Abstract
Several RNA silencing pathways in plants restrict viral infections and are suppressed by distinct viral proteins. Here we show that the endogenous trans-acting (ta)siRNA pathway, which depends on Dicer-like (DCL) 4 and RNA-dependent RNA polymerase (RDR) 6, is suppressed by infection of Arabidopsis with Cauliflower mosaic virus (CaMV). This effect was associated with overaccumulation of unprocessed, RDR6-dependent precursors of tasiRNAs and is due solely to expression of the CaMV transactivator/viroplasmin (TAV) protein. TAV expression also impaired secondary, but not primary, siRNA production from a silenced transgene and increased accumulation of mRNAs normally silenced by the four known tasiRNA families and RDR6-dependent secondary siRNAs. Moreover, TAV expression upregulated DCL4, DRB4 and AGO7 that mediate tasiRNA biogenesis. Our findings suggest that TAV is a general inhibitor of silencing amplification that impairs DCL4-mediated processing of RDR6-dependent double-stranded RNA to siRNAs. The resulting deficiency in tasiRNAs and other RDR6-/DCL4-dependent siRNAs appears to trigger a feedback mechanism that compensates for the inhibitory effects.
INTRODUCTION
Multiple RNA silencing pathways in plants downregulate gene expression at transcriptional and post-transcriptional levels by generating small (s)RNAs, 21–24 nt in length (1–4). These sRNAs act either by guiding the cleavage of cognate RNAs, blocking productive translation of these RNAs, or inducing methylation of cognate DNA targets. The major pathways in Arabidopsis differ in the nature of sRNA precursors and requirements for Dicer-like (DCL) and RNA-dependent RNA polymerase (RDR) proteins. Micro (mi)RNAs are generated by DCL1 from hairpin structures of primary-miRNA transcripts. In contrast, endogenous small interfering (si)RNAs are generated by different DCLs from double-stranded (ds)RNA precursors produced by RDRs. For example, heterochromatic and repeat-associated siRNAs are generated by DCL3 from RDR2-dependent precursors, whereas trans-acting siRNAs (tasiRNAs) by DCL4 from RDR6-dependent precursors. Both miRNAs and siRNAs bind Argonaute (AGO) proteins and guide the resulting RNA-induced silencing complexes (RISCs) to the complementary region of target nucleic acids.
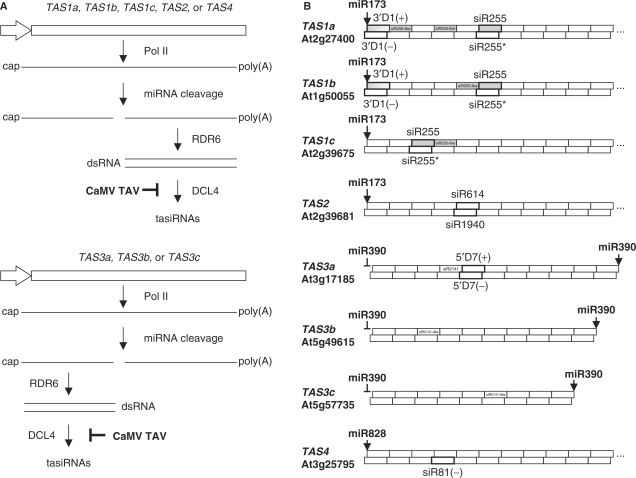
The present study focuses on effects of viral infection on the function and regulation of the tasiRNA-generating (TAS) pathways. Genetic evidence and limited biochemical data suggest that tasiRNA biogenesis is initiated by miRNA-mediated cleavage of a primary TAS transcript to generate a substrate for RDR6 that lacks the cap and/or the poly(A) tail. The presumed long dsRNA product of RDR6 activity is then processed by DCL4 from the miRNA-cleaved terminus into 21-nt siRNA duplexes with 2 nt 3′-overhangs (Figure 1). TasiRNAs are encoded by both strands of dsRNA precursors and at different in-phase positions, 2–12 21 nt processing cycles from the site of miRNA cleavage (5–10).
Figure 1.
Models for the biogenesis of tasiRNAs from two distinct types of Arabidopsis tasiRNA-generating (TAS) loci. (A) For each type of tasiRNA biogenesis the TAS gene structure is shown schematically with the promoter indicated with an arrow shape. Pol II transcription driven by this promoter generates capped and polyadenylated transcript. This transcript is targeted for cleavage by miRNA. In the case of TAS1, TAS2 and TAS4 tasiRNA families (top), the 3′-product of this cleavage event is converted into dsRNAs by RDR6 activity. In the case of the TAS3 tasiRNA family (bottom), the 5′-product is converted into dsRNA by RDR6. In both cases, the resulting dsRNA is processed by DCL4 into 21-nt tasiRNAs. Suppression of the latter step by the CaMV TAV protein is inferred from the present study.(B) The hypothetical dsRNAs derived from the Arabidopsis Col-0 tasiRNA-generation loci TAS1a, TAS1b, TAS1c, TAS2, TAS3a, TAS3b, TAS3c and TAS4 are shown schematically. In each case, position of the miRNA cleavage site is indicated by arrows; for TAS3 loci, position of the second, non-cleavable miR390-binding site is also shown. The 21-nt tasiRNAs of both sense (+) and antisense (–) polarities produced by DCL4-mediated, in-phase processing of the dsRNA are shown by boxes. The tasiRNAs probed in this study are in bold-lined boxes with their names indicated. The TAS1 loci-derived siR255 and siR255-like species are in dark boxes. TAS1a- and TAS1b-derived siRNAs D1(+) species differ only at 5′-terminal nucleotide (U and A, respectively).
Four Arabidopsis TAS families with up to three distinct loci have been identified (6,9,11). TAS1 and TAS2 tasiRNAs are initiated by miR173, TAS3 ones by miR390 and TAS4 ones by miR828 (Figure 1). The TAS3 family is special in that the two functional tasiRNAs are encoded upstream of the miR390 cleavage site and that a second miR390 target site further upstream is not cleaved (12). The two miRNA target sites are thought to delimit the region of TAS3 primary transcripts converted into dsRNA. Although target mRNAs for all four TAS families have been identified (9), biological functions have only been elucidated for TAS3-tasiRNAs that target the auxin-response factor genes ARF3 and ARF4 to control the juvenile-to-adult transition in leaf development (13,14).
Recent evidence indicates that the AGO7 protein specifically associates with miR390 to target the TAS3a transcript and thereby initiate tasiRNA biogenesis (15). However, only the TAS3 pathway genetically requires AGO7 (13) and the biogenesis of tasiRNAs of other TAS families appear to be initiated by AGO1-RISC that associates with most miRNAs (16,17). DRB4, a dsRNA binding (DRB) protein, has been implicated in tasiRNA biogenesis as a DCL4-binding partner (18). However, DRB4-deficient mutants still accumulate substantial levels of 21 nt tasiRNAs from TAS1, TAS2 and TAS3 loci (13,19–21), suggesting that DCL4 does not absolutely require DRB4 for processing dsRNA precursors of tasiRNAs or that DRB4 acts downstream of the processing step.
Certain protein coding genes give rise to siRNAs that depend on RDR6 for their production. Interestingly, many of these genes are targets of tasiRNAs and, in some cases, of miRNAs (9,12). A common feature of such secondary siRNA-generating loci is that they are targeted by more than one small RNA. It has been proposed that successive amplification of secondary siRNAs by an RDR6-/DCL4-dependent mechanism might function to achieve efficient silencing of paralogous genes in expanding gene families (9). The same mechanism involving RDR6 and DCL4 appears to generate secondary siRNAs from silenced transgenes (22), which contribute to amplification and cell-to-cell spread of silencing (20,23).
By analogy, RDR6-dependent production of secondary siRNAs might contribute to anti-viral defense based on RNA silencing (24). Indeed, RDR6 was implicated in defense against an RNA cucumovirus (25) and in exclusion of an RNA potexvirus from the shoot apical meristem (26).
Cauliflower mosaic virus (CaMV) is a pararetrovirus with a dsDNA genome and a polycistronic pregenomic RNA with seven genes (27). The transactivator/viroplasmin (TAV) is produced from a subgenomic monocistronic CaMV RNA, and this protein activates polycistronic translation of the remaining six proteins (28). TAV is multifunctional and equipped with the nuclear targeting and export signals (29), as well as ssRNA- and dsRNA- and protein-binding domains. It forms large cytoplasmic inclusion bodies, within which virus particles accumulate (30). TAV also determines symptom severity and host range (31) and, as a transgene, causes virus-like symptoms and late flowering (32,33). Recently it was shown to act as a suppressor of transgene silencing (34).
We and others have previously reported that four Arabidopsis DCL proteins generate CaMV-derived siRNAs, 21, 22 and 24 nt in length (35,36). The relative contribution of DCL4, known to restrict RNA virus infection (37) was minor, however, and RDR6 was not required for CaMV siRNA biogenesis (35), suggesting that RDR6-/DCL4-dependent amplification of viral secondary siRNAs is suppressed.
Here we report that CaMV infection interferes with endogenous RDR6-dependent tasiRNA and secondary siRNA pathways and demonstrate that TAV is solely responsible for this interference. Furthermore our work uncovers novel mechanistic features of tasiRNA-generating pathways and suggests feedback regulation of tasiRNA levels through activation of the tasiRNA biogenesis machinery.
MATERIALS AND METHODS
Arabidopsis mutant and transgenic lines
Arabidopsis DCL- and RDR-deficient mutant lines dcl2-5, dcl3-1, dcl4-2, dcl23, dcl34, dcl234, rdr6-15 and rdr2-1 (all in Col-0 background) used in this study were described previously (35 and references therein). The CaMV TAV transgenic Arabidopsis lines CM-2, CM-6 and D4-2 (in Col-0 background) were constructed by Yu et al. (38). The silenced GFP transgenic Arabidopsis line GF-FGxGFP was described by Moissiard et al. (22)
Plant growth and virus infection
Arabidopsis wild-type, silencing-deficient mutant and TAV transgenic plants were raised from seeds in soil and maintained in a Phytochamber (Sanyo) at 20°C with 12-h light and 12-h dark. Four to five weeks post-germination, seedlings were inoculated with CaMV using particle bombardment as we described previously (35,39): 1 μm gold particles were coated with 1-μg plasmid DNA of the CaMV CM1841 infectious clone pCa122 (40) and delivered at 1100 psi. CaMV symptoms started to develop about 3 weeks post-inoculation. One month post-inoculation, unless otherwise stated, the virus-infected plants were harvested and used for RNA extraction.
RNA extraction and analysis
Total RNA was extracted from 1-g plant tissue, derived from the pools of plants grounded into fine powder in liquid nitrogen, using Trizol reagent (Invitrogen, AG, Basel, Basel land, Switzerland) according to the manufacturer's protocol. About 20 or 30 μg total RNA were resuspended in 10 μl loading buffer (0.10% bromophenol blue in 100% de-ionized formamide), heated at 95°C for 2 min and loaded on 15% (or 5%) polyacrylamide denaturing gel (a 19 : 1 ratio of acrylamide to bis-acrylamide, 8 M urea). The gel was run using the SE 600 electrophoresis apparatus (Hoefer Inc., Holliston, MA, USA) at 300 V for 4 h and then RNAs were transferred to a Hybond N+ membrane by electroblotting in 1× TBE buffer at 10 V overnight. The hybridization was performed at 35°C for 18 to 24 h in UltraHyb-oligo buffer (Ambion, Warrington, Cheshire, UK) using as a probe, short DNA oligos (Table S1) end-labeled with 32P by polynucleotide kinase (New England Biolabs/Bioconcept, Allschwil, Basel land, Switzerland) and purified through MicroSpin™ G-25 columns (GE Healthcare, Glattbrugg, Zurich, Switzerland) according to the manufacturers’ recommendations. The blot was washed two times with 2 × SSC, 0.5% SDS for 30 min at 37°C. The signal was detected after exposure to a phosphor screen using a Molecular Imager (BioRad). For repeated hybridization, the membrane was stripped with 0.5 × SSC, 0.5% SDS for 30 min at 80°C and then with 0.1 × SSC, 0.5% SDS for 30 min at 80°C.
Microarray analysis
All microarray analyses were done using two replicates. For each replicate, three to four CaMV-infected or mock-inoculated plants and, in the case of TAV transgenic lines, nine transgenic or wt (Col-0) seedlings grown in soil as described above were harvested 1 month post-inoculation and 15 days post-germination, respectively. Total RNA was prepared by the Trizol method as described above and further purified using an RNeasy Plant Mini Kit (Qiagen, AG, Hombrechtikon, Zurich, Switzerland) following the manufacturer's recommendations. The quality of the isolated RNA was determined with a NanoDrop ND 1000 (NanoDrop Technologies) and a Bioanalyzer 2100 (Agilent Technologies). Only those samples with a 260 nm/280 nm ratio between 1.8–2.1 and a 28S/18S ratio within 1.5–2 were further processed. Total RNA samples (2 µg) were reverse-transcribed into double-stranded cDNA, in vitro transcribed in presence of biotin-labeled nucleotides using an IVT Labeling Kit (Affymetrix), purified and quantified using BioRobot Gene Exp—cRNA Target Prep (Qiagen). The labeled cRNA quality was determined using Bioanalyzer 2100. For array hybridization, biotin-labeled cRNA samples (15 µg) were fragmented randomly to 35–200 bp at 94°C according to the Affymetrix protocol. Samples were hybridized to GeneChip® Arabidopsis ATH1 Genome arrays for 16 h at 45°C. Arrays were then washed using an Affymetrix Fluidics Station 450 FS450 0004 protocol. An Affymetrix GeneChip Scanner 3000 (Affymetrix Inc.) was used to measure the fluorescent intensity emitted by the labeled target. Raw data processing was performed using the Affymetrix AGCC software. The normalized data were analysed using GeneSpring GX* 7.3 (Agilent Technologies).
RESULTS
TAV is responsible for CaMV infection-mediated interference with siR255 biogenesis
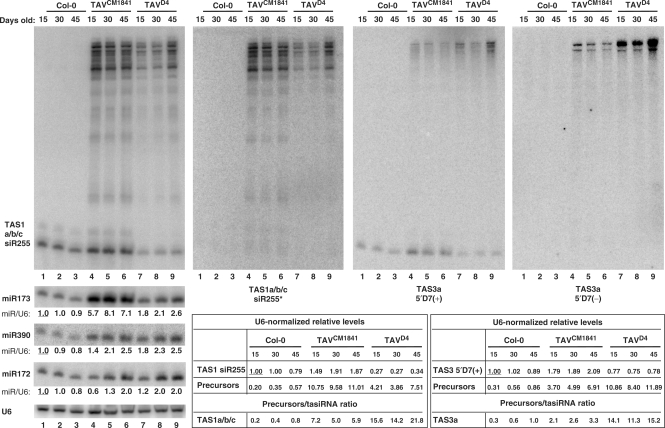
CaMV infection in Arabidopsis thaliana causes overaccumulation of several RDR6-dependent precursors of TAS1-derived tasiRNA siR255 (35) (Figure 2). To test whether the CaMV TAV protein implicated in silencing suppression contributes to this effect, we analyzed two Arabidopsis transgenic lines that constitutively express TAV proteins from the CaMV strains CM1841 and D4 (lines CM-2 and D4-2, respectively) (38). Both lines accumulate comparable levels of TAV, but the CM1841 TAV plants develop extensive chlorotic symptoms, whereas the D4 plants are nearly symptom-less (38). Blot hybridization analysis of total plant RNA using the siR255-specific probe showed that both transgenic lines accumulate, in addition to the 21-nt siR255, several precursors ranging in size from ∼35 to ∼600 nt. The sizes and relative abundances of these RNAs were very similar in both transgenic lines as well as in control Col-0 (wt) plants infected with CaMV strain CM1841. Moreover, CaMV infection of the TAV transgenic lines did not alter these patterns of RNA accumulation (Figure 2). Notably, these long precursors of siR255 are also present in non-infected wt plants, albeit at very low levels (Figure 2 and data not shown). The levels of siR255 normalized for the U6 loading control were reduced to 55% of the wild-type levels in CaMV-infected plants. Of the two TAV transgenic lines, significant reduction in siR255 levels was observed only in the D4 line (see Figures 2 and 3). The stronger interference with siR255 biogenesis in the D4 line than in the CM1841 line was also apparent from a higher ratio of the siR255 precursors (a total of several RNAs ranging from ∼35 to ∼600 nt) to the 21 nt siR255 product, compared to the CM1841 line (Figure 2, see the imbedded table). Nonetheless, the CM1841 line accumulated somewhat higher amounts of the siR255 precursors.
Figure 2.
Accumulation of tasiRNA precursors in CaMV-infected and TAV transgenic plants. Total RNA from pools of CaMV-infected (+) or mock-inoculated (–) wild-type plants (Col-0) and TAV transgenic lines CM1841 and D4 harvested 1 month post-inoculation were analyzed by RNA blot hybridization (using 15% PAGE). Membranes were successively hybridized with DNA oligo probes for the tasiRNAs TAS1a/b/c siR255, TAS3a 5′D7(+), the miRNAs miR173, miR390 and miR828, CaMV-derived siRNAs and the small nuclear RNA U6 (see Table S1 for probe details). Positions of ssRNA markers are indicated. U6 signal serves as a loading control. The U6-normalized, relative levels of miRNAs are shown under the respective scan. The U6-normalized, relative levels of the tasiRNAs and the tasiRNA precursors (a total of several RNAs ranging from ∼35 to 600 nt) as well as the precursor/tasiRNA ratios are shown in the table. For each miRNA and tasiRNA, its accumulation in the uninfected, nontransgenic Arabidopsis (lane 1) was set to 1.0 (underlined). For the tasiRNA precursors, their accumulation was related to the corresponding tasiRNA.
Figure 3.
Differential impact of TAV proteins from CaMV strains CM1841 and D4 on TAS1 versus TAS3 tasiRNAs and their precursors. Total RNA from pools of noninfected, wild-type (Col-0) plants and TAV transgenic lines CM1841 and D4 harvested at 15, 30 and 45 days post-germination were analyzed by RNA blot hybridization (15% PAGE). Membranes were successively hybridized with DNA oligo probes for the tasiRNAs TAS1a/b/c siR255, TAS1a/b/c siR255*, TAS3a 5′D7(+), TAS3a 5′D7(–), the miRNAs miR173, miR390 and miR172, and U6 (see Table S1 for probe details). U6 signal serves as a loading control. The U6-normalized, relative levels of the tasiRNAs and the tasiRNA precursors (a total of several RNAs ranging from ∼35 to 600 nt) as well as the precursor/tasiRNA ratios are shown in the two tables. The U6-normalized, relative levels of miRNAs are shown under the respective scan. For each miRNA and tasiRNA, its accumulation in the nontransgenic Arabidopsis at day 15 (lane 1) was set to 1.0 (underlined). For the tasiRNA precursors, their accumulation was related to the corresponding tasiRNA.
We made a time-course analysis of the TAV effect on the siR255-generating pathway using pools of the transgenic and the wt plants harvested 15, 30 and 45 days post-germination. Wt plants bolted and started to flower 30–45 days post-germination, whereas the transgenic lines displayed slower growth and flowered 1 (CM1841) or 2 (D4) months later, confirming earlier observations (32). In the CM1841 line, the siR255 precursors were equally abundant at all three time points. In contrast, the D4 line accumulated lower amounts of the precursors at the two earlier time points (Figure 3). Despite the difference in accumulation of the precursors, at each time point the precursors/tasiRNA ratio was about two to three times higher in the D4 line than in the CM1841 line (Figure 3), confirming that TAV expression in the D4 line has a stronger negative impact on tasiRNA processing. Interestingly, the levels of miR173 that initiates the siR255-generating pathway (Figure 1) were increased pronouncedly in the CM1841 line (Figure 3), thus correlating with higher accumulation of the siR255 precursors.
Further analysis of infected and TAV transgenic plants using probes specific for both strands of a presumed tasiRNA duplex (siR255/siR255*) detected longer RNAs identical in size and relative abundance but negligible levels of 21-nt siR255* (Figure 3). These results, and our earlier findings for CaMV-infected wt plants (35) (Figure 4) suggest that most larger-sized RNAs are potentially double-stranded molecules. Taken together, we conclude that the CaMV TAV protein is solely responsible for the CaMV-mediated interference with siR255 biogenesis.
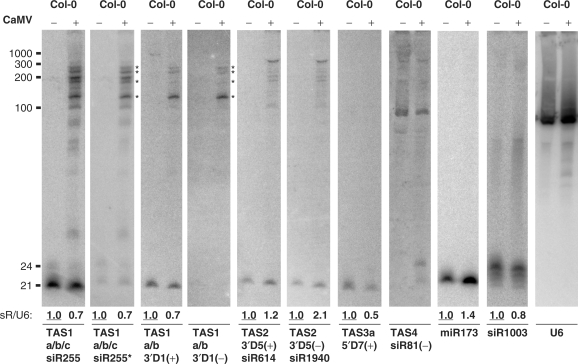
Figure 4.
Impact of CaMV infection on the biogenesis of tasiRNAs from different TAS loci. Total RNA from pools of CaMV-infected (+) and mock-inoculated wild type (Col-0) plants harvested 1 month post-inoculation were analyzed by RNA blot hybridization (15% PAGE). Membranes were successively hybridized with DNA oligo probes for the tasiRNAs TAS1a/b/c siR255, TAS1a/b/c siR255*, TAS1a/b 3′D1(+), TAS1a/b 3′D1(–), TAS2 siR612, TAS2 siR1940, TAS3a 5′D7(+), the miRNA miR173, the 5S rDNA repeat-associated siRNA siR1003 and U6 (see Table S1 for probe details). U6 signal serves as a loading control. Positions of ssRNA markers are indicated. Asterisks indicate the dsRNAs hybridized to both the internal (TAS1a/b/c siR255- and siR255*-specific) and the miR173-terminal [TAS1a/b 3′D1(+)- and 3′D1(–)-specific] probes.
Analysis of dsRNA precursors of TAS1 tasiRNAs
siR255 is encoded by three TAS1 family loci TAS1a, TAS1b and TAS1c at processing positions 3′D6(+), 3′D6(+) and 3′D3(+), respectively, located six, six and three 21-nt cycles downstream of the miR173 cleavage site (6). Four additional siR255-like species are encoded by TAS1a, TAS1b and TAS1c (Figure 1B, filled boxes). To further characterize the long RNAs that hybridize to the siR255- and the siR255*-specific probes in CaMV-infected plants (35), we re-probed the respective blot with P-32 labeled, 21- and 19-nt-long DNA oligonucleotides (Table S1) complementary to the predicted TAS1a 21-nt siRNAs 3′D1(+) and 3′D1(–) (Figure 1B). These probes are expected to hybridize to the termini of presumed dsRNAs from TAS1a and TAS1b with only 1-nt mismatch at the end but not to those from TAS1c and TAS2 with >4 mismatches. Such dsRNAs could be either the hypothetical full-length products of RDR6 activity or partially processed products from an end opposite to the miR173 cleavage site. We detected 21-nt TAS1a 3′D1(+) species in both mock control and CaMV-infected plants, but no 3′D1(–) species (Figure 4). Both TAS1a 3′D1 (+) and 3′D1(–) specific probes also strongly hybridized to four of the 12 longer RNAs detectable with siR255- and siR255*-specific probes in CaMV-infected plant samples (indicated with asterisks in Figure 4).
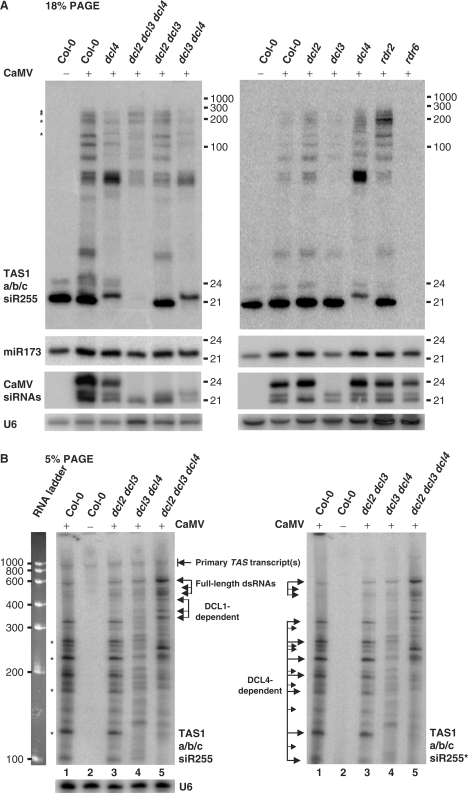
We also analyzed the siR255 precursors in DCL-deficient single, double and triple mutants dcl2, dcl3, dcl4, dcl23, dcl34 and dcl234 as well as RDR-deficient mutants rdr2 and rdr6, described earlier (35). To better resolve longer precursors of tasiRNAs and identify single-stranded (ss) TAS1 transcripts of 800–1000 nt, total RNA was separated using 5% PAGE (Figure 5B) and low-molecular-weight RNAs were separated using 18% PAGE (Figure 5A). The size range of the single-stranded, siR255-containing RNAs we detected (Figure 5B) corresponded to TAS1a, 1b and 1c transcripts of 929, 838 and 989 nt in length, respectively. Strikingly, accumulation of most TAS1-derived RNAs was perturbed in the dcl4 mutant backgrounds relative to the wt and the other dcl mutants (Figure 5A and B). Thus, these RNAs may represent intermediates of DCL4-mediated processing of the longest (full-length) dsRNA precursors of 500–600 bp, which accumulated equally in the wt and all the dcl mutant plants infected with CaMV (Figure 5B). In the absence of DCL4, other DCLs appear to compensate for its deficiency: abundant 22-nt sRNA and several longer intermediates accumulating in dcl4 and dcl34 are DCL2-dependent (Figure 5A and B). The 24- and 25-nt sRNAs and two RNAs migrating at the ca. 80-nt position are DCL3-dependent (Figure 5A). The RNA pattern in dcl234 indicates that full-length precursors of siR255 are partially processed by DCL1 resulting in a distinct pattern of RNA intermediates but no detectable siRNAs (Figure 5). Finally, none of the precursors or mature TAS1 tasiRNAs was detected in rdr6 but all were detected in rdr2 (Figure 5A), showing that RDR6 is required for biogenesis of these tasiRNA precursors. Similar results were obtained for precursors and tasiRNAs encoded by TAS2 and TAS3 loci (data not shown).
Figure 5.
Dissection of the siR255-generating pathway in CaMV-infected DCL- and RDR-deficient mutants. RNA samples from CaMV-infected wild type (Col-0), the single (dcl2-5, dcl3-1 and dcl4-2), double (dcl2 dcl3, dcl3 dcl4; 35) or triple (dcl2 dcl3 dcl4; 35) DCL-deficient and the RDR2- (rdr6-15) and the RDR6-deficient (rdr2-1) mutants harvested 1 month post-inoculation were analyzed by RNA blot hybridization using 18% PAGE for low-molecular-weight RNA (A) and 5% PAGE for total RNA (B). Membranes were successively hybridized with DNA oligo probes for TAS1a/b/c siR255, TAS1a/b/c siR255*, miR173, CaMV siRNAs and U6 (see Table S1 for probe details). U6 signal serves as a loading control. Positions of ssRNA markers are indicated. The dsRNAs hybridized to the miR173-terminal [TAS1a/b 3′D1(+)- and 3′D1(−)-specific] probes in Figure 4 are indicated by asterisks.
CaMV infection and TAV expression interfere with the pathways generating the four known tasiRNA families
We also investigated if expression of the TAS2-TAS4 families was affected by CaMV infection. The single TAS2 locus encodes siR614 and siR1940 species as the fifth cycle duplex [3′D5(+) and 3′D5(–)] downstream of the miR173 site (6) (Figure 1B). In uninfected plants, the probes specific for these tasiRNAs identified the expected 21-nt molecules. CaMV infection induced the accumulation of four additional longer RNAs, whose sizes were similar in both polarities (Figure 4). A similar pattern of TAS2 long dsRNAs was found in the two TAV transgenic lines, with higher levels accumulating in the CM1841 line and a higher precursor/tasiRNA ratio in the D4 line (Figure S1). These findings indicate that CaMV infection and expression of TAV proteins interfere in a similar manner with miR173-initiated, TAS1 and TAS2 pathways.
The miR390-initiated tasiRNAs are derived from three loci of the TAS3 family (Figure 1), with TAS3a giving rise to most of the sRNA reads (9). The TAS3a-specific tasiRNA 5′D7(+) but not its passenger strand 5′D7(–) was detected. Both sense and antisense probes detected long precursors of TAS3a tasiRNAs in CaMV-infected, but not mock-inoculated, wt plants and in both TAV transgenic lines (Figures 2 and 3). Note that these precursors were much less abundant in CaMV-infected plants than in the TAV transgenic lines (Figure 2). In contrast to TAS1 and TAS2, the TAS3a tasiRNA precursors were more abundant in the D4 line than in the CM1841 line and were inversely correlated in abundance with their mature tasiRNAs (Figure 3). Nonetheless, the precursors/tasiRNA ratio was comparable for both TAS3a and TAS1/TAS2 in the D4 line, whereas it was higher for TAS1/TAS2 than TAS3a in the CM1841 line (Figure 3). This suggests that TAV expression in the D4 line exerts strong and comparable effects on both the miR173- and the miR390-initiated tasiRNA biogenesis, whereas TAV expression in the CM1841 line has generally weaker effects on both but a more pronounced effect on the miR173-initiated tasiRNA biogenesis.
The miR828-initiated tasiRNAs derived from the single TAS4 locus (11) are of very low abundance (9). We did not detect any small RNA in control Col-0 plants with the probes specific for the cloned 21 nt TAS4 siR81(–) species or its passenger strand. In both CaMV-infected and TAV transgenic plants, however, ∼24–26 nt small RNA of one polarity was detected using the siR81(–)-specific probe (Figures 4 and S1). This probe, but not its reverse complement (data not shown), detected long RNAs that were equally abundant in control and CaMV-infected plants, but overaccumulated in TAV transgenic plants. At least one distinct long RNA appears to be CaMV TAV-specific (indicated with asterisk in Figure S1). Thus, CaMV infection and TAV expression both interfere with the TAS4 biogenesis.
CaMV infection and TAV expression upregulate both tasiRNA targets and components of tasiRNA biogenesis
Our finding that TAV expression dramatically increased the accumulation of tasiRNA precursors and the ratio of precursors to tasiRNA suggests that TAV impairs processing of RDR6-dependent dsRNA to tasiRNA by DCL4. However, the levels of tasiRNAs themselves are not drastically reduced in CaMV-infected or TAV D4 plants, and, in some samples of the CM1841 plants, are even slightly increased (Figures 2 and 3). This could be due to incomplete spatial and/or temporal overlap of viral infection (TAV protein production) and the tasiRNA biogenesis and activity pathways. Additionally, a feedback regulation may partially or fully restore normal levels of tasiRNAs. We therefore investigated if the action of tasiRNAs in knocking down their target gene transcripts is affected and whether the genes mediating tasiRNA biogenesis are regulated in CaMV-infected and TAV transgenic plants. Using the ATH1 whole-genome microarray, we profiled total RNA from CaMV-infected versus mock-inoculated wt (Col-0) and tasiRNA-deficient dcl234 plants (harvested 1 month post-inoculation) as well as from non-infected transgenic versus wt (Col-0) plants harvested 15 days post-germination. Confirming the above time-course experiment (Figure 3), the siR255 precursors were more abundant in the CM1841 than in the D4 line samples (Figure S2; note that, of the two independent, CM1841 TAV-expressing lines shown, CM-2 was profiled). Notably, the TAV transgene transcript levels were higher in the D4 line than in the two CM1841 lines CM-2 and CM-6 (Figure S2), which is consistent with slightly higher accumulation of TAV protein and its breakdown products in this D4 line (D4-2) compared to the CM1841 lines (38; and data not shown). The higher expression of TAV protein in the D4 line might account for the stronger negative impact on tasiRNA biogenesis in this line as inferred above from the dsRNA precursors/tasiRNA ratios. However we cannot exclude that 5% difference at the amino acid level between the two TAV proteins is responsible for the observed effects.
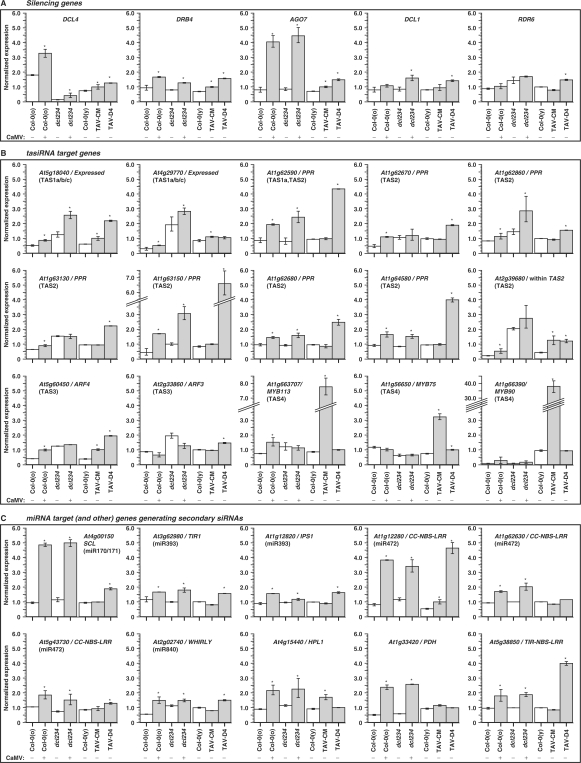
The microarray analysis showed that the key genes involved in tasiRNA biogenesis including DCL4, DRB4 and AGO7 were upregulated by both CaMV infection and TAV expression (Figure 6A, Table S2), thus supporting the feedback regulation hypothesis. Notably, the D4 line exhibited a more pronounced upregulation of these genes than the CM1841 line. Additionally, DCL1 and RDR6 were found to be modestly upregulated in the D4 line (Figure 6A, Table S2). The transcript levels of DRB4 and AGO7 were elevated to similar levels in both CaMV-infected wild-type and dcl234 plants (Figure 6A, Table S2). Interestingly, residual levels of a DCL4 transcript were detected in dcl234 carrying a T-DNA insertion in the 23rd exon of the DCL4 gene and those were also slightly elevated upon CaMV infection. Furthermore, DCL1 was also modestly upregulated in CaMV-infected dcl234 (Figure 6A, Table S2). Other genes presently implicated in the miRNA- and tasiRNA-generating pathways, namely AGO1, HYL1/DRB1, HEN1, SE, SGS3 and SDE5 were not affected by the CaMV challenges. Of the silencing-related genes not implicated in the tasiRNA pathway so far, notable CaMV-mediated upregulation was found for AGO2, TOUSLED, NRPD1a, DDM1, DME, HDA18, RPA2 and FAS2, whereas DRB3 was downregulated (data not shown).
Figure 6.
Response of the Arabidopsis transcriptome to CaMV infection and TAV transgene expression reveals the interference with the tasiRNA and secondary siRNA pathways. The microarray analysis data for CaMV-infected (+) and mock-inoculated (–) Arabidopsis wt (Col-O) and the dcl2 dcl3 dcl4 triple mutant (dcl234) plants as well as for the CaMV TAV transgenic lines CM1841 and D4 are presented as the bar charts with error bars showing standard deviation from mean value of the normalized expression levels (Tables S2–S4) of the genes upregulated in response both to CaMV infection (in Col-0 and dcl234) and TAV expression (in CM1841 and/or D4). The data are for two samples, each representing a pool of three (or, in the case of TAV transgenic lines, nine) plants. The statistically significant changes are indicated by asterisks. (A) shows the genes mediating tasiRNA biogenesis (DCL4, DRB4, AGO7, DCL1 and RDR6), (B) the genes targeted by tasiRNAs of the four TAS families (indicated in brackets) and (C) the miRNA targets (the miRNA indicated in brackets) and other genes silenced by secondary siRNAs.
Further analysis of the microarrays revealed that several predicted tasiRNA target genes were modestly upregulated by CaMV infection (Figure 6B, Table S3). These included (i) two ‘expressed protein’ genes targeted by siR255 and three other tasiRNAs encoded by the TAS1 a/b/c loci, (ii) six PPR genes targeted by single or multiple siRNAs derived from TAS1a or TAS2 loci, (iii) AFR4 targeted by two TAS3a siRNAs and (iv) MYB113 targeted by the TAS4 siR81(–). Thus, subsets of the targets for all the known TAS families were elevated. Most of these were also upregulated in noninfected dcl234 plants, consistent with the tasiRNA deficiency. Interestingly, the target genes unaffected in the noninfected dcl234 plants were upregulated when these plants were infected with CaMV (Figure 6B, Table S3). Subsets of targets for tasiRNA in all four TAS families were also elevated in TAV transgenic lines (Figure 6B, Table S3). The target genes affected and the degree of upregulation depended on both the TAV line and the TAS family. The CM1841 TAV expression had the most pronounced effect on the TAS4 targets with all three known targets of siR81(–) being strongly upregulated, while its effect on the TAS1 and TAS3 targets was less pronounced and on the TAS2 targets rather negligible. In contrast, the D4 TAV expression had almost no effect on the TAS4 targets, whereas most of the TAS2 targets were strongly upregulated (Figure 6B, Table S3). Furthermore, one of the two TAS1 targets and the TAS3 target ARF4 were more pronouncedly upregulated in the D4 line than in the CM1841 line, and the D4 line showed slightly elevated levels of the second TAS3 target, ARF3 (Figure 6B, Table S3).
The gene At2g39680 imbedded within TAS2 (At2g39681) in opposite orientation (7) was upregulated upon CaMV infection and in both TAV transgenic lines (Figure 6B, Table S3). Together with the earlier report on its upregulation in rdr6 (41), our data suggest that At2g39680 is a target of TAS2-derived tasiRNAs, the biogenesis of which is impaired by CaMV TAV.
Many targets of tasiRNAs give rise to RDR6-dependent secondary siRNAs, likely to be involved in amplification of silencing (9). Thus, CaMV TAV-mediated interference with secondary siRNA production could account for upregulation of some of the tasiRNA targets (Table S3). Our further inspection of the transcriptome profiles revealed that both CaMV infection and TAV expression (in one or both lines) cause upregulation of several other Arabidopsis genes (Figure 6C, Table S4) that normally give rise to RDR6-dependent secondary siRNAs (9). Interestingly, many of these genes are targets of miRNAs, like the TAS genes themselves. Notably, TAV expression in the D4 line elevated transcript levels for most of these genes, whereas the CM1841 line showed elevated levels for only a few of them (Figure 6C), which is similar to the above observations for the TAS2 targets (Figure 6B). The vast majority of other miRNA targets, which does not give rise to secondary siRNAs, was not upregulated in CaMV-infected or TAV transgenic plants (data not shown), suggesting that TAV does not interfere with miRNA action. The notable exception is AGO1, which represents an miRNA target gene generating secondary siRNAs that was not upregulated by TAV (data not shown). However, expression of this gene is auto-regulated at both post-transcriptional and transcriptional levels by a complex fine-tuning mechanism (42), which could fully compensate for the inhibitory effect of TAV.
CaMV interferes with production of RDR6-dependent secondary siRNAs from a silenced transgene
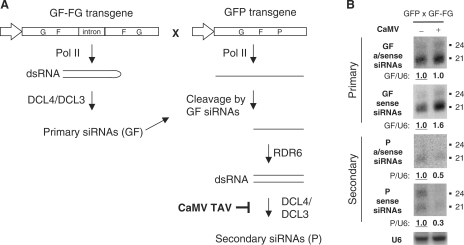
Our transcriptome analysis suggests that TAV interferes with RDR6-dependent secondary siRNA pathways. To test this hypothesis we used the Arabidopsis GFP × GF-FG transgenic line (22), which carries a green fluorescent protein (GFP) transgene driven by the CaMV 35S promoter and a second transgene expressing an inverted repeat cognate to the GF portion of the GFP coding region (GF-FG). In this system, the 21-nt primary siRNAs produced by DCL4-mediated processing of the GF-FG dsRNA are thought to target the GFP transcript for cleavage and degradation. The latter process triggers RDR6-dependent production of secondary siRNAs from the P region of the GFP transgene (Figure 7A). The rdr6 mutation introduced into the GFP × GF-FG line has been shown to eliminate the secondary siRNAs without affecting accumulation of primary siRNAs or GFP silencing (22). CaMV infection of the GFP × GF-FG line did not result in re-appearance of GFP fluorescence under UV light (data not shown), indicating that GFP silencing was not suppressed. Consistent with the silenced GFP phenotype, RNA blot hybridization of total plant RNA with probes specific to the GF region showed that accumulation of primary siRNAs of both polarities was not reduced but rather slightly increased (Figure 7B). In contrast, accumulation of secondary siRNAs from the P region was substantially reduced during viral infection (Figure 7B). These results suggest that CaMV infection selectively interferes with the biogenesis of RDR6-dependent secondary siRNAs without any substantial effect on the production of primary siRNAs and their action in silencing GFP expression.
Figure 7.
CaMV infection interferes with the biogenesis of secondary siRNAs derived from the GFP transgene silenced by GF-FG dsRNA. (A) Model for the biogenesis of the primary (GF) and the secondary (P) siRNAs derived from the GF-FG and the GFP transgenes, respectively, in the transgenic GF-FG × GFP line. The structures of the two transgenes present in this line as shown schematically, with the CaMV 35S promoter indicated by an arrow shape. In the GF-FG transgene the inverted repeat of the GF region of the GFP coding sequence is equipped with an intron. The Pol II transcription generates a GF-FG transcript that folds back into perfect dsRNA. The latter is processed by DCL4 (and DCL3) into primary siRNAs (GF). These siRNAs target in trans a GFP mRNA transcribed from the GFP transgene for cleavage at multiple positions within the GF region. The 3′-product of these cleavage events retaining an intact P region is converted by RDR6 into dsRNA. The resulting dsRNA is processed by DCL4 (and DCL3) into secondary siRNAs (P). The later step is selectively impaired by CaMV infection as inferred from the following experiment.(B) Total RNA from CaMV-infected (+) and mock-inoculated (–) transgenic GF-FG X GFP line plants harvested 1 month post-inoculation were analyzed by RNA blot hybridization (15% PAGE). Membranes were successively hybridized with DNA oligo probes specific for sense and antisense siRNAs derived from the GF and P regions of the GFP coding sequence and U6 (see Table S1 for probe details). U6 signal serves as a loading control. The U6-normalized, relative levels of siRNAs are shown under the respective scans. In each case, the siRNA accumulation in the noninfected Arabidopsis was set to 1.0 (underlined).
DISCUSSION
TAV interferes with RDR6-/DCL4-dependent silencing systems
We established that the CaMV TAV protein, whether expressed from the viral genome or from a transgene, interferes with Arabidopsis tasiRNA-generating pathways. This interference is associated with (i) overaccumulation of unprocessed, RDR6-dependent precursors of tasiRNAs; (ii) increased levels of some Arabidopsis mRNAs normally silenced by tasiRNAs of four known TAS families; and (iii) upregulation of genes encoding DCL4, DRB4 and AGO7, which are involved in tasiRNA biogenesis. We also found that CaMV TAV interferes with RDR6-dependent secondary siRNA pathways. The latter finding is consistent with the ability of TAV to suppress amplicon transgene-induced silencing (34) that depends on RDR6 (43). Furthermore, CaMV siRNAs accumulating in infected plants are largely RDR6- and DCL4-independent (35; Figure 5A). Taken together, these findings suggest that TAV suppresses silencing by preventing RDR6-/DCL4-dependent amplification of secondary siRNAs.
TAV might act at the step of DCL4-mediated processing of RDR6-dependent dsRNA
The available evidence suggests that TAV impairs processing of RDR6-produced dsRNA to small RNAs. In the case of tasiRNAs, this is likely to occur in an RDR6/DCL4 processing center believed to recruit TAS transcripts targeted for cleavage by miRNAs (15). This processing center might also generate secondary siRNAs from some genes targeted by miRNAs or primary siRNAs (9). Exactly how TAV inhibits processing is not known. One possibility, which we favor, is that TAV interacts with a protein or proteins in the processing center downstream of RDR6 (Figure 1A). Potential targets include dsRNA-binding proteins, such as DRB4, which is known to interact with DCL4 (18), or DCL4 itself. Another possibility, which is not mutually exclusive, is that TAV binds dsRNA precursors and thereby inhibits their processing to siRNAs by DCL4. While this hypothesis is consistent with the presence of a conserved dsRNA-binding motif in TAV (44,45), we have not been able to detect binding of TAV to either long dsRNA and short RNA duplexes (Shivaprasad,P.V. and Burgyan,J., unpublished data) using a sensitive assay (46). Moreover, several lines of evidence suggest that binding of TAV to dsRNAs per se is not sufficient to block downstream processing. Thus, we found that, in the presence of TAV, DCLs can still efficiently process non-RDR-derived dsRNAs such as a fold-back structure of the GF-FG transcript (Figure 7). Similarly, RDR6-/RDR2-independent primary CaMV siRNAs presumably processed from dsRNAs derived from sense and antisense viral transcripts (35) were not affected by TAV overexpression in transgenic plants (Figure 2). In future studies, it will be interesting to analyze how TAV-mediated suppression selectively impairs processing of RDR6 products and identify which DCL4 co-factor could be responsible for selective action of DCL4 on such dsRNA substrate.
Novel features of tasiRNA pathways
Our functional characterization of TAV uncovered novel mechanistic and regulatory features of tasiRNA-generating pathways. RDR6-dependent, potentially double-stranded precursors of tasiRNAs, which overaccumulated in the presence of TAV, had been previously inferred from small RNA cloning and genetic evidence (6) but escaped detection so far. Using dcl mutants and RNA blot hybridization analysis, we verified that these precursors are preferentially processed by DCL4 and identified intermediates (and/or byproducts) of this processing. Notably, the estimated sizes of the longest (full-length) TAS1-derived RDR6-dependent sense and antisense RNAs fall into a range of 450–600 nt (Figure 5B), suggesting that complete products of miR173-guided cleavage of TAS1 transcripts are converted by RDR6 into dsRNA. It was proposed that TAS1- and TAS2-derived dsRNAs are preferentially processed from the miRNA cleavage site terminus (6). Unexpectedly, we discovered that some abundant, DCL4-dependent, shorter-than-full-length TAS1 sense and antisense RNAs retain the miR173-cleavage site terminus (Figures 4 and 5). This might be due to processing of TAS1 dsRNA precursors from the opposite side. Alternatively, at a first round of tasiRNA biogenesis, certain antisense tasiRNAs produced from the full-length dsRNAs might target the miR173-cleaved ssTAS transcript and thus create secondary, shorter substrates for RDR6. Such a two hit trigger mechanism has been proposed for biogenesis of TAS3-derived tasiRNAs as well as for secondary siRNAs derived from some Arabidopsis protein-coding genes (9,12). Our work in progress aims to precisely map the termini of the full-length precursors of tasiRNAs and their derivatives described here, which should lead to new insight in the biogenesis of tasiRNAs and other RDR6-dependent siRNAs.
Our transcriptome analysis suggests that the tasiRNA pathway is subject to negative auto-regulation. TAV impairs processing of tasiRNAs and secondary siRNAs, leading to increased accumulation of their target transcripts. But it also upregulates genes coding for proteins that mediate siRNA biogenesis, including DCL4, DRB4 and AGO7. Together with the rather modest effects of TAV expression on target genes, this implies that deficiencies in the tasiRNA pathway can lead to a compensatory activation of the key pathway components that partially or fully restores tasiRNAs and target mRNA levels. This hypothesis explains the difference observed for the two distinct TAV transgenic lines, one of which (D4) accumulating higher levels of target mRNAs and lower levels of tasiRNAs than the other (CM1841), at least for TAS1, TAS2 and TAS3 pathways (see Figures 3, 6 and S2, respectively). This would imply more efficient, feedback-mediated restoration of the tasiRNA and mRNA target levels in the CM1841 line than in the D4 line. Nonetheless in both lines, as a net effect of the ongoing TAV-mediated inhibition of tasiRNA processing and the feedback upregulation of tasiRNA biogenesis, the unprocessed precursors of TAS1, TAS2 and TAS3 tasiRNAs overaccumulate. The miRNA pathway genes DCL1 (11,47) and AGO1 (42) are known to be auto-regulated. Our study supports the view that negative feedback regulation is a general feature of small RNA silencing pathways in plants. The mechanism of feedback regulation of the tasiRNA pathway remains to be investigated. Like in the case of miRNA-mediated regulation of DCL1 and AGO1 (11,42, 47), the mRNAs of DCL4, DRB4 and AGO7 may be targeted by some tasiRNAs and/or secondary siRNAs, the biogenesis of which is mediated by the DCL4, DRB4 and AGO7 proteins. Therefore, TAV-mediated interference with the biogenesis of such siRNAs would elevate the levels of the respective mRNAs. To our knowledge, no RDR6-dependent siRNA cognate to DCL4, DRB4 or AGO7 sequences have been reported.
TAV might suppress RNA-silencing based antiviral defense
Virus infection involves a complex interplay of host defense mechanisms and counter-defense mechanisms of the virus (24,48). We believe that one function of TAV is to suppress host defense mechanisms concerned with restricting virus spread by systemic RNA silencing. RDR6-/DCL4-dependent mechanisms generating secondary siRNAs from silenced transgenes (22) and endogenous genes (9), which are inhibited by TAV, appear to contribute to the amplification and spread of transgene silencing (20,23). RDR6 is required for exclusion of potato virus X from shoot apical meristem (26) and defense against cucumber mosaic virus (25). We propose that by interfering with the amplification of CaMV secondary siRNAs, TAV reduces the effectiveness of systemic silencing. This is consistent with the finding that secondary viral siRNAs do not appear to accumulate in CaMV-infected tissues: most CaMV siRNAs are RDR6-independent (35; Figure 5A), while DCL4, which restricts RNA-virus infection through production of 21-nt viral siRNA (37), makes only a minor contribution to production of CaMV siRNAs (35; Figure 5A).
Many viruses overcome host defenses by expressing viral proteins that suppress RNA silencing (48). Overexpression of these suppressors in transgenic plants often results in developmental abnormalities that result from the interference of these proteins with the miRNA pathway (49). Thus, some abnormalities associated with virus infection may be ‘collateral’ effects of the viruses counter-defense to RNA silencing. Finally, the functional redundancy of components of the miRNA pathway and their negative auto-regulation by miRNAs might be additional layers of host defense. We assume that a similar interplay between host and virus defenses affects tasiRNA and other secondary siRNA pathways targeted by TAV. While our data show that TAV does not generally affect miRNA pathways, overexpression of TAV in transgenic plants leads to various abnormalities including delayed flowering (32,33,38; and this study) that appear to be correlated with inhibition of tasiRNA processing. Interestingly, recent studies using the GF-FG × GFP transgenic line established that suppressor proteins of two RNA viruses, the potyvirus HC-Pro and the carmovirus P38, interfere with the accumulation of secondary, but not primary, GFP siRNAs and endogenous TAS1-derived tasiRNAs (22). Feedback regulation in the TAS pathway may well explain conflicting findings that HC-Pro transgene expression does (22) or does not (50) affect accumulation of TAS1 tasiRNAs.
Endnote
While the manuscript of this paper was under review, Haas et al. (51) reported that transgene expression of TAV protein from the CaMV Cabb B-JI strain suppresses silencing through physical interaction with DRB4 protein, which leads to reduction of siR255 levels. They also reported that siR255 precursors accumulate in a drb4 mutant line. Thus, their work confirms our findings for the CaMV D4 TAV line and further suggests that TAV binding to DRB4 might interfere with DCL4-DRB4 co-operation in processing tasiRNAs, which would in turn trigger the feedback upregulation of tasiRNA biogenesis components including DCL4, AGO7 and DRB4 itself as reported here.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Indo-Swiss Collaboration in Biotechnology and Swiss National Science Foundation. PVS was in part supported by a short-term EMBO fellowship. Funding for open access charge: Novartis Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Nachelli Malpica for technical assistance, Marzanna Künzli (Zürich Centre for Functional Genomics) for microarray hybridization and normalization, Wilhelm Gruissem (Institute of Plant Sciences, ETH, Zürich) for providing the transcriptomics platform, and Olivier Voinnet for providing seed of the GF-FG/GFP line. We are grateful to Thomas Boller, Andres Wiemken and Christian Körner for hosting the group at the Botanical Institute.
REFERENCES
- 1.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. RNA-directed DNA methylation and Pol IVb in Arabidopsis. Cold Spring Harb. Symp. Quant. Biol. 2006;71:449–459. doi: 10.1101/sqb.2006.71.028. [DOI] [PubMed] [Google Scholar]
- 2.Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 3.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 4.Hohn T, Akbergenov R, Pooggin MM. Production and transport of the silencing signal in transgenic and virus-infected plant systems. In: Waigmann E, Heinlein M, editors. Viral Transport in Plant. Berlin, Germany: Springer; 2007. pp. 127–157. [Google Scholar]
- 5.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 6.Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curaba J, Chen X. Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J. Biol. Chem. 2008;283:3059–3066. doi: 10.1074/jbc.M708983200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 13.Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouché N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 14.Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 15.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 Interaction and Dual Functionality in TAS3 Trans-Acting siRNA Formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 16.Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 17.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 19.Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T. The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol. Biol. 2007;63:777–785. doi: 10.1007/s11103-006-9125-8. [DOI] [PubMed] [Google Scholar]
- 20.Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- 21.Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 2008;582:2753–2760. doi: 10.1016/j.febslet.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Moissiard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brosnan CA, Mitter N, Christie M, Smith NA, Waterhouse PM, Carroll BJ. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl Acad. Sci. USA. 2007;104:14741–14746. doi: 10.1073/pnas.0706701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 26.Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothnie HM, Chapdelaine Y, Hohn T. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv. Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- 28.Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell. 2001;106:723–733. doi: 10.1016/s0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- 29.Haas M, Geldreich A, Bureau M, Dupuis L, Leh V, Vetter G, Kobayashi K, Hohn T, Ryabova L, Yot P, et al. The open reading frame VI product of Cauliflower mosaic virus is a nucleocytoplasmic protein: its N terminus mediates its nuclear export and formation of electron-dense viroplasms. Plant Cell. 2005;17:927–943. doi: 10.1105/tpc.104.029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Himmelbach A, Chapdelaine Y, Hohn T. Interaction between cauliflower mosaic virus inclusion body protein and capsid protein: implications for viral assembly. Virology. 1996;217:147–157. doi: 10.1006/viro.1996.0102. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi K, Hohn T. The avirulence domain of Cauliflower mosaic virus transactivator/viroplasmin is a determinant of viral virulence in susceptible hosts. Mol. Plant Microbe. Interact. 2004;17:475–483. doi: 10.1094/MPMI.2004.17.5.475. [DOI] [PubMed] [Google Scholar]
- 32.Zijlstra C, Scharer-Hernandez N, Gal S, Hohn T. Arabidopsis thaliana expressing the cauliflower mosaic virus ORF VI transgene has a late flowering phenotype. Virus Genes. 1996;13:5–17. doi: 10.1007/BF00576974. [DOI] [PubMed] [Google Scholar]
- 33.Cecchini E, Gong Z, Geri C, Covey SN, Milner JJ. Transgenic Arabidopsis lines expressing gene VI from cauliflower mosaic virus variants exhibit a range of symptom-like phenotypes and accumulate inclusion bodies. Mol. Plant Microbe. Interact. 1997;10:1094–1101. doi: 10.1094/MPMI.1997.10.9.1094. [DOI] [PubMed] [Google Scholar]
- 34.Love AJ, Laird J, Holt J, Hamilton AJ, Sadanandom A, Milner JJ. Cauliflower mosaic virus protein P6 is a suppressor of RNA silencing. J. Gen. Virol. 2007;88:3439–3444. doi: 10.1099/vir.0.83090-0. [DOI] [PubMed] [Google Scholar]
- 35.Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F., Jr, Hohn T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moissiard G, Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc. Natl Acad. Sci. USA. 2006;103:19593–19598. doi: 10.1073/pnas.0604627103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 38.Yu W, Murfett J, Schoelz JE. Differential induction of symptoms in Arabidopsis by P6 of Cauliflower mosaic virus. Mol. Plant Microbe. Interact. 2003;16:35–42. doi: 10.1094/MPMI.2003.16.1.35. [DOI] [PubMed] [Google Scholar]
- 39.Akbergenov R, Si-Ammour A, Blevins T, Amin I, Kutter C, Vanderschuren H, Zhang P, Gruissem W, Meins F., Jr., Hohn T, et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34:462–471. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobayashi K, Tsuge S, Stavolone L, Hohn T. The cauliflower mosaic virus virion-associated protein is dispensable for viral replication in single cells. J. Virol. 2002;76:9457–9464. doi: 10.1128/JVI.76.18.9457-9464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 44.Cerritelli SM, Fedoroff OY, Reid BR, Crouch RJ. A common 40 amino acid motif in eukaryotic RNases H1 and caulimovirus ORF VI proteins binds to duplex RNAs. Nucleic Acids Res. 1998;26:1834–1840. doi: 10.1093/nar/26.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi K, Hohn T. Dissection of cauliflower mosaic virus transactivator/viroplasmin reveals distinct essential functions in basic virus replication. J. Virol. 2003;77:8577–8583. doi: 10.1128/JVI.77.15.8577-8583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, López-Moya JJ, Burgyán J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 48.Burgyán J. Role of silencing suppressor proteins. Methods Mol. Biol. 2008;451:69–79. doi: 10.1007/978-1-59745-102-4_5. [DOI] [PubMed] [Google Scholar]
- 49.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mlotshwa S, Pruss GJ, Peragine A, Endres MW, Li J, Chen X, Poethig RS, Bowman LH, Vance V. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS ONE. 2008;3:e1755. doi: 10.1371/journal.pone.0001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haas G, Azevedo J, Moissiard G, Geldreich A, Himber C, Bureau M, Fukuhara T, Keller M, Voinnet O. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J. 2008;27:2102–2112. doi: 10.1038/emboj.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.