, these two techniques have not been systematically compared to evaluate their sensitivity and specificity.
To evaluate techniques for the rapid screening for homologous recombinants, we compared q-PCR based on the copy number of genomic regions deleted by Southern analysis screening for the detection of homologous recombination using conventional positive–negative gene-targeting constructs (13). We further compared q-PCR (11) to a previously described FISH protocol (12) for the screening of BAC-based constructs that cannot be screened by Southern analyses. We demonstrate here that screening by q-PCR is a sensitive tool for screening for homologous recombinants generated either from conventional gene-targeting constructs or from whole BAC-based constructs designed to disrupt either one or two genes simultaneously. Importantly, we find that although BAC-based constructs can appear to have high rates of homologous recombination when evaluated by FISH, q-PCR analyses revealed that many of these homologous recombinants were actually false positives. Our results raise important concerns both for the screening and use of whole BAC-based constructs for homologous recombination.
MATERIALS AND METHODS
Construction of the conventional positive–negative selection Tec targeting vector
A PstI fragment of 4.6 (5′ arm) and a Psi–HindIII fragment of 4.9 kb (3′ arm) flanking the targeted exons 5 and 6 in Tec were subcloned in the vector pPNT-double loxP, a derivative of pPNT (14) in which a PGK-driven neomycin-resistance gene, neo, is flanked by loxP sites (15,16) (gift of A. Wynshaw-Boris, University of California, San Francisco, CA, USA and S. Hirotsune, Osake City University School of Medicine, Osake, Japan) to generate a 1276 bp deletion. For the Rlk gene-targeting construct, a XbaI fragment of 3.1 kb (5′ arm) and an EcoRV–BglII fragment of 5.6 kb (3′ arm) that flank exon 10 were subcloned into pPNT-double loxP to generate a 1147 bp deletion.
Construction of the BAC-targeting vector for Tec and Rlk
Both the Tec and Rlk genes present in the BAC RPCI-23-6518 (isolated from RPCI-23 C57BL/6 library) were modified sequentially by recombineering (17). Exons 5 and 6 in Tec and 10 in Rlk (18) were disrupted in the identical location as conventional targeting constructs and replaced with neo and blasticidin resistance (blasticidin S deaminase, bsd) genes, respectively, both driven by the PGK promoter. DNA fragments of 500 bp surrounding the targeted sequences were subcloned in the plasmids PL452 (17) for Tec and PL514 for Rlk (PL514 is a variant of PL452 in which the neo gene was replaced with the bsd gene by recombineering; Liu,P., unpublished data) to make the cassettes for recombineering. BACs were purified on CsCl gradients and linearized with NotI.
Construction of the TLT-1 targeting vectors
The conventional and whole BAC TLT-1 targeting vectors were generated by recombineering as described (A.V.W., J.G.R., P.L.S and D.W.M., submitted for publication).
Electroporation and selection of embryonic stem cells
The modified-BAC and the conventional targeting constructs were linearized with NotI, cleaned with phenol and choloroform, ethanol precipitated and resuspended at 1 μg/μl in 5 mM Tris. Linearized DNA preps were analyzed by agarose gel electrophoresis and, for BAC constructs, pulse-field gel electrophoresis (Chef-DR III, BioRad, Hercules, CA, USA). DNA was electroporated into 2 × 107 HGTC-8 C57Bl/6 ES cells (19) in 1 ml of phosphate-buffered saline (without Ca2+ and Mg2+) using a BioRad Genepulser at 600 V and 25 μF (using 10–25 μg DNA for conventional constructs and 50 μg for BAC constructs) in a 0.4 cm gap cuvette. ES cells electroporated with the modified-Tec/Rlk BAC were seeded at 3.3 × 106 cells/plate on neo, bsd-resistant feeder cells (20) in 10 plates. One day later, cells were fed with media containing G418 (140 μg/ml, Invitrogen, Carlsbad, CA, USA) and blasticidin (5 μg/ml, Invitrogen). Antibiotic concentrations were determined by titration on the HGTC-8 C57Bl/6 ES cell line. A portion of the transfected ES cells (3.3 × 106 cells) were also selected in only one of the two antibiotics (one plate equivalent per antibiotic). Media with antibiotics were changed daily until clones were ready to be picked (after ∼1 week in selection). The HGTC-8 C57Bl/6 ES line was maintained as previously described (19).
Southern blot screening of ES clones
ES cells were expanded without feeders and lysed overnight at 37°C in 10 mM Tris–HCl pH 8, 100 mM EDTA, 100 mM NaCl, 0.5% SDS with 250 μg/ml proteinase K, and then treated with RNase (400 μg/ml) for 1 h at 37°C. Genomic DNA from ES clones was purified by phenol/chloroform/isoamyl alcohol using Phase Lock Gel Heavy tubes (Eppendorf, Hamburg, Germany), precipitated with ethanol at room temperature and resuspended in 5 mM Tris pH 8.0. Southern blotting was performed according to standard protocols (21).
PCR screening of ES clones
ES clones electroporated with BAC DNA were initially screened for the presence of both neo and bsd resistance markers by conventional PCR. The primers were designed to detect the targeted loci and the resistance marker. Primers for Tec–neo: SCTecF: 5′ TCA TGA CTC TCA CGG TAA GGG 3′ and neo: 5′ GTG CAA TCC ATC TTG TTC AAT GG 3′; and for Rlk–bsd: SCRlkR: 5′ GCA GTG GAA TGA GAA CAG TAG G 3′ and bsdF 5′ ATG GCC TTT GTC TCA AGA AGA ATC C 3′. PCR was performed using Platinum Taq DNA polymerase High Fidelity (Invitrogen) under the following conditions: 95°C for 3 min, followed by 35 cycles of 94°C for 45 s, 60°C for 45 s and 68°C for 2 min.
FISH
Metaphase preparation from ES cell lines were prepared by standard air-drying technique and FISH was performed with BAC DNA labeled by Nick Translation technique with Spectrum Orange as described (22). The full-length BACs used to generate the gene-targeting constructs (i.e. RPCI-23-6518 for the Tec/Rlk constructs) were used as probes. A minimum of 10 cells were analysed per clone. In all cases, two pairs of hybridization signals were observed on chromosome 5 for the Tec/Rlk construct. In cases where a third pair was observed, the third pair of signals was always observed on the same chromosome within a given clone (but on different chromosomes in the different clones). No evidence of aneuploidy was observed in these clones.
q-PCR for screening of targeted ES clones
Real-time q-PCR was used to screen targeted ES cells, by discriminating the difference between control WT ES (containing two copies of the targeted exon) and correctly targeted heterozygous ES clones (having one copy or the targeted exon). The q-PCR was performed using ABI 7500 (Applied Biosystems, Foster City, CA, USA) and Platinum SYBR Green q-PCR SuperMix (Invitrogen). DNA concentration of the ES clones was measured by UV spectrophotometer and adjusted to 2.5 ng/μl. The reaction was set up using 12.5 ng of DNA per reaction. To normalize DNA concentration between ES clones and control (WT ES or ES cells resistant to either G418 or blasticidin), primers for known diploid genes such as Itk were used as endogenous controls (all amplicons range between 250 and 400 bp). For accurate results, threshold cycles (CT) for the endogenous control gene for all the ES clones had CT values ±0.5. Primers for Itk locus: IF (5′ GCCGTAAATGAACAGGTGGTGA 3′) and IR (5′ TGCTCCAGACTGTGAGAGTCG 3′. To determine correct integration of the targeting constructs, primers were designed corresponding to the deleted sequence in Tec: TF 5′ TTGTTCTAGGTTGTTCATGATGC 3′ and TR 5′ GCAAGAGCTATGTTAGCAATGC 3′; Rlk: RF 5′ GTCTGAAGAAGACTTCATTGAGG 3′ and RR 5′ ACTCACACAGCGATCCAATGTC 3′; or TLT-1: TLTF 5′ ATC ACA GAT GCC ATA GCT ACC 3′ and TLTR 5′ AAC TGG CAC CAC ACC TTG AG 3′. The number of insertions of the targeting construct was evaluated by using primers for either neo: NF (5′ AGCACGTACTCGGATGGAAGC 3′) and NR (5′ CAGAAGAACTCGTCAAGAAGGC 3′) or bsd genes BF 5′ ATGCCTTTGTCTCAAGAAGAATCC and BR 5′ TTAGCCCTCCCACACATAACC 3′. All primer sets had efficiencies of amplification between 83% and 105% relative to Itk, which are considered acceptable for comparative purposes (Applied Biosystems’ Guide to Relative Quantification of Gene Expression, Manual 2) (amplification efficiencies: 95.7% for the Tec primers, 89.6% for the Rlk primers, 103.6% for the neo primers and 96.1% for the bsd primers). As reference for the number of copies integrated, we used control ES cell lines that contain one copy of either neo or bsd markers determined by Southern blot.
PCR reactions were performed in duplicate in 25 μl using 96-well plates (MicroAmp Fast optical Applied Biosystems), containing primers for the endogenous control (Itk) and for either of the deleted exons or resistance markers. Data were only deemed acceptable if SDs were <0.1. The following conditions were used: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 94°C for 15 s, 60°C for 1 min. The CT and relative quantification (RQ) were calculated by using the ABI 7500 SDS 1.3.1 software [2−ΔΔCT method (23)]. For analysis, the PCR reactions were calibrated to WT ES cells (for deleted exons) setting two copies (WT) as the calibrator, having an RQ value of 1. For quantification of the selectable markers, values were compared to those of G418 or blasticidin-resistant ES cells containing one copy of the resistant marker (the RQ value of which was also set to 1).
RESULTS
Validation of q-PCR as a screen for gene targeting
To evaluate q-PCR for screening for homologous recombinants, we utilized a SYBR-green-based variation on a technique involving three primer sets (11). The first primer set was designed to amplify a region that would be deleted by a correct gene-targeting event. Following correctly targeted homologous recombination, the template would only be present as a single copy in the genome. However, in the case of non-targeted clones, two copies of the template should be present from the two endogenous WT alleles. A second set of primers was designed to detect the selectable marker used in the targeting construct and was used to evaluate copy number of the inserts, by comparing to DNA from a clone containing one copy of the same selectable marker. Finally, a third set (or multiple sets) of control primers that amplify different reference autosomal loci was used to normalize DNA concentration. Correctly targeted clones that delete a portion of an autosomal gene would therefore show only one copy of the endogenous sequence and one copy of the selectable marker. Non-homologously integrated clones where the construct is randomly inserted would show two copies of the targeted region and one or more copies of the selectable marker. The rare case of a correctly targeted clone that also contained multiple inserts would show only one copy of the endogenous sequence, but multiple copies of the selectable marker. Clones where there was an aberrant duplication of the region to be deleted (24,25) would also not be scored as positive.
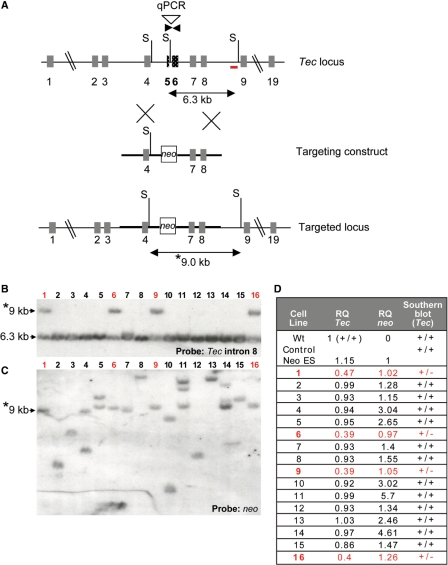
To evaluate the ability of q-PCR to detect homologous recombinants, we examined DNA from ES cells that had been transfected with a gene-targeting construct designed to delete 1.2 kb including exons 5 and 6 of the gene encoding the Tec tyrosine kinase, which were replaced with a PGK-driven neo selectable marker (Figure 1A). DNA from these ES clones had been previously screened by Southern blotting with external and internal probes to determine which clones were correctly targeted and whether there were multiple inserts of the targeting vector (Figure 1B and C). For q-PCR, DNA samples were first quantitated and diluted so equivalent amounts of DNA were used. The copy number of the targeted exon was then evaluated by q-PCR using the ΔΔCT method (23) with a reference gene (Itk) as a control to normalize the amount and quality of ES genomic DNA. All primer sets had similar efficiencies of amplification as tested on control DNA and all samples had CT values within 0.5 for the reference gene. Wild-type (WT) ES DNA (with two endogenous copies of Tec) was used as a calibrator for the copy number of the targeted sequence (setting the RQ to a value of 1). The copy number of the selectable marker was also evaluated by comparing RQ values to that from DNA from a calibrator ES cell clone previously identified as containing one copy of the gene encoding neomycin resistance, neo, (the RQ of which was also arbitrarily set to a value of 1). Thus, correctly targeted clones would have RQ values close to 0.5 for the targeted sequence in Tec and 1 for neo (Figure 1D).
Figure 1.
Screening by Southern blot and q-PCR of ES clones targeted with a conventional construct. (A) Schematic showing Tec targeting vector designed to delete 1.2 kb including exons 5 and 6. The primers used for q-PCR are indicated in dark inverse arrows. The 3′ Tec probe for Southern blotting is indicated as the red bar (in intron 8). S: SspI sites in the construct and the endogenous gene. Double arrows indicated the endogenous and targeted SspI fragments detected in Southern analyses. (B, C) Southern blot analysis of ES DNA digested with SspI and hybridized with (B) the 3′ probe (located in intron 8 of the Tec gene). The migration of the endogenous 6.3 kb SspI band and the targeted 9.0 kb fragment (asterisk) are indicated. (C) The same membrane was stripped and re-hybridized with a neo probe. The targeted 9.0 fragment (asterisk) is indicated. (D) q-PCR was performed using ES DNA from clones previously examined by Southern blotting and primers for the deleted region of Tec, neo and a reference gene Itk. As calibrators, DNA from WT ES cells (containing two copies of the deleted Tec region) and from an ES cell clone harboring one copy of neo were used with RQ values set to 1. Results are compared to Southern analyses. Targeted clones have RQ values close to 0.5 for Tec and 1 for neo. Clones containing a disruption of Tec are indicated in red.
In all cases, q-PCR accurately detected the homologous recombinants originally found by Southern blotting, correctly identifying all 15/108 homologously targeted clones and no others, and confirming that a single copy insertion of the selectable marker occurred (Figure 1D, see clones 1, 6, 9 and 16). Similar results were obtained with a second targeting construct disrupting a different gene, Rlk/Txk, where all three homologous recombinants were correctly identified out of 153 screened (Supplementary Figure S1 and data not shown). In contrast, a conventional PCR strategy designed to cross the region of homology in the Tec targeting construct (generating a 5 kb product for one arm and 6 kb for the other) only detected 50% of correctly targeted events in an initial screen, perhaps due to the long arm length. Thus, q-PCR showed a high degree of sensitivity and specificity for the detection of homologous recombinants.
Evaluation of q-PCR for the detection of homologous recombination of BAC-based constructs
Having confirmed that q-PCR was able to correctly identify homologous recombination at the Tec and Rlk genes, we then used q-PCR for detection of homologous recombination using full-length BAC vectors. Although both q-PCR (11) and FISH (12) have been reported to screen successfully for homologous recombinants generated from whole BAC-based constructs, these two techniques have not been systematically evaluated for sensitivity and specificity.
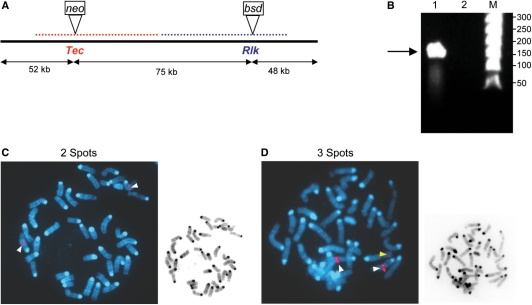
To evaluate screening of full-length BAC-based targeting constructs, we compared FISH versus q-PCR for screening of recombinants generated from a BAC-based construct designed to simultaneously introduce the mutations described above into the linked Tec and Rlk genes, which lie adjacent on mouse chromosome 5 (Figure 2). Thus, we were able to use the same q-PCR design that we had previously validated. While the first exon of the Rlk gene lies only 3 kb downstream from the last exon of Tec, the sites of our mutations lie 75 kb apart (Figure 2A). In our BAC-based construct, the Tec gene was disrupted by recombineering (26–29) introducing a PGK-driven neo selectable marker in the same location as the conventional construct described above. The Rlk gene was disrupted on the same BAC using a PGK-driven blasticidin resistance gene (bsd) as the marker for recombineering. Both mutations were in equivalent locations to those generated by our conventional constructs and were located ∼50 kb from the ends of the BAC. The BAC construct was linearized (Figure 2B) and transfected into ES cells, which were selected in either G418 or blasticidin, or in both antibiotics. Selection in both antibiotics gave ∼10- to 20-fold fewer clones than either single selection (Table 1 and data not shown). Similarly, only 4–8% of clones selected in either single antibiotic had integrated the other selectable marker as detected by PCR.
Figure 2.
Screening ES clones targeted by a Tec/Rlk modified-BAC by FISH. (A) Schematic diagram of the BAC targeting construct in RP23 61-85. Location of the Tec gene is indicated in red. Location of the Rlk gene is indicated in blue. The locations of the respective mutations introduced in Tec and Rlk are indicated by the inserted neo and bsd genes, respectively. (B) Pulse field gel electrophoresis of (1) the BAC targeting construct (175 kb) digested with NotI is indicated with an arrow; (2) empty lane and (M) Lambda Ladder PFG Marker (New England BioLabs, Ipswich, MA, USA). (C, D) Representative FISH images using the BAC RPCI-23-6518 probe containing Tec and Rlk genes (red on chromosome 5). Two pairs of spots in chromosome 5 (white arrows) suggest homologous integration (C) whereas a third set of spots (yellow arrow) in another chromosome indicates non-homologous integration (D).
Table 1.
Summary of screening of ES clones electroporated with the Tec/Rlk-modified BAC
| Antibiotic selection | Number of clones | Positives by PCR |
Positives by FISH (%) |
Positives by real-time PCR (%) |
|||
|---|---|---|---|---|---|---|---|
| neo (Tec) | bsd (Rlk) | 2 spots | 3 spots | Tec (±) | Rlk (±) | ||
| G418 | 25/1 plate | 25 | 2 | – | – | 5 (20) | 0 |
| Blasticidin | 23/1 plate | 1 | 23 | – | – | 0 | 3 (13) |
| G418+blasticidin | 11/10 plates | 11 | 10 | 4 (36) | 7 | 3 (27) | 1 (9) |
ES cells were electroporated with BAC RPCI-23 65-18 modified by recombineering and selected in G418 (one plate), blasticidin (one plate) or G418 and blasticidin (10 plates). Clones were screened by FISH, PCR for the presence of the neo and bsd markers and q-PCR (examining the copy number of the targeted Tec and Rlk exons, as well as the neo and bsd resistance markers). DNA from WT ES cells (containing two copies of the deleted Tec and Rlk region) was used as a calibrator and from ES cells that were either targeted in the same position in Tec and Rlk locus by conventional constructs used as both calibrators and positive controls. Only three clones had disrupted Tec and one clone (indicated in red) had disrupted both genes, although four clones appeared to be positive when screened by FISH.
To screen for homologous recombination using this BAC-based targeting vector, we initially used the technique of Seed and colleagues (12). In this technique, we first confirmed integration of the selectable marker and loss of the BAC vector ends using PCR, and then used FISH (using the entire BAC as a probe) to determine whether two or more (generally three) pairs of signals hybridizing to the BAC were present (Figure 2C and D). Clones showing two pairs of signals (both on chromosome 5) were presumed to be targeted homologously with the integrated BAC replacing one of the endogenous alleles (Figure 2C). Clones showing more than two pairs of signals contained extra copies of the BAC that were not targeted to the endogenous gene (Figure 2D). The proper structures of the targeted alleles were also confirmed by Southern analyses. We first noted that according to our results with FISH, the use of BAC-based vectors appeared to increase targeting frequency compared to conventional targeting vectors (36% targeting frequency versus 14% for our conventional Tec construct). However, we also noted that in some clones the third pair of spots detected by FISH had reduced intensity compared to the two endogenous signals (Figure 2D), suggesting that only a piece of the BAC had integrated into the genome.
To further examine these targeting events, we examined the Tec and Rlk loci by q-PCR, using the same strategy outlined above (with primers for Tec, neo, Rlk, bsd and a reference gene Itk). A representative example of two similar electroporations is shown (Tables 1 and 2, Supplementary Tables S1 and S2). Re-examination of the BAC-transfected clones revealed that in multiple cases, clones that were scored as having two spots on FISH did not show disruption of the endogenous locus (Table 2, clones GB5 and 8). These results suggest there may have been breakage of the BACs, leading to random insertion of small pieces of DNA that included the selectable marker but were below the detection level of FISH. Moreover, some clones showed disruption of only one of these genes despite two selectable markers having been integrated (Table 2, clones GB2 and 7). For one clone, GB7, although an extra insertion was identified by FISH, q-PCR revealed that one of the genes (Tec) had actually been homologously targeted, arguing that multiple inserts of BAC DNA had occurred. Importantly, these results suggest that q-PCR identified that there were both false positives and false negatives in the original screening by FISH.
Table 2.
Comparison of results for individual ES clones electroporated with the Tec/Rlk-modified BAC
| ES selected with G418/blasticidin | RQ Tec | RQ Rlk | FISH screening |
|---|---|---|---|
| WT | 1.0 | 1.0 | |
| Positive Tec+/− | 0.6 | 1.1 | |
| Positive Rlk+/− | 1.2 | 0.5 | |
| GB 1 | 0.3 | 0.4 | 2 spots |
| GB 2 | 0.6 | 1.0 | 2 spots |
| GB 3 | 1.2 | 1.0 | 3 spots |
| GB 4 | 1.2 | 1.0 | 3 spots |
| GB 5 | 1.3 | 1.0 | 2 spots |
| GB 6 | 1.3 | 1.1 | 3 spots |
| GB 7 | 0.6 | 0.9 | 3 spots |
| GB 8 | 1.1 | 1.1 | 2 spots |
| GB 9 | 1.3 | 1.0 | 3 spots |
| GB 10 | 1.2 | 1.1 | 3 spots |
| GB 11 | 1.2 | 1.1 | 3 spots |
Summary of results for individual ES cells selected with both G418 and blasticidin and screened by q-PCR for deletion of Tec- and Rlk-targeted exons. Results are compared to those obtained by FISH using the BAC probe. Clones that are positive by q-PCR are in bold. Clones in red contained a gene-disruption of Tec. Clones indicated in blue contained a gene-disruption for Rlk. Results of FISH screening that are false positives are italicized.
To confirm that these results were not specific to the Tec and Rlk genes, we examined another independently generated set of constructs that disrupted a distinct locus encoding TLT-1 on mouse chromosome 17 (A.V.W., J.G.R., P.L.S and D.W.M., submitted for publication). Again, a conventional targeting construct disrupting the TLT-1 gene with a PGK-driven neo resistance marker was compared to an equivalent whole BAC-based construct. Examination of the conventional targeting construct gave 17% targeting frequency, as confirmed by Southern blotting (data not shown). Initial screening of the BAC-based construct by PCR and FISH using the TLT-1 BAC as a probe suggested that 8 of 12 clones were correctly targeted (positive for neo, negative for the BAC ends and having two spots on FISH). Thus, the BAC-based construct again appeared to have high rates of homologous recombination. However, we also noted again that some of the clones had small third spots when screened by FISH; one clone also had an altered structure as determined by Southern analyses of the targeted region. Importantly, reexamination of these G418-resistant clones by q-PCR revealed that only half of these clones (four of eight) had actually disrupted the TLT-1 gene (Table 3). One of these clones was used to generate chimeric mice that have transmitted the mutation through the germline to generate mice deficient in TLT-1, confirming that the mutation disrupted the TLT-1 gene (A.V.W., J.G.R., P.L.S and D.W.M., submitted for publication). Thus, q-PCR revealed that multiple clones initially thought to have undergone homologous recombination as detected by FISH, were actually false positives.
Table 3.
Screening of ES clones electroporated with the TLT-1-modified BAC
| Clones | RQ TLT-1 | FISH |
|---|---|---|
| WT | 1.01 | |
| 1 | 0.92 | 3 spots |
| 2 | 0.91 | 3 spots |
| 3 | 1.11 | 2 spots |
| 4 | 1.01 | 3 spots |
| 5 | 0.52 | 2 spots |
| 6 | 0.49 | 2 spots |
| 7 | 0.55 | 2 spots |
| 8 | 1.02 | 2 spots |
| 9 | 0.93 | 2 spots |
| 10 | 0.53 | 2 spots |
| 11 | 0.85 | 2 spots |
| 12 | 1.23 | 3 spots |
ES cells were electroporated with BAC TLT-1 modified by recombineering. ES cells selected with G418 were screened by q-PCR for deletion of TLT-1 exon and by FISH using the labeled TLT-1 BAC as a probe. Clones that are positive by q-PCR are in bold. Results of FISH screening that are false positives are italicized.
DISCUSSION
In this article, we compare q-PCR to other screening strategies for detection of gene targeting. Our data suggest that q-PCR using a three primer set assay is a sensitive and reproducible tool that can reduce the number of potential conventional gene-targeted clones that need to be screened by Southern analyses, thereby greatly facilitating screening of large numbers of clones. In our hands, we find this technique to be superior to PCR screening for a number of reasons. First, it is not limited by false negatives that can sometimes occur when trying to amplify long pieces of DNA from genomic DNA preparations. Thus, it does not require one arm of the construct to be a relatively short length, which can potentially decrease the frequency of homologous recombination (4). Second, false positives, which can result from annealing of long PCR products or low levels of plasmid contamination, should also not be a problem with this technique. Third, q-PCR can also detect multiple inserts based on the copy number of the selectable marker. Finally, q-PCR should eliminate the rare case of clones in which the deleted part of the gene is aberrantly duplicated, as it occurs occasionally (24,25) and can be missed on some Southern analyses screens. In such cases, q-PCR would not indicate a loss of copy number of the region to be deleted. Although q-PCR cannot replace the use of Southern analyses, which can detect additional altered recombination structures that can occur during gene targeting, we find that q-PCR provides a rapid screening alternative with superior results to conventional PCR prior to confirmation with Southern analyses. A recent report has reached similar conclusions using a related TaqMan-based q-PCR technique to detect insertions based on disruption of products annealing to the TaqMan probe (30). Together our findings support the utility of using q-PCR to detect distinct types of gene-targeting events.
We further find that q-PCR is superior to FISH for the detection of whole BAC-based constructs that cannot be screened by Southern analyses. A previous study suggested that FISH can be a useful tool for screening for homologous recombination using full-length BAC constructs (12). However, in our hands, we have found that screening of BAC-based constructs by FISH can be prone to false positives. Indeed, although we initially thought we obtained clearly higher rates of homologous recombination using full-length BAC constructs when we screened by FISH alone (2.5- to 5-fold higher than our equivalent conventional constructs), re-evaluation with q-PCR demonstrated that the rates of gene targeting were similar to those obtained with conventional constructs (the same to 2-fold higher).
Presumably, false positives (indicated by two pairs of spots on FISH) can occur when small pieces of DNA are integrated that are below the limits of detection by fluorescent hybridization (∼10 kb in our hands). Our observations that in multiple cases the third spot detected by FISH was reduced in intensity compared to the endogenous loci, supports the notion that only a portion of the BAC had integrated. We observed this complication both while using a BAC construct designed to disrupt only one gene, as well as when we attempted to disrupt two adjacent genes using a single BAC-targeting construct. Indeed, when we initially attempted to disrupt both Tec and Rlk using a BAC construct in which the Rlk gene only contained a deletion but no selectable marker, we failed to obtain any double-targeted clones (from 86 clones screened, unpublished observations). We also note that we detected some altered structures by Southern analyses in clones from two of our BAC constructs, also suggesting that breakage of the construct occurred. These results are consistent with deletions that have been observed in BAC-based transgenesis using recombination-mediated cassette exchange (31) and can also be seen in gene conversion.
While breakage of the BAC may have occurred during preparation of the DNA, we observed these findings using three different constructs prepared in two different laboratories and pulse-field analyses did not reveal gross breakage of the linearized BACs prior to electroporation (Figure 2B). Alternatively, these observations may result from breakage of the BACs secondary to electroporation. [We note that we performed electroporation under our standard conditions, which differ from those of Seed and Stewart and their colleagues (10,12). We also note that stability of BAC DNA can be improved with buffers containing spermine and spermidine, which we did not try in these studies (32)]. Nonetheless, our results argue that q-PCR is a superior technique for detection of recombination of BAC-based vectors in part, because it is not subject to false positives that can result when small pieces of BAC constructs integrate. Conversely, the fact that in one clone q-PCR revealed that one of the genes (Tec) had actually been homologously targeted despite an extra insertion identified by FISH, argues that q-PCR can also reveal false-negative results from FISH. Our findings also raise the concern that it may be difficult to monitor breakage and integration of smaller pieces of DNA when using whole BAC-based constructs for gene targeting. For that reason, it may be optimal to screen initially with q-PCR and use FISH as a complementary technique for examining clones derived from BAC-based targeting vectors.
Our results are also of interest for the generation of multiple mutations using a single whole BAC-based construct, which can be desirable when trying to minimize handling of ES cells prior to injection into blastocysts, and for the introductions of two mutations on the same sister chromatid. Stewart and colleagues (10) have previously reported the use of whole BAC-based constructs for the generation of gene-targeted alleles at two adjacent loci using modified BACs. In their work, the genes were disrupted close to the ends of the BAC (within 5 kb) so that recombination could be detected by Southern hybridization. However, this group also observed that one of the four clones expressing both selectable markers only disrupted one gene. While this was seen at a lower frequency than in our experiments, it is possible that our constructs, which did not limit the length of one arm for detection by Southern analyses, may have required breakage to obtain efficient integration of the selectable marker. Nonetheless, these findings also support the idea that BAC-based constructs may still only recombine over short distances. Given our observations, it is not clear that disruption of both genes occurred via integration of one piece of DNA via a single double cross-over event. Our results therefore raise the possibility that double knockouts might be equivalently generated from simultaneous transfection of two conventional targeting constructs.
Together, these data suggest that q-PCR is a useful tool for the detection of both conventional and BAC-based gene-targeting constructs. Our data suggest that q-PCR can greatly reduce the numbers of clones from conventional constructs requiring further screening by Southern analyses. Moreover, our data strongly argue that q-PCR provides a more accurate assessment of gene targeting using full BAC constructs than using FISH alone, but also raise concerns about the use of whole BAC-based constructs where it may be difficult to monitor partial integration products.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health, National Human Genome Research Institute and National Cancer Institute. Wellcome Trust (P.L.). Funding for open access charge: Intramural Research Program of the National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank L. Garrett and A. Kimmel for helpful comments.
REFERENCES
- 1.van der Weyden L, Adams DJ, Bradley A. Tools for targeted manipulation of the mouse genome. Physiol. Genomics. 2002;11:133–164. doi: 10.1152/physiolgenomics.00074.2002. [DOI] [PubMed] [Google Scholar]
- 2.Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 3.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 4.Deng C, Capecchi MR. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell. Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmer A, Gruss P. Production of chimaeric mice containing embryonic stem (ES) cells carrying a homoeobox Hox 1.1 allele mutated by homologous recombination. Nature. 1989;338:150–153. doi: 10.1038/338150a0. [DOI] [PubMed] [Google Scholar]
- 6.Joyner AL, Skarnes WC, Rossant J. Production of a mutation in mouse En-2 gene by homologous recombination in embryonic stem cells. Nature. 1989;338:153–156. doi: 10.1038/338153a0. [DOI] [PubMed] [Google Scholar]
- 7.Kim HS, Smithies O. Recombinant fragment assay for gene targetting based on the polymerase chain reaction. Nucleic Acids Res. 1988;16:8887–8903. doi: 10.1093/nar/16.18.8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lay JM, Friis-Hansen L, Gillespie PJ, Samuelson LC. Rapid confirmation of gene targeting in embryonic stem cells using two long-range PCR techniques. Transgenic Res. 1998;7:135–140. doi: 10.1023/a:1008876526826. [DOI] [PubMed] [Google Scholar]
- 9.McDermott J, Zhao Y, Sauer B. A simple polymerase chain reaction screen for homologous targeting in embryonic stem cells. Anal. Biochem. 2004;332:401–403. doi: 10.1016/j.ab.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 10.Testa G, Zhang Y, Vintersten K, Benes V, Pijnappel WW, Chambers I, Smith AJ, Smith AG, Stewart AF. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat. Biotechnol. 2003;21:443–447. doi: 10.1038/nbt804. [DOI] [PubMed] [Google Scholar]
- 11.Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Seed B. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat. Biotechnol. 2003;21:447–451. doi: 10.1038/nbt803. [DOI] [PubMed] [Google Scholar]
- 13.Thomas KR, Capecchi MR. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990;346:847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- 14.Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 15.Crabtree JS, Scacheri PC, Ward JM, Garrett-Beal L, Emmert-Buck MR, Edgemon KA, Lorang D, Libutti SK, Chandrasekharappa SC, Marx SJ, et al. A mouse model of multiple endocrine neoplasia, type 1, develops multiple endocrine tumors. Proc. Natl Acad. Sci. USA. 2001;98:1118–1123. doi: 10.1073/pnas.98.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, Muramatsu M, Miyagawa S, Hiraiwa N, Yoshiki A, Wynshaw-Boris A, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol. Cell. Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stalker J, Gibbins B, Meidl P, Smith J, Spooner W, Hotz HR, Cox AV. The Ensembl web site: mechanics of a genome browser. Genome Res. 2004;14:951–955. doi: 10.1101/gr.1863004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng J, Dutra A, Takesono A, Garrett-Beal L, Schwartzberg PL. Improved generation of C57BL/6J mouse embryonic stem cells in a defined serum-free media. Genesis. 2004;39:100–104. doi: 10.1002/gene.20031. [DOI] [PubMed] [Google Scholar]
- 20.Chen YT, Liu P, Bradley A. Inducible gene trapping with drug-selectable markers and Cre/loxP to identify developmentally regulated genes. Mol. Cell. Biol. 2004;24:9930–9941. doi: 10.1128/MCB.24.22.9930-9941.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. Vol. 1. NY, USA: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 22.Dutra AS, Mignot E, Puck JM. Gene localization and syntenic mapping by FISH in the dog. Cytogenet. Cell Genet. 1996;74:113–117. doi: 10.1159/000134395. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Moens CB, Auerbach AB, Conlon RA, Joyner AL, Rossant J. A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes Dev. 1992;6:691–704. doi: 10.1101/gad.6.5.691. [DOI] [PubMed] [Google Scholar]
- 25.Hasty P, Rivera-Perez J, Chang C, Bradley A. Target frequency and integration pattern for insertion and replacement vectors in embryonic stem cells. Mol. Cell. Biol. 1991;11:4509–4517. doi: 10.1128/mcb.11.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotta-de-Almeida V, Schonhoff S, Shibata T, Leiter A, Snapper SB. A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 2003;13:2190–2194. doi: 10.1101/gr.1356503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Testa G, Vintersten K, Zhang Y, Benes V, Muyrers JP, Stewart AF. BAC engineering for the generation of ES cell-targeting constructs and mouse transgenes. Methods Mol. Biol. 2004;256:123–139. doi: 10.1385/1-59259-753-X:123. [DOI] [PubMed] [Google Scholar]
- 28.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 29.Sawitzke JA, Thomason LC, Costantino N, Bubunenko M, Datta S, Court DL. Recombineering: in vivo genetic engineering in E. coli, S. enterica, and beyond. Methods Enzymol. 2007;421:171–199. doi: 10.1016/S0076-6879(06)21015-2. [DOI] [PubMed] [Google Scholar]
- 30.Soliman GA, Ishida-Takahashi R, Gong Y, Jones JC, Leshan RL, Saunders TL, Fingar DC, Myers M.G., Jr A simple qPCR-based method to detect correct insertion of homologous targeting vectors in murine ES cells. Transgenic Res. 2007;16:665–670. doi: 10.1007/s11248-007-9110-2. [DOI] [PubMed] [Google Scholar]
- 31.Prosser HM, Rzadzinska AK, Steel KP, Bradley A. Mosaic complementation demonstrates a regulatory role for myosin VIIa in actin dynamics of stereocilia. Mol. Cell. Biol. 2008;28:1702–1712. doi: 10.1128/MCB.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schedl A, Larin Z, Montoliu L, Thies E, Kelsey G, Lehrach H, Schutz G. A method for the generation of YAC transgenic mice by pronuclear microinjection. Nucleic Acids Res. 1993;21:4783–4787. doi: 10.1093/nar/21.20.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.