Abstract
Single molecule analysis of individual enzymes can require oriented immobilization of the subject molecules on a detection surface. As part of a technology development project for single molecule DNA sequencing, we faced the multiple challenges of immobilizing both a DNA polymerase and its DNA template together in an active, stable complex capable of highly processive DNA synthesis on a nonstick surface. Here, we report the genetic modification of the archaeal DNA polymerase 9°N in which two biotinylated peptide ‘legs’ are inserted at positions flanking the DNA-binding cleft. Streptavidin binding on either side of the cleft both traps the DNA template in the polymerase and orients the complex on a biotinylated surface. We present evidence that purified polymerase–DNA–streptavidin complexes are active both in solution and immobilized on a surface. Processivity is improved from <20 nt in the unmodified polymerase to several thousand nucleotides in the engineered complexes. High-molecular weight DNA synthesized by immobilized complexes is observed moving above the surface even as it remains tethered to the polymerase. Pre-formed polymerase–DNA–streptavidin complexes can be stored frozen and subsequently thawed without dissociation or loss of activity, making them convenient for use in single molecule analysis.
INTRODUCTION
Significant interest in the sequencing of single DNA molecules dates to 1989 when Keller and colleagues began experimenting with ‘sequencing by degradation’. In their experiments, isolated fully labeled DNA molecules are degraded by an exonuclease, and individual labeled bases are detected as they are sequentially cleaved from the DNA (1–3). This approach was ultimately compromised by poor DNA solubility caused by the densely packed dye labels (R. Rigler, personal communication). More recently, alternative single-molecule approaches have been investigated, including ‘sequencing by synthesis’, where bases are detected one at a time as they are sequentially incorporated into DNA by a polymerase (4–6); and nanopore sequencing where electrical signals are detected while single DNA molecules pass through protein or solid-state nanopores (7–9). So far, only sequencing by synthesis has been successful. In the method of Quake and colleagues (4), base-labeled nucleotide triphosphates (dNTPs) are incorporated into DNA immobilized on a microscope coverglass. Each type of dNTP is applied separately in a fluidic cycle, and incorporated bases are imaged on the surface after washing away the excess of free nucleotides. While the obtained sequence reads are short, high sequencing rates can potentially be achieved by analyzing billions of different, individual molecules in parallel (http://www.helicosbio.com), with applications in resequencing and gene expression profiling. Other applications such as denovo sequencing or cancer genome sequencing would benefit from longer reads.
To obtain long single-molecule reads, potentially tens of kilobases, we and others (5) (http://www.pacificbiosciences.com, http://visigenbio.com) are developing sequencing-by-synthesis approaches using phosphate-labeled nucleotides. These nucleotides are labeled with a fluorophore on the terminal phosphate instead of on the base. Labeled nucleotides are detected while bound to polymerase during the catalytic reaction. The label is released with pyrophosphate as the nucleotide is incorporated into DNA. An advantage is that the DNA remains label-free and fully soluble. Individual polymerase enzymes immobilized on a microscope coverglass would be monitored in real time to detect the sequence of incorporated nucleotides. In order to achieve long reads, the polymerase, but not the DNA, should be attached to the coverglass. Polymerase attachment facilitates detection because it keeps the active site at a single position on the coverglass surface. In the alternative format, with the polymerase in solution and the DNA attached, the enzyme active site would be a moving target for detection, diffusing up to several microns from the DNA attachment point as the primer strand is extended from long templates.
Our approach using immobilized polymerases places three system requirements on the enzyme. First, it should efficiently utilize phosphate-labeled nucleotides. Starting with the Therminator™ variant of 9°N DNA polymerase (10,11) (http://www.neb.com), we used directed evolution to improve activity with phosphate-labeled dNTPs 26-fold, from 0.8 to 21 nt/s measured at 74°C (manuscript in preparation). This mutant polymerase was the ‘parent’ enzyme for the work presented here (see Supplementary Material). The second requirement is polymerase attachment to a nonstick coverglass surface, oriented to permit access to the DNA and nucleotide substrates while allowing for the normal protein conformational changes associated with the catalytic cycle (12). The third requirement is for extreme processivity, with the polymerase holding on to the same template DNA molecule for the duration of the sequencing run. Early release would terminate the run and limit the read length. Therminator polymerase, which we initially chose for its native tolerance of phosphate-modified dNTPs, has low processivity, <20 nt (see Supplementary Material). In this report, we describe a protein structural modification meeting the dual requirements of immobilization and processivity.
In natural systems, most DNA polymerases depend on accessory proteins to enhance processivity. For example, bacteriophage T7 DNA polymerase encircles DNA by binding with Escherichia coli thioredoxin (13,14); herpes simplex virus DNA polymerase associates with a DNA-binding protein (15); and both prokaryotic and eukaryotic replicative polymerases attach to toroidal DNA sliding clamp proteins (16–21). In contrast, φ29 DNA polymerase achieves processivity without accessory proteins by bridging the DNA-binding cleft with an integral protein ‘finger’ called TPR2 (12,22). Several groups have now engineered processivity into polymerases by incorporating aspects of these natural mechanisms. For example, to mimic the T7 DNA polymerase-thioredoxin mechanism, processivity was enhanced in E. coli DNA polymerase I by fusing it with an E. coli thioredoxin-binding domain (23). A similar approach fused Taq DNA polymerase with the thioredoxin-binding domain of phage T3 DNA polymerase (24). An archaeal PCNA-like sliding clamp was recruited to improve the processivity of Taq polymerase by fusing it with cognate archaeal-binding domain (25). Wang et al. (26) fused the DNA-binding protein Sso7d to Taq and Pfu DNA polymerases, thereby improving processivity in both enzymes without involving accessory proteins.
In this report, we describe an artificial processivity complex that both traps the template DNA on the polymerase and facilitates oriented immobilization on biotinylated surfaces. Starting with the parent polymerase (above) adapted to phosphate-labeled dNTPs, we inserted AviTag™ peptide ‘legs’ at two surface-exposed locations flanking the DNA-binding cleft. The AviTag peptides provide highly specific sites for enzymatic biotinylation of the polymerase by E. coli biotin–protein ligase. Processivity is enhanced with streptavidin binding the AviTag legs, retaining the template in the DNA-binding cleft. We show that the template DNA is stably associated with the polymerase, and that the polymerase–DNA–streptavidin complexes are active both in solution and when immobilized on biotinylated coverglass surfaces. We demonstrate that the clamp converts a naturally nonprocessive DNA polymerase into a highly processive one capable of incorporating thousands of nucleotides without dissociating from the template.
MATERIALS AND METHODS
Materials
Buffer C was used for protein dilutions and polymerase assays: 10 mM Tris–Cl pH 8.0, 50 mM KCl, 0.1% Tween-20, 0.1 mM EDTA; ‘10×’ buffer C is 10-fold concentrated buffer C. Isoelectric focusing gels were from Invitrogen, Carlsbad, CA (Novex pH 3-10, Cat No. EC6655A5). Alexa Fluor-680-labeled streptavidin was from Invitrogen and unlabeled NeutrAvidin was from Thermo Scientific/Pierce Biotechnology, Rockford, IL. Biotin-coated magnetic beads used in purifying the complexes were from Bangs Laboratories, Fishers, IN (BioMag beads cat #BM552, 1.5 μm diameter, concentration 5.2 mg/ml, binding capacity 3.5 mg streptavidin/ml).
Polymerase AviTag constructs
The starting enzyme was a mutant of Therminator DNA polymerase (http://www.neb.com) adapted by directed evolution for efficient utilization of phosphate-labeled nucleotides (manuscript in preparation). AviTag is a peptide substrate for E. coli biotin–protein ligase which, when fused to a target protein, provides a site for efficient enzymatic biotinylation (http://www.avidity.com). The overlapping primer PCR method of Chiu et al. (27) was used to insert AviTag in the mutant polymerase at two positions (Therminator coordinates K53-V54 and K229-F230). The 21-amino acid insertion ssGLNDIFEAQKIEWHEgass comprises AviTag (upper case) flanked by arbitrarily chosen amino acids (lower case); enzymatic biotinylation occurs at the epsilon-amine of the lysine (K). The starting plasmid was a 6.4-kb pBAD-HisC plasmid (Invitrogen) containing the mutant polymerase gene. Details of the construction procedures are given in Supplementary Material.
Polymerase purification
The His-tagged polymerases were expressed from a pBAD plasmid vector (Invitrogen) either in E. coli TOP-10 (for nonbiotinylated polymerase; Invitrogen) or in E. coli AVB100 (for in vivo biotinylation; http://www.avidity.com). Briefly, 25 ml cultures were inoculated from an overnight culture and grown under ampicillin selection to an absorbance at 600 nm of 0.6–0.8 at 37°C. The cells were then induced by adding arabinose (0.04% final) and grown for an additional 4 h at 37°C. After harvesting by centrifugation, the induced cells were incubated with 2.5 mg/ml lysozyme in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.05% Tween-20) containing protease inhibitors for 20 min on ice. Following two freeze/thaw cycles, the lyzed cells were sonicated to decrease viscosity, heated to 75°C for 15 min to denature E. coli enzymes, and precleared by centrifugation. The lysate was incubated for 1 h at 4°C with Ni-NTA-agarose (Qiagen, Valencia, CA, USA) to capture the His-tagged proteins. The resin was washed with 20 mM imidazole buffer and protein eluted with buffer containing 200 mM imidazole. The elution buffer was exchanged in one of two ways, depending on whether or not the purified polymerase was to be further biotinylated in vitro. For the parent polymerase without AviTag legs, the preparation was dialyzed overnight against storage buffer (10 mM Tris–HCl pH 8.0, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT). With the imidazole now removed, the protein concentration was determined by UV absorbance (extinction coefficient calculated by amino acid composition at http://expasy.org/tools/protparam.html, giving a molar extinction coefficient for the parent polymerase of 144 000/cm at 280 nm), IGEPAL CA-630 detergent (Sigma-Aldrich, St. Louis, MO) was added to 0.02% w/v, and an equal volume of glycerol was added to a final concentration of 50% for storage at −20°C. Typical final protein concentrations were 1–5 μM in polymerase. Alternatively, to further biotinylate those polymerase variants having AviTag legs, the protein eluted in imidazole buffer was washed three times on a Microcon YM-30 centrifugal filter (Millipore, Billerica, MA) by first centrifuging up to 1 ml of sample at 14 000g for 12 min. The sample was washed three times by rediluting the concentrated protein in 500 μl of 10 mM Tris–Cl pH 8.0 and centrifuging. Protein concentration was measured as above and the sample was biotinylated in vitro using a kit from Avidity, Aurora, CO (cat #BIRA500) at ≤12 μM in polymerase. After incubating at 30°C for 4 h, the sample was washed five times in storage buffer using a YM-30 centrifugal filter. Protein concentration was measured by UV absorbance, then IGEPAL and glycerol were added as above. In vitro biotinylation was found not to improve the level of biotinylation of polymerase expressed in E. coli AVB100 (data not shown, but measured as in Figure 3). Still, the in vivo-biotinylated polymerase preparations were routinely biotinylated further, in vitro, in order to always ensure maximum levels of biotinylation.
Figure 3.
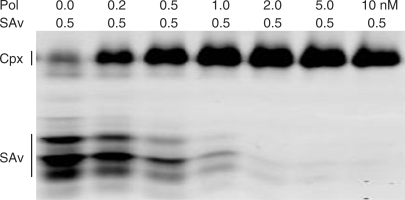
Binary complexes of polymerase and streptavidin. Complexes were formed by incubating 0.5 nM Alexa Fluor-680 labeled streptavidin (Invitrogen) plus 0.2–10.0 nM purified DBio polymerase (i.e. two legs) at 37°C for 10 min. The complexes were resolved by isoelectric focusing and the gel was scanned using a LI-COR Odyssey infrared imager. The positions of the binary complexes (Cpx) and unbound streptavidin (SAv) are indicated. A small fraction of the unbound streptavidin appears to have the same isoelectric point as the binary complexes (first lane).
Primed M13 DNA
The DNA (20 nM) and primer (30 nM) were annealed in a 400 μl volume containing 272 μl of water, 40 µl of 10× buffer salts (100 mM Tris–Cl pH 7.5, 500 mM KCl), 76 μl of M13 single-stranded DNA (104 nM; New England Biolabs, Ipswich, MA) and 12 μl of either an unlabeled primer (1 μM of 5′-cgcctgcaacagtgccacgctgagagcc, desalted grade, Integrated DNA Technologies, Inc., Coralville, IA) or a 5′-labeled primer (1 μM of IRDye™-700 5′-cacgacgttgtaaaacgacggccagtgc, LI-COR Biosciences Inc.). The samples (4 each of 100 μl aliquots) were heated in a PCR thermocycler (Bio-Rad Laboratories/MJ Research, Hercules, CA) at 95°C for 2 min, 60°C for 10 min, ramp 0.1°C/s to 30°C and were stored at −20°C.
Therminator processivity
A gel-based primer extension assay was used to determine processivity in the parent polymerase (see Supplementary Material). Processivity values were calculated as the inverse of the decay constant in exponential fits to lane profiles of electrophoretic band intensities. Native Therminator processivity was <20 nt as measured under conditions similar to those used in experiments with complexes.
Ternary complexes
A 50-μl sample containing 14 nM of primed M13 ssDNA, 28 nM of polymerase and 56 nM of streptavidin was prepared in two steps by first mixing 8 μl of water, 5 μl of 10× buffer C, 35 μl of primed M13 ssDNA and 1.4 μl of 1 μM polymerase; incubating at 55°C for 2 min; adding 0.56 μl of 5 μM streptavidin and incubating at 37°C for 15 min. Required protein dilutions were in buffer C. Complexes were purified from excess streptavidin (total streptavidin 2.8e-12 moles) by adding 10 μl (52 ng) of BioMag beads (streptavidin binding capacity 3.1e-12 moles/52 ng; Bangs Labs) and inverting the sample on a rotating wheel at 12 r.p.m. for 10 min, which allows time for fast binding of free streptavidin but not for the much slower binding of the stearically encumbered complexes. The beads were removed with a magnet (Promega, Madison, WI), the 60 μl supernatant was transferred to a new tube and the bead-binding procedure was repeated as before. The purified complexes (70 μl supernatant) were stored at 4°C for up to a week, or stored frozen at −80°C for future use. Assuming 100% conversion of the M13 DNA into complexes, the final concentration would be 10 nM in complexes.
Polymerization kinetics of ternary complexes
Primer extension on purified complexes was quantified as a function of dNTP concentration. Purified ternary complexes were prepared as described (see Ternary complexes section) using the 5′-labeled primed M13 template (see Primed M13 DNA section). Primer extension reactions were initiated by mixing 6 μl of 2× concentrated component mix (below) plus 6 μl of 10 nM complexes. Incubation was at 54°C for 60 s and reactions were terminated by adding 12 μl of formamide–EDTA gel loading buffer (LI-COR cat# 830-04997). The final concentration of all reaction components was 20 mM Tris–HCl pH 9.2, 50 mM KCl, 5 mM MgSO4, 0.02% IGEPAL and 1–400 μM each of the four unlabeled dNTPs. Control samples with uncomplexed polymerase were as above, but replacing complexes with a mixture of 11 nM of M13 DNA, 10 nM of labeled primer and a saturating amount of polymerase (as shown by doubling the polymerase amount with no effect on the kinetic measurements). Primer extension products were resolved by electrophoresis in a 10% polyacrylamide TBE–Urea slab gel and were detected by fluorescence (LI-COR 4200 DNA Analyzer, Lincoln, NE, USA). A size standard was included on the gel to calibrate the lengths of the primer extension products. The number-average of nucleotides added per second per complex was determined by image analysis using Image J (http://rsb.info.nih.gov/ij/), where the intensity of each band was weighted by its molecular size (nt); in this approach, it is not necessary to know the concentration of the complexes. Plots of v versus v/s were used to determine Km (−slope) and Vmax (y-intercept), where v is nucleotides per second and s is the molar concentration of each dNTP.
Immobilization
Microscope coverglass (Corning No. 1-1/2, Corning Inc., Lowell, MA) was coated with indium-tin oxide (ITO, 140 ohm/square, ZC&R Inc., Torrance, CA) film as a binder for a polyethylene oxide (PEG) nonstick coating (below). Chambers were formed on the coverglass by first applying an adhesive Mylar™ tape (70 μm thick, 3 × 4 array of 1/32" diameter holes, Grace Bio Labs, Bend, OR) to the ITO, then applying an adhesive silicone rubber mat (1/16" thick, 1/8" diameter holes, Grace Bio Labs) in array-register to the tape. The ITO surface in each 1/8" diameter chamber was coated with the polyethylene-oxide-DOPA3 block copolymers mPEG-2000-DP3 and biotin-mPEG-3400-DP3 (Supplementary Figure S2) from Nerites Corporation, Madison, WI (http://www.nerites.com) as follows. Each compound was dissolved under a nitrogen atmosphere at 1 mg/ml in 0.6 M K2SO4, 0.1 M MOPS-KOH buffer, pH 6 and stored in 200 μl single-use aliquots in sealed tubes at −80°C. A 1:8 mixture of the unbiotinylated and biotinylated compounds, respectively, was applied to the chambers. The chambers were sealed with a plastic coverslip, placed in a humid sealed jar, and incubated for 20 h in a 60°C oven to allow the tri-DOPA moiety to bind to the metal oxide surface. The chambers were rinsed with water and were stored dry, in the dark and in the ambient atmosphere, for up to 2 months before use. Purified complexes, nominally 10 nM in buffer C, were applied in 1 μl volumes to the coated chambers. The chambers were sealed with a plastic coverslip, placed in a sealed humid jar, and incubated at 50°C for 90 min to allow the complexes to bind to the biotinylated surface. The chambers were rinsed with water and stored filled with water at 4°C for up to 12 h before use. The chambers are shown in Supplementary Figure S3.
Microscopy
Movies and images of immobilized complexes were obtained with a Roper Cascade 512 electron-multiplying CCD camera (Roper/Photometrics, Tucson, AZ) on an Olympus IX-70 inverted microscope fitted with the Olympus TIRF accessory. An Ar-Kr mixed gas laser was used for illumination at 488 nm (Spectra-Physics, Mountain View, CA; Stabilite 2018).
RESULTS
Processivity clamp design
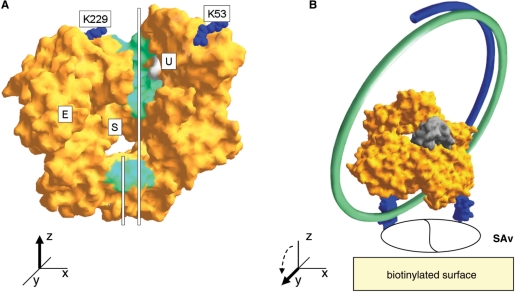
Our objectives were to form an irreversible, catalytically active complex between a polymerase and a DNA template, and to orient the complexes on a nonstick surface for single DNA molecule sequencing. We would start with a mutant of Therminator DNA polymerase already selected by directed evolution for the efficient utilization of phosphate-labeled nucleotides (see Introduction section). Like other family B DNA polymerases, Therminator binds DNA in a cleft intersecting the nucleotide binding site (28,29) (Figure 1A). We reasoned that DNA could be trapped in the polymerase by bridging the DNA-binding cleft with streptavidin and binding the complex to a substrate (Figure 1B). This scheme requires specific biotinylation at two surface locations on the polymerase, one on each side of the cleft. The tetravalent binding capacity of streptavidin and its strong affinity for biotin (30) could be utilized to bind the biotinylated polymerase to a biotinylated surface. For this architecture to succeed, the polymerase should be specifically biotinylated at the two locations flanking the DNA-binding cleft. This would be achieved by engineering the polymerase gene with two AviTag peptide ‘legs’ inserted at the surface-exposed amino acid positions K53 and K229 flanking the DNA-binding cleft (Figure 1B). AviTag is an artificial peptide substrate efficiently biotinylated by E. coli biotin–protein ligase (31) (http://www.avidity.com). The two AviTag legs would not only facilitate biotinylation but, in projecting up to 25 Å outward from the polymerase surface, should also allow ample clearance for the single-stranded template DNA to feed into the DNA-binding cleft. In an animated model of the structurally related RB69 DNA polymerase (http://www.csb.yale.edu/people/wang/rb69/Movies.html), the structurally homologous locations of the two AviTag insertions in Therminator polymerase appear to be relatively rigid, with little or no participation in the major conformational changes accompanying the polymerization catalytic cycle (12). As such, surface attachment at these points should compatible with polymerase activity.
Figure 1.
Concept. (A) DNA binding face of 9°N DNA polymerase [1QHT.pdb;(29)], the naturally occurring progenitor of the modified polymerase used in this report. The primer (short white line) and template DNA (long line) strands are drawn in the DNA-binding cleft, the floor of which is colored green. A pocket in one wall of the groove, shown in archaeal family B polymerases to bind deoxyuracil in the template strand (28), is colored gray and marked ‘U’; the existence of this pocket provides evidence that the template tracks in the indicated cleft. AviTag peptides were inserted C-terminal to the marked amino acids K53 and K229. The exonuclease domain (E) and nucleotide binding active site (S) are noted. Amino acids defining the floor of the groove (green) are D4, T5, D6, Y7, I8, R17, K118, D235, M244, D251, K253, W342, D343, R346, S347, S348, N351, W356, V389, T590, K591, K592 and K593, and for the uracil binding pocket (gray) V93, E111, I114, P115, P116 and R119. (B) Alternate view rotated 90° about the x-axis. Circular template DNA (green line) is shown passing through the tunnel formed by polymerase and surface-bound streptavidin (SAv; the divided white oval denotes ambiguity in the number of bound streptavidins, 1 or 2). The primer strand (blue line) is shown hybridized to the template. Two AviTag legs (blue; at bottom of the polymerase body) are modeled at positions K53 and K229; in the model, the length of each AviTag leg is about 25 Å from base to tip. The highly mobile ‘N’ and ‘O’ helices associated with the open–closed conformational change characteristic of polymerases are colored gray (29).
Polymerase modification
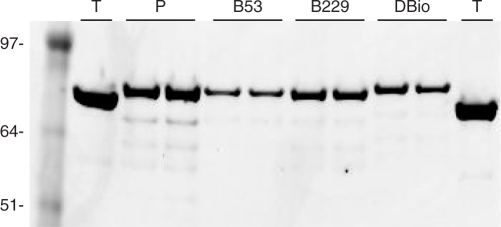
An oligonucleotide encoding the AviTag peptide was inserted in the parent polymerase gene at one or both of the targeted locations (see Materials and Methods section). The progenitor gene for Therminator DNA polymerase had originally been obtained from New England Biolabs and then was recloned into a pBAD expression vector (Invitrogen), giving it a C-terminal hexahistidine tag to enable affinity purification with Ni-NTA beads. Five rounds of directed evolution resulted in seven new mutations adapting the polymerase to the efficient utilization of phosphate-labeled nucleotides (T.M. Urlacher, manuscript in preperation). Starting with this polymerase, we prepared constructs having an AviTag leg inserted between K53-V54 (construct ‘B53’), between K229-F230 (‘B229’) or at both locations (‘DBio’). Candidate constructs were screened by PCR from E. coli colonies and were confirmed by DNA sequencing. The modified proteins were expressed in E. coli AVB100 in order to biotinylate the AviTag legs in vivo (http://www.avidity.com). The engineered polymerases were affinity purified using Ni-NTA beads and then further biotinylated in vitro with biotin-protein ligase (see Materials and Methods section). The biotinylated polymerases were evaluated for purity on an SDS–PAGE gel (Figure 2).
Figure 2.
Purified polymerases with AviTag legs. (T) Therminator protein purchased from New England Biolabs. (P) The parent enzyme (a mutant of Therminator selected for improved utilization of phosphate-labeled dNTPs) has a higher molecular weight compared to Therminator because it has a C-terminal Myc-His fusion used for purification. (B53) Polymerase ‘P’ with AviTag leg at position K53. (B229) Polymerase ‘P’ with AviTag leg at position 229. (DBio; ‘dual biotin’) Polymerase ‘P’ with two AviTag insertions at K53 and K229. Duplicate samples (5 μl each) of the Ni-NTA purified proteins were resolved by SDS–PAGE [Invitrogen NuPAGE Bis–Tris (MOPS) 4–12%]. The gel was stained with Coomassie blue and imaged with a LI-COR Odyssey infrared imager (application note http://www.licor.com/bio/Odyssey2/Odyssey13.jsp). Marker sizes (kDa) are indicated.
Binary complexes of polymerase and streptavidin
To estimate the extent of biotinylation, we mixed increasing amounts of the DBio polymerase (0–10 nM) with a fixed amount of fluorophore-labeled streptavidin (0.5 nM). The obtained complexes were resolved in an isoelectric focusing gel and bands containing labeled streptavidin were imaged using a LI-COR infrared fluorescence imager (Figure 3). The amount of bound streptavidin increased with polymerase concentration, with nearly all of the streptavidin bound by a 2-fold molar excess of polymerase. This indicates that at least half of the polymerase proteins are biotinylated and capable of binding streptavidin, which is sufficient to proceed with DNA-binding experiments.
Ternary complexes of polymerase, DNA and streptavidin
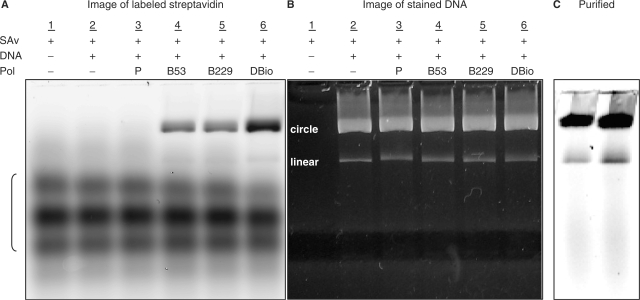
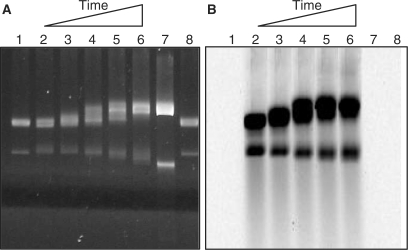
All four polymerase variants were tested for the ability to form stable ternary complexes with DNA and streptavidin. In this experiment, primed M13 DNA, polymerase and labeled streptavidin were mixed in a molar ratio of 1:2:4, respectively. The DNA and polymerase were mixed first, then streptavidin was added with the idea of trapping the DNA in the polymerase. The products were resolved by agarose gel electrophoresis and the infrared fluorescence signal was imaged to reveal the labeled streptavidin (Figure 4A). The DNA was separately imaged by staining the gel with SYBR Gold and photographing under UV illumination (Figure 4B). Complexes were identified as labeled streptavidin comigrating with either the circular or linear M13 DNA. The important result is that ternary complexes are observed and that their formation depends on the presence of a polymerase with at least one AviTag leg. So, although the polymerase is unlabeled and thus not directly detectable, the requirement for biotinylated polymerase in binding labeled streptavidin to DNA establishes that the complexes comprise all three components. These results leave unresolved the question of whether ternary complexes comprise one streptavidin binding across both legs of the DBio polymerase, or if instead there are two streptavidins, one binding to each leg (Figure 1B). Approximately twice (2.1-fold) as much streptavidin was bound by the DBio polymerase as compared to the single-leg variants (Figure 4A), suggesting that there is one streptavidin bound per leg. That is, the protein configuration may prevent a single streptavidin spanning the two legs.
Figure 4.
Ternary complexes made with primed M13 DNA, polymerase (zero, one or two AviTag legs) and Alexa Fluor-680-streptavidin (lanes 3–6), with controls omitting either DNA or polymerase (lanes 1 and 2). Complexes were separated by electrophoresis in a 2.2% agarose gel (10 v/cm, 1 h). (A) The tested polymerases were the parent enzyme without AviTag legs (P); the two single-leg variants (B53 and B229); and the dual-leg polymerase (DBio). The labeled streptavidin (SAv) was detected using a LI-COR Odyssey infrared imager. The relative quantities of SAv associated with both circular and linear DNA were determined by integrating pixel intensities: lane 3 (SAv = 0), lane 4 (SAv = 1.0), lane 5 (SAv = 1.0) and lane 6 (SAv = 2.1). (B) Gel from (A), but stained with SYBR Gold (Invitrogen) to visualize the DNA by UV transillumination (312 nm). (C) Purified complexes of DBio polymerase (infrared image), showing that unbound labeled streptavidin has been removed (compare to the unpurified complexes in A, lane 6).
Purification
To minimize competitive binding to biotinylated surfaces, the ternary complexes were purified from the excess of streptavidin used in preparation. We developed a simple procedure based on the faster binding of streptavidin to biotinylated magnetic beads as compared to the slower binding of the bulky ternary complexes. In this procedure, biotinylated magnetic beads are added to unpurified complexes in sufficient capacity to bind all of the streptavidin present. The sample is mixed for 10 min at room temperature, the beads are magnetically removed, and the process is repeated once. These purified complexes are largely free of excess streptavidin (Figure 4C). No additional label is removed by further cycles of purification, indicating that essentially all of the functional streptavidin is removed by the two cycles of bead purification. With the complexes being formed from a 1 : 2 : 4 mixture of DNA: polymerase: streptavidin, most of the polymerase not associated with DNA is likely to be in binary complex with streptavidin. As such, the kinetics-based purification procedure would remove not only excess streptavidin, but also excess streptavidin–polymerase complexes.
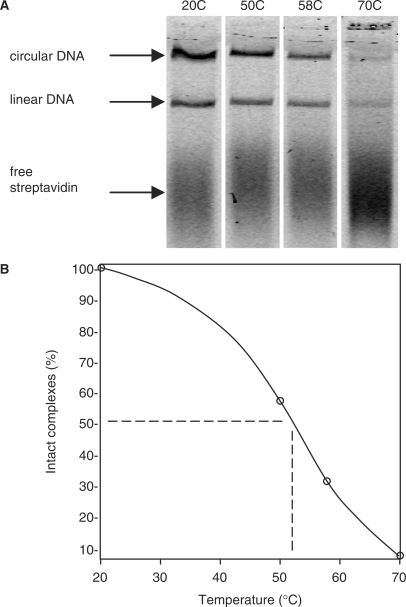
Thermal stability
We determined the thermal stability of ternary complexes to see if they could be used near the 74°C temperature optimum of the polymerase. Purified ternary complexes made with labeled streptavidin were incubated between 20°C and 70°C for 2 h and the fraction of complexes surviving intact was determined by agarose gel electrophoresis (Figure 5A). Incubation at 70°C resulted in nearly complete dissociation of streptavidin from the DNA, which seems to rule out using the complexes at the optimal 74°C polymerization temperature. Interpolating the plot in Figure 5B, we identify 54°C as optimal for testing the polymerization activity of ternary complexes, where about 45% of the complexes survived intact after 2 h incubation and where polymerase activity is about one-third the maximum at 74°C (not shown). The apparent stabilities measured here could have been affected by reassociation occurring in the time period (∼10 min) between cooling the samples and separating the components by electrophoresis.
Figure 5.
Thermal stability of ternary complexes. Purified complexes (3 nM) were incubated for 2 h at the indicated temperatures and samples were resolved by electrophoresis in 2.2% agarose. (A) The labeled streptavidin component was imaged using an Odyssey infrared imager. (B) For each lane, the fluorescence signals comigrating with the two DNA bands (circular, linear) were summed and the results were plotted normalized to the 20°C sample. The data points were connected by a piecewise spline curve (Maple 11, http://www.maplesoft.com).
Activity of ternary complexes in solution
To see if the ternary complexes are catalytically active in solution, purified complexes were incubated with unlabeled dNTPs to allow for primer extension on the associated M13 templates. Samples were incubated at 54°C for times up to 90 min and primer extension products were resolved on an agarose gel. DNA products were imaged by staining with SYBR Gold™. The complexes appear capable of highly processive DNA synthesis, as indicated by the full-length products obtained at the longer incubation times (Figure 6A, lanes 2–6). The majority of the labeled streptavidin remained associated with the DNA product bands, which suggests that the complexes remained mostly intact for the 90-min duration of the reaction (Figure 6B). There was little or no evidence for strand displacement synthesis by the complexes, which would be revealed as high molecular weight DNA trapped in the wells as seen, for example, with the strand-displacing DNA polymerase φ-29 (Supplementary Figure S4).
Figure 6.
DNA synthesis by ternary complexes. Complexes (1.2 nM) made with labeled streptavidin were mixed with 200 μM each of the four unlabeled dNTPs and 5 mM MgCl2 in buffer C, and were incubated at 54°C for 0, 3, 10, 30 and 90 min (lanes 2–6). The samples were resolved by electrophoresis in a 2.2% agarose gel. As pointed out in Figures 4 and 5, the upper DNA band comprises complexes with circular DNA, while the lower (i.e. faster migrating) band comprises complexes with linear DNA. (A) The DNA component was stained with SYBR Gold™ and imaged under UV transillumination. (B) The streptavidin component was imaged by fluorescence using a LI-COR Odyssey infrared imager. Controls include M13 DNA alone (lanes 1 and 8), and M13 fully extended with a saturating amount of Taq DNA polymerase (lane 7).
Polymerization kinetics in solution
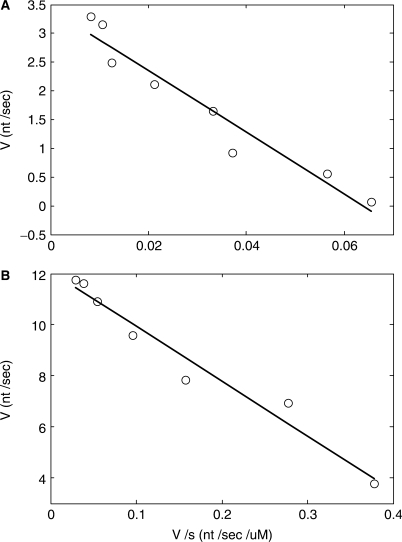
Kinetic constants of purified complexes were determined in a primer extension assay. In this experiment, the streptavidin was unlabeled and an infrared dye-labeled primer was used to the detect primer extension products with single-base resolution. Samples were incubated for 60 s at 54°C with unlabeled dNTP concentrations of from 1 to 400 μM each. Primer extension products were resolved by electrophoresis in an automated sequencer (LI-COR 4200). Each lane on the gel was analyzed to quantify the average number of nucleotides incorporated per template molecule (see Materials and Methods section), and polymerization rates were calculated at each dNTP concentration. A reciprocal plot gave Km[dNTP] = 54 μM and Vmax = 3.4 nt/s for the purified complexes (Figure 7). By comparison, a control experiment showed that free, uncomplexed polymerase had a greater affinity for nucleotides and a faster polymerization rate (Km[dNTP] = 21 μM and Vmax = 12.0 nt/s measured at 54°C).
Figure 7.
Polymerization kinetics of purified complexes (see Materials and Methods section). DNA-synthesis rates (v, nucleotides per second) were determined for various nucleotide substrate concentrations (s, micromolar). (A) Purified complexes: Km = 54 μM, Vmax = 3.4 nt/s. (B) Control sample of uncomplexed polymerase and DNA: Km = 21 μM, Vmax = 12.0 nt/s.
Activity of immobilized complexes
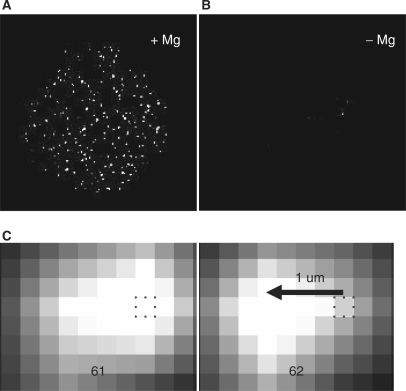
The thermal stability and activity of ternary complexes in solution suggests they are capable of processive DNA synthesis (Figures 5 and 6). To confirm and extend this result, we determined whether the complexes are active when immobilized on a surface. Microscope coverglass chambers were coated with a 1:8 mixture of biotinyl-PEG3400 and PEG2000 polymers to provide a nonstick surface capable of specifically binding streptavidin (see Materials and Methods section). A solution of unlabeled ternary complexes was applied to the chambers and allowed to bind for 90 min. The chambers were washed to remove unbound complexes and a nucleotide cocktail containing base-labeled AlexaFluor-488-dUTP plus unlabeled dATP, dCTP and dGTP was added. After incubation at 54°C for 2½ h, the chambers were washed to remove free nucleotides and the coverglass surface was imaged by TIRF microscopy. A field of bright fluorescent spots was seen (Figure 8A). Each spot is believed to mark a single, multiply labeled DNA molecule synthesized by surface-attached complexes. This conclusion is supported by two additional observations ruling out the possibility that the bright spots are nonspecifically adsorbed clusters of labeled dUTP. First, no spots were observed in a negative control omitting Mg++, which is required for base incorporation (Figure 8B). Second, the spots were seen moving freely back and forth while still remaining tethered to the surface (Figure 8C, and Supplementary Movie M1). This behavior is consistent with individual, labeled DNA molecules retained by individual polymerases, as illustrated in Figure 1B.
Figure 8.
DNA synthesis by immobilized complexes. Purified ternary complexes made with unlabeled streptavidin were immobilized in a reaction chamber on a PEG–biotin coated coverglass (see Materials and methods section). (A) The reaction chamber was filled with 1 μl of buffer C containing 5 mM MgCl2, 100 μM each of dATP, dCTP, dGTP and base-labeled Alexa Fluor-488-dUTP (Invitrogen). The chamber was sealed with a plastic coverslip and incubated in a humid jar at 54°C for 90 min. The chamber was rinsed with water to remove unincorporated nucleotides and the coverglass surface was imaged by TIRF microscopy (see Materials and methods section). The labeled complexes were seen waving back and forth under Brownian motion, while remaining tethered to the surface (Supplementary Movie M1). (B) Control reaction inhibiting polymerase activity by replacing Mg++ with 0.1 mM EDTA. (C) Zoomed-in view of a single DNA spot from Movie M1 showing movement of about 1 μm leftward occurring between frames 61 and 62. A pixel is marked for reference (x–y coordinate 386, 534). Exposure time was 80 ms and the pixel dimension 0.27 μ. Movie M1 was acquired in a replicate experiment as in (A).
Complexes can be stored frozen for subsequent use
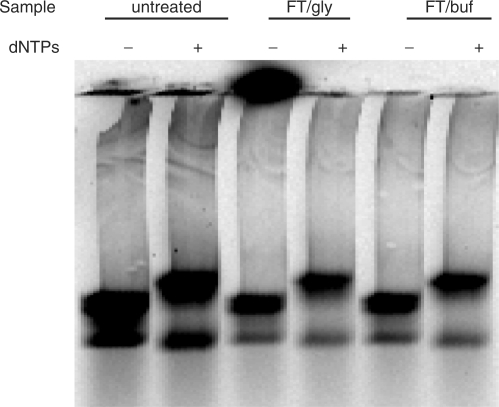
In the course of doing this work, we found that complexes could be stored for up to 1 week at 4°C with minimal loss in activity. For purposes of single-molecule DNA sequencing, however, it would be more convenient to prepare complexes with genomic DNA samples in advance and store them indefinitely until needed. To see if preformed complexes could be frozen without losing activity, we prepared samples in buffer alone or in buffer plus 25% glycerol. Aliquots were frozen in liquid nitrogen, stored overnight at −80°C and thawed at ambient temperature. Samples were tested by incubating with unlabeled dNTPs at 54°C for 30 min and analyzed on an agarose gel (Figure 9). Both of the frozen samples not only survived intact, but also showed full activity indistinguishable from a not-frozen control. Since damage to complexes is most likely to occur during freezing and thawing, we believe that these engineered complexes could be frozen indefinitely until needed.
Figure 9.
Complexes are stable to freezing and thawing. Purified complexes (3 nM) made with Alexa Fluor-680 streptavidin were treated by freezing 20 μl aliquots in liquid nitrogen, storing overnight at −80 and thawing at 20°C. Treated samples (FT) were in buffer C alone (buf) or in buffer C plus 25% glycerol (gly). The thawed complexes were tested for activity by incubating 2 μl of complexes in the presence or absence (±) of 200 μM each of dATP, dCTP, dGTP and dTTP in a final volume of 10 μl buffer C at 55°C for 30 min. Primer extension products were resolved by electrophoresis in a 1.5% agarose gel and the labeled streptavidin component was imaged using a LI-COR Odyssey infrared imager. An unfrozen control was held overnight at 4°C (untreated).
DISCUSSION
We have shown that immobilization of a polymerase–DNA complex on a solid surface enhances processivity. This was accomplished by engineering the polymerase with two biotinylated AviTag peptide legs located on either side of the DNA-binding cleft. Both insertions were compatible with the protein structure of the polymerase, allowing it to be overexpressed and biotinylated in E. coli. Stable ternary complexes of polymerase, primed template DNA and streptavidin could be immobilized on a biotinylated, nonstick surface. The expected architecture of immobilized complexes suggests that the template DNA would thread through a tunnel formed by the body of the polymerase, the AviTag legs and the surface (Figure 1B).
The ternary complexes were first assembled in solution, then purified from excess streptavidin and finally were immobilized on the surface. In theory, an alternative assembly pathway might have been used, where binary complexes of the biotinylated polymerase and DNA would be directly attached to a surface precoated with streptavidin. This potentially simpler route was not attempted in the present work, however, because of the known slower binding of protein–DNA complexes to surfaces as compared to the binding of free protein. This was the basis of our purification method. Since the DNA exists in a random coil configuration, polymerase bound to DNA would be relatively inaccessible and binding to the surface would be slow. With free polymerase and DNA in equilibrium with complexes, transiently unbound polymerase molecules would quickly bind to the surface, resulting in a decreased number of immobilized complexes.
Experiments with ternary complexes in solution provided estimates of kinetic constants and processivity. We found that the complexes are somewhat compromised in DNA synthesis, showing an increased requirement for dNTPs and a slower catalytic rate (2.6-fold higher Km and 3.5-fold lower Vmax) compared to the free polymerase (Figure 7). In solution, processivity was apparently enhanced from the <20 nt characteristic of the Therminator parent polymerase (see Supplementary Material), to >7 kb in the ternary complexes (Figure 6). Whereas the processivity of stable complexes was estimated in a simple primer extension reaction (Figure 6), a more elaborate assay was required to quantify processivity of the Therminator parent (see Supplementary Material). The difference is that for complexes, each individual polymerase is stably associated with an individual DNA template, so that processivity can be estimated by simple primer extension. On the other hand, the Therminator parent is free to exchange among different template molecules during the primer extension reaction, preventing direct assessment of processivity by simple primer extension.
Fidelity of the immobilized complexes was not determined. An analysis of fidelity would ideally require amplifying, cloning and sequencing in an unbiased manner the small population of individual DNA molecules polymerized on the surface, taking into account that the original M13 template is also present on the surface. In the worst case, which is ruled out, we believe, by the results in Figure 6, the complexes could be synthesizing DNA by terminal transferase polymerization, independent of the template. Figure 6 shows that primer extension products of circular DNA complexes (upper bands) migrated slower in the gel relative to the single-stranded circular template, while extension products of the linear complexes (lower bands) migrated faster than the linear template. This observed template specificity suggests that the primer extension products are from templated polymerization, and are not mere products of template-independent terminal transferase activity peculiar to the complexes.
Enhanced processivity in the complexes was further demonstrated by direct observation of the primer extension products of individual surface-attached complexes. Immobilized complexes were exposed to a cocktail of unlabeled dATP, dCTP and dGTP plus base-labeled dUTP. After incubation for 90 min, the surface was rinsed to remove unincorporated nucleotides. DNA released from dissociated complexes would also be removed, so that the only remaining DNA would be that associated with immobilized complexes (Figure 8). Individual labeled DNA spots were seen moving back and forth even as they remained tethered to the surface (Supplementary Movie M1), consistent with the concept of Figure 1. The extent of this DNA motion allows for a rough estimate of processivity. For example, Figure 8C shows an example where a DNA spot appears to have moved about 1 μ between consecutive movie frames. Assuming the polymerase attachment point was at the center of this particular motion, the DNA tether fully stretched would be a minimum of 0.5 μ in length, which corresponds to 1.5 kb of DNA. Such a stretched tether, plus the apparently larger amount of DNA in the bulk fluorescent spot, argues for a processivity of several thousand nucleotides achieved by this engineered, immobilized polymerase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health (3R44HG002292-04S1 and5P01HG003015-02, to J.W.). Funding for open access charge: LI-COR Biosciences, Inc.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Bill Jack and Andy Gardner (New England Biolabs) for providing the 9°N and Therminator DNA parent polymerase genes used in this work, as well as for helpful advice and comments on the article. The authors acknowledge Janice Aschoff and Kaitlyn Williams for assistance with the illustrations.
REFERENCES
- 1.Jett JH, Keller RA, Martin JC, Marrone BL, Moyzis RK, Ratliff RL, Seitzinger NK, Shera EB, Stewart CC. High-speed DNA sequencing: an approach based upon fluorescence detection of single molecules. J. Biomol. Struct. Dyn. 1989;7:301–309. doi: 10.1080/07391102.1989.10507773. [DOI] [PubMed] [Google Scholar]
- 2.Stephan J, Dorre K, Brakmann S, Winkler T, Wetzel T, Lapczyna M, Stuke M, Angerer B, Ankenbauer W, Foldes-Papp Z, et al. Towards a general procedure for sequencing single DNA molecules. J. Biotechnol. 2001;86:255–267. doi: 10.1016/s0168-1656(00)00417-x. [DOI] [PubMed] [Google Scholar]
- 3.Werner JH, Cai H, Jett JH, Reha-Krantz L, Keller RA, Goodwin PM. Progress towards single-molecule DNA sequencing: a one color demonstration. J. Biotechnol. 2003;102:1–14. doi: 10.1016/s0168-1656(03)00006-3. [DOI] [PubMed] [Google Scholar]
- 4.Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc. Natl Acad. Sci. USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 6.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 7.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys. J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagerqvist J, Zwolak M, Di Ventra M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 2006;6:779–782. doi: 10.1021/nl0601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhee KJ, Burney RE, Mackenzie JR, Flora J. Predicting the utilization of helicopter emergency medical services: an approach based on need. Ann. Emerg. Med. 1984;13:916–923. doi: 10.1016/s0196-0644(84)80670-8. [DOI] [PubMed] [Google Scholar]
- 10.Gardner AF, Jack WE. Acyclic and dideoxy terminator preferences denote divergent sugar recognition by archaeon and Taq DNA polymerases. Nucleic Acids Res. 2002;30:605–613. doi: 10.1093/nar/30.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Southworth MW, Kong H, Kucera RB, Ware J, Jannasch HW, Perler FB. Cloning of thermostable DNA polymerases from hyperthermophilic marine Archaea with emphasis on Thermococcus sp. 9 degrees N-7 and mutations affecting 3′-5′ exonuclease activity. Proc. Natl Acad. Sci USA. 1996;93:5281–5285. doi: 10.1073/pnas.93.11.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steitz TA. Visualizing polynucleotide polymerase machines at work. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 14.Modrich P, Richardson CC. Bacteriophage T7 Deoxyribonucleic acid replication in vitro. A protein of Escherichia coli required for bacteriophage T7 DNA polymerase activity. J. Biol. Chem. 1975;250:5508–5514. [PubMed] [Google Scholar]
- 15.Weisshart K, Chow CS, Coen DM. Herpes simplex virus processivity factor UL42 imparts increased DNA-binding specificity to the viral DNA polymerase and decreased dissociation from primer-template without reducing the elongation rate. J. Virol. 1999;73:55–66. doi: 10.1128/jvi.73.1.55-66.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker TA, Bell SP. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 17.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 18.Nossal NG. Protein-protein interactions at a DNA replication fork: bacteriophage T4 as a model. FASEB J. 1992;6:871–878. doi: 10.1096/fasebj.6.3.1310946. [DOI] [PubMed] [Google Scholar]
- 19.Sexton DJ, Berdis AJ, Benkovic SJ. Assembly and disassembly of DNA polymerase holoenzyme. Curr. Opin. Chem. Biol. 1997;1:316–322. doi: 10.1016/s1367-5931(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 20.von Hippel PH. A ring to bind eukaryotic DNA polymerase. Structure. 1995;3:123–124. doi: 10.1016/s0969-2126(01)00140-x. [DOI] [PubMed] [Google Scholar]
- 21.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez I, Lazaro JM, Blanco L, Kamtekar S, Berman AJ, Wang J, Steitz TA, Salas M, de Vega M. A specific subdomain in phi29 DNA polymerase confers both processivity and strand-displacement capacity. Proc. Natl Acad. Sci. USA. 2005;102:6407–6412. doi: 10.1073/pnas.0500597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedford E, Tabor S, Richardson CC. The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc. Natl Acad. Sci. USA. 1997;94:479–484. doi: 10.1073/pnas.94.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson JF, Fox R, Harris DD, Lyons-Abbott S, Loeb LA. Insertion of the T3 DNA polymerase thioredoxin binding domain enhances the processivity and fidelity of Taq DNA polymerase. Nucleic Acids Res. 2003;31:4702–4709. doi: 10.1093/nar/gkg667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motz M, Kober I, Girardot C, Loeser E, Bauer U, Albers M, Moeckel G, Minch E, Voss H, Kilger C, et al. Elucidation of an archaeal replication protein network to generate enhanced PCR enzymes. J. Biol. Chem. 2002;277:16179–16188. doi: 10.1074/jbc.M107793200. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Prosen DE, Mei L, Sullivan JC, Finney M, Vander Horn PB. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res. 2004;32:1197–1207. doi: 10.1093/nar/gkh271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu J, March PE, Lee R, Tillett D. Site-directed, Ligase-Independent Mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004;32:e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fogg MJ, Pearl LH, Connolly BA. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez AC, Park HW, Mao C, Beese LS. Crystal structure of a pol alpha family DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. 9 degrees N-7. J. Mol. Biol. 2000;299:447–462. doi: 10.1006/jmbi.2000.3728. [DOI] [PubMed] [Google Scholar]
- 30.Jung LS, Nelson KE, Stayton PS, Campbell CT. Binding and dissociation kinetics of wild-type and mutant streptavidins on mixed biotin-containing alkylthiolate monolayers. Langmuir. 2000;16:9421–9432. [Google Scholar]
- 31.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.