Abstract
Metnase is a human SET and transposase domain protein that methylates histone H3 and promotes DNA double-strand break repair. We now show that Metnase physically interacts and co-localizes with Topoisomerase IIα (Topo IIα), the key chromosome decatenating enzyme. Metnase promotes progression through decatenation and increases resistance to the Topo IIα inhibitors ICRF-193 and VP-16. Purified Metnase greatly enhanced Topo IIα decatenation of kinetoplast DNA to relaxed circular forms. Nuclear extracts containing Metnase decatenated kDNA more rapidly than those without Metnase, and neutralizing anti-sera against Metnase reversed that enhancement of decatenation. Metnase automethylates at K485, and the presence of a methyl donor blocked the enhancement of Topo IIα decatenation by Metnase, implying an internal regulatory inhibition. Thus, Metnase enhances Topo IIα decatenation, and this activity is repressed by automethylation. These results suggest that cancer cells could subvert Metnase to mediate clinically relevant resistance to Topo IIα inhibitors.
INTRODUCTION
Transposases mediate DNA movement in lower organisms by excising defined segments of DNA and then reinserting them at other locations in the genome, a process that can be repeated multiple times for a given segment (1,2). While transposase activity probably accounts for half of the present organization of the human genome, almost all of these sequences are pseudogenes, as unregulated DNA mobility would be deleterious to human cells, causing genome instability (1–4).
Recently we identified and characterized Metnase (also called SETMAR), a human protein with a transposase domain derived from Mariner transposons fused to a SET domain. Metnase is expressed in most tissues, methylates histone H3, promotes foreign DNA integration and enhances nonhomologous end-joining (NHEJ) of DNA double-strand breaks (DSBs) (5). Metnase is present only in primates, and it possesses partial transposase activity, including sequence-specific DNA binding, assembly of paired end complexes, cleavage of the 5′-end of the Mariner terminal inverted repeat and promotion of integration at a TA dinucleotide target site (6–8).
We found that Metnase has endonuclease activity that nicks and linearizes but does not degrade supercoiled DNA (9). Therefore, we postulated that Metnase plays a role in decatenating DNA. DNA replication results in intertwined sister chromatids that must be untangled, or decatenated, to ensure proper chromatid segregation in mitosis and prevent chromatid breaks during anaphase. Topoisomerase IIα (Topo IIα) is the critical decatenating enzyme. It functions by creating transient DSBs through which it passes a second double-stranded DNA and then religates the broken ends (10).
In human cells, chromosome catenation status is actively monitored, and at two points in the cell cycle, catenated DNA inhibits cycle progression. One decatenation checkpoint prevents progression from G2 to M (11), and the other prevents progression from metaphase to anaphase (12–15). The decatenation checkpoints are highly conserved in plants and animals and have been observed in many tissue and cell types (16,17), including yeast (18). Emerging data indicate that decatenation checkpoints are impaired in human cancers and in both embryonic and hematopoietic stem cells (16,19,20). The decatenation checkpoints are activated when ATM- and Rad3-related (ATR) senses catenated chromosomes and then signals through BRCA1 to inhibit cyclin B1 and Cdk1 to halt cell cycle progression toward mitosis (17,21). In addition, ATR may inhibit PLK1 when chromatids remain catenated, thereby preventing progression to mitosis (21).
While decatenation checkpoint signaling is becoming clearer, the precise biochemical mechanism of decatenation is less well defined. We report here an interaction between Metnase and Topo IIα and show that Metnase promotes Topo IIα decatenation activity and enhances progression through the metaphase decatenation checkpoint in vivo. This enhancement is inhibited by the automethylation of Metnase.
MATERIALS AND METHODS
Immunoprecipitation
Cells were harvested and lysed with radio immuno precipitation assay (RIPA) buffer. Protein extracts were precleared with 40 μl of protein A/G beads. Two milligrams of protein extract was used for each immunoprecipitation. Two microliters of anti-V5 (Invitrogen, Carlsbad, CA), 2 µl of anti-Metnase or 2.5 μl of anti-Topo IIα (Topogen, Port Orange, FL) antibodies was used in a total volume of 500 μl. Lysates were incubated overnight in 500 µl RIPA buffer with antibody at 4°C in the presence of 1 U DNase I. Thirty microliters of protein A/G beads was added and incubated for 2 h at 4°C. Beads were then washed three times with RIPA buffer and twice with phosphate-buffered saline (PBS). The beads were boiled, and samples were run on a SDS–PAGE gel and analyzed by western blot using appropriate anti-sera. Input represented 1% of the total lysate for all experiments.
Manipulating Metnase expression
HEK-293 derivatives over-expressing Metnase-V5 were generated by stably transfecting pcDNA-Metnase (neomycin selectable), and derivatives under-expressing Metnase were stably transfected with U6-siRNA Metnase (hygromycin selectable), as described (5). Controls were transfected with pcDNA or U6 vector that makes a nonsense siRNA transcript. HEK-293T cells were stably transfected with the puromycin-selectable pCAPP-Metnase vector or empty pCAPP. Total Metnase levels were measured by western blot using anti-Metnase serum (5). Expression levels were measured before each experiment to confirm appropriate Metnase expression levels. Cells with forced under-expression of Metnase grew more slowly than vector-transduced cells, and this was controlled for in all experiments.
Metaphase arrest analysis
ICRF-193 (MP Biomedicals, Solon, OH) reversibly inhibits Topo IIα without inducing DNA DSBs, thereby allowing assessment of the decatenation checkpoint (11,17,22). ICRF-193 and VP-16 were dissolved in DMSO and further diluted in growth media. Unsynchronized cells were treated with 10 μM ICRF-193, 5 μM VP-16 or equivalent volume of DMSO (vehicle control) for 4 or 18 h, harvested, fixed and stained with an anti-tubulin antibody and 4′-6-diamidino-2-phenylindole (DAPI, Vectashield). These concentrations were chosen by dose–response experiments. Interphase cells and cells at metaphase were counted by immunofluorescence microscopy to assess ICRF-193- and VP16-induced metaphase arrest. The fractional increase in ICRF-193 or VP-16 metaphase-arrested cells over vehicle controls was due to failure to traverse the M phase decatenation checkpoint (12–15). Each experiment was performed at least three times with 850–1800 cells counted per condition; statistics were calculated by using t-tests, and standard deviation (SD) is shown.
Localization of Topo IIα and Metnase
After appropriate treatment, 2 × 104 cells were cytospun onto microscope slides, air dried for 5 min at room temperature and fixed in 100% methanol for 10 min at −20°C. The remaining steps were carried out at room temperature. Slides were dried in air for 15 min, rehydrated with PBS for 5 min, blocked for 30 min with 1% bovine serum albumin and 0.5% Tween 20 in PBS and washed three times with PBS. Antibodies were diluted in blocking buffer and incubated on the slides for 1 h, and then washed three times with PBS. Slides were incubated in 1.5 μg/ml DAPI for 15 min, the solution was aspirated and coverslips were mounted with Mowiol mount media. Cells were visualized with a Zeiss fluorescence microscope with ×63 or ×100 objectives. The following antibodies were used: rabbit anti-Topo IIα (Topogen) and mouse anti-V5 (Invitrogen). The secondary antibodies used were Alexa Fluor 555 labeled goat anti-rabbit IgG (Invitrogen) and Alexa Fluor 488 labeled goat anti-mouse IgG (Invitrogen). To obtain the relative DNA content of cells with immunofluorescent co-localization of Metnase and Topo IIα, Vision Assistant version 2.6 from National Instrument was used. This program digitally searches for cells with specific pixel intensity, allowing for cells to be grouped according to DAPI fluorescent intensity into bimodal Go/G1 and G2/M peaks, with S phase cells having intensities in between. At least 300 cells were digitally quantified for each population, and a one-tailed Student's t-test was used to assess differences in the cell cycle phase populations.
Plasmid cleavage and kinetoplast DNA decatenation
The vector pFlag-Metnase was stably transfected into HEK-293 cells, and Metnase was purified from lysates using anti-FLAG beads followed by elution from a Heparin-Sepharose 6 Fast Flow column (Amersham Biosciences, Piscataway, NJ). Silver staining showed no other protein contaminants (9). Purified recombinant Topo IIα (GE Healthcare) and catenated kinetoplast DNA (kDNA; Topogen) were used according to the manufacturer's instructions. For plasmid DNA cleavage, 50 ng/μl of plasmid pAdTrack (chosen for its large size) was incubated with various Metnase concentrations at 37°C for 60 min in fresh 25 mM Tris–HCl (pH 7.5), 5 mM MnCl2 and 1 mM dithiothreitol (DTT), and the products were analyzed by 1% agarose gel electrophoresis. kDNA are catenated minicircles that can be used as an in vitro substrate for decatenation experiments (23,24). Buffer and kDNA from the DNA Gyrase Assay kit (Topogen) were used to assess kDNA decatenation. Metnase is much less active in Mg2+ than in Mn2+, but Topo IIα is not active in Mn2+. At the MgCl2 concentration in this kit (8 μM), there is no measurable Metnase nuclease activity. kDNA (200 ng/µl) was incubated in the manufacturer's buffer with increasing concentrations of Metnase and/or Topo IIα for 4 h at 37°C per the manufacturer's instructions and then analyzed by 1% agarose gel electrophoresis. When Metnase was tested alone for its effect on kDNA, MnCl2 at 10 μM replaced the MgCl2. Each experiment was performed at least three times, with densitometric analysis, and averages and SD are shown.
Nuclear extracts
Nuclear extracts were prepared as we described (25) from HEK-293T cells transduced with pcDNA-Metnase or empty vector control. A final 4-h dialysis step in 50 mM Tris–HCl (pH 8.0), 120 mM KCl, 20 mM MgCl2, 10% glycerol and 0.5 mM DTT at 4°C was included to reduce the salt concentration. This buffer was also used in the kDNA decatenation assays with nuclear extracts. Each experiment was performed at least three times, with densitometric analysis, and averages and SD are shown.
Mass spectroscopy
Samples were digested with trypsin (Sequencing Grade Modified from Promega, Madison, WI) overnight. The digested peptides were extracted and placed in ZipTip pipette tips (Millipore) containing C18 reverse-phase media to concentrate and purify peptides. Samples were spotted on a MALDI plate, and peptide mass fingerprinting was done in MS mode on the Applied Biosystems 4700 MALDI Tof-Tof. The peptides were identified by searching the peptide masses against the National Center for Biotechnology Information and the Swissprot databases. Results were confirmed by sequencing the tryptic peptides in MS/MS mode and using the database-searching algorithm MASCOT to match the amino acid m/z. Each experiment was performed twice with independent lots of Metnase and Topo IIα.
RESULTS
Metnase interacts with Topo IIα
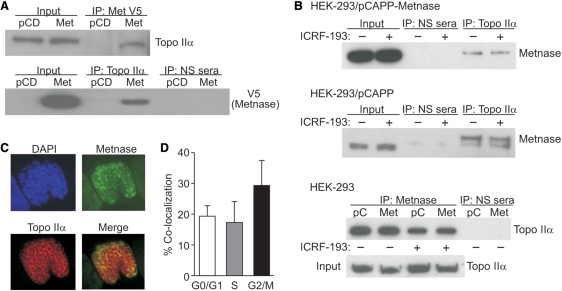
Since Topo IIα is the critical decatenating enzyme, the physical interaction between Metnase and Topo IIα was first investigated using co-immunoprecipitation assays. In reciprocal co-immunoprecipitation experiments shown in Figure 1A, Topo IIα was detected in Metnase immunoprecipitates, and Metnase was present in Topo IIα immunoprecipitates. Using anti-sera against native Metnase (5), endogenous Metnase was also found to be present in Topo IIα immunoprecipitates (Figure 1B). This co-immunoprecipitation was observed in the presence of DNase I, indicating that Metnase and Topo IIα interact independently of DNA. The presence of ICRF-193 did not delete the cellular interaction between Topo IIα and Metnase (Figure 1B). By western analysis using synchronized cells, Metnase expression is nearly equivalent in all phases of the cell cycle (not shown).
Figure 1.
Physical interaction between Metnase and Topo IIα. (A) V5-tagged and endogenous Metnase immunoprecipitated (IP) from HEK-293 cells co-immunoprecipitates Topo IIα, detected by western blotting. PCD, expression vector control; NS, non-specific anti-sera. (B) The Topo IIα inhibitor ICRF-193 does not block the interaction between either endogenous or transduced Metnase and Topo IIα. (C) Immunofluorescence co-localization of Metnase (green) with Topo IIα (red) during chromosome condensation indicated by orange color in merged photomicrograph. (D) Co-localization is maximal in G2/M cells on condensed chromosomes.
Next, we examined Metnase and Topo IIα intracellular co-localization using immunofluorescence microscopy. This analysis revealed that Metnase and Topo IIα frequently co-localize, and co-localization was particularly strong on condensed chromosomes (Figure 1C). When cells were segregated by DNA content, there was a 2-fold greater co-localization of Metnase and Topo IIα in G2/M cells than in G1 or S phase cells (Figure 1D).
Metnase promotes progression through the metaphase decatenation checkpoint
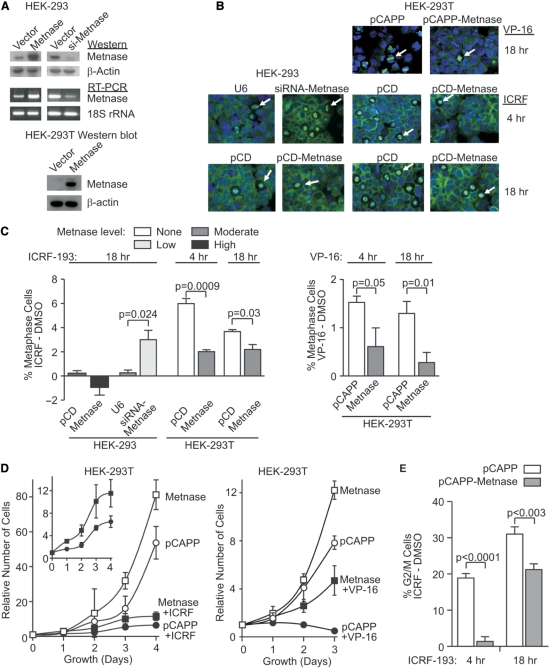
The decatenation checkpoints can be triggered by exposure to ICRF-193, which blocks Topo IIα function without damaging DNA and therefore does not activate the G2/M DNA damage checkpoint (11–17). Progression through the metaphase decatenation checkpoint, the last point at which chromatids can decatenate before aberrant separation, was analyzed by measuring the percentage of cells arrested in metaphase by ICRF-193. Increases in the percentage of ICRF-193-induced metaphase cells reflect arrest at the metaphase decatenation checkpoint. We examined HEK-293 cells expressing normal, 3-fold increased or 5-fold decreased levels of Metnase (Figure 2A), after treatment with ICRF-193, VP-16 or vehicle control (Figure 2B and C). HEK-293 cells express moderate levels of both Topo IIα and Metnase and typically display an attenuated metaphase decatenation checkpoint. These cells were chosen because they provide enhanced sensitivity for detecting increased decatenation checkpoint arrest with Metnase under-expression. Most cell types express Metnase (5), but HEK-293T cells do not express Metnase at detectable levels, perhaps due to their transformation with T antigen (Figure 2A, 24). They have an intact metaphase decatenation checkpoint, which allowed us to analyze the decatenation checkpoint in matched cells that either express or do not express Metnase.
Figure 2.
Metnase promotes progression through the metaphase decatenation checkpoint. (A) Manipulated Metnase levels in HEK-293 and HEK-293T cells examined using western analysis with anti-Metnase antibody and RT–PCR as described (5). β-Actin and 18S rRNA served as loading controls for western blots and RT–PCR, respectively. HEK-293, but not HEK-293T, cells express endogenous Metnase. Stable Metnase over-expression was achieved in HEK-293 cells transfected with pCD-Metnase, and HEK-293T cells transfected with pCAPP-Metnase; siMetnase indicates stably transfected siRNA Metnase knockdown in HEK-293. (B) Decatenation checkpoint arrest induced by ICRF-193 was monitored by the increase in metaphase cells using immunofluorescence microscopy of cells stained with anti-tubulin antibodies (green) and DAPI (blue). pCD-vector control, U6-siRNA control. (C) Quantitative analysis of ICRF-193 or VP-16 induces mitotic checkpoint arrest after subtraction of vehicle controls. (D) HEK-293T cells stably transfected with pCAPP-Metnase or empty vector were seeded in medium with or without 0.5 μM ICRF-193 on Day 0 and counted daily. HEK-293T cells stably transfected with pCAPP-Metnase or empty vector were seeded in medium with or without 5 μM VP-16 on Day 0 and counted daily thereafter. (E) Flow cytometric analysis of G2/M fractions in HEK-293T cells stably transfected with empty or Metnase expression vectors after 10 µM ICRF-193 treatment.
When cells were treated with ICRF-193, the percentage of cells arrested in metaphase was inversely proportional to Metnase levels. Thus, Metnase expression in HEK-293T cells resulted in a 3-fold decrease in metaphase cells compared to vector controls after a 4-h ICRF-193 treatment, and a 2.5-fold decrease after an 18-h treatment. Reducing Metnase expression in HEK-293 cells resulted in a 12-fold increase in metaphase cells after ICRF-193 exposure compared to control cells, and increasing Metnase expression in HEK-293 cells resulted in a 5-fold decrease in the percentage of metaphase cells. We also observed that ICRF-193 treatment yields a significantly higher percentage of metaphase cells in untransduced HEK-293T cells, which lack Metnase, compared to untransduced HEK-293 cells, which express moderate levels of Metnase. Thus, resistance to ICRF-193-induced metaphase arrest correlates with Metnase expression level. Interestingly, increased Metnase also reduced metaphase arrest induced by VP-16 (Figure 2B and C), although such an arrest could also occur from DSBs. Metnase over-expression allowed cells to continue to proliferate in the presence of cytotoxic concentrations of VP-16 by 4-fold (Figure 2D). In addition, Metnase over-expression promoted proliferation in the presence of ICRF-193 by 2-fold (Figure 2D).
We confirmed the effects of Metnase expression on progression through the decatenation checkpoints by analyzing cell cycle profiles in the presence or absence of ICRF-193 using flow cytometry. This measures both G2/M and metaphase decatenation checkpoints (15). When HEK-293T cells were treated for 4 h with 10 μM ICRF-193, the increase in the G2/M fraction was 13.5-fold higher in control cells compared to cells transfected with a Metnase expression vector (Figure 2E). After an 18-h treatment with ICRF-193, the difference between control and Metnase-expressing cells was smaller, but there were still significantly less cells arrested in G2/M with increased Metnase levels (Figure 2E). Based on these in vivo data, we proposed that increased expression of Metnase enhances Topo IIα activity, increasing the efficiency of chromosome decatenation and thereby inhibiting initiation of the decatenation checkpoint arrest.
Metnase enhances Topo IIα decatenation activity
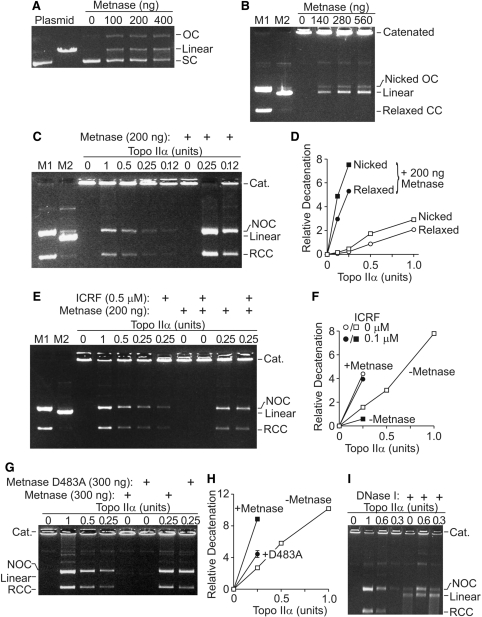
To understand the molecular mechanism by which Metnase promotes decatenation, we characterized the activity of purified Metnase on supercoiled and catenated DNA in vitro. Metnase converts supercoiled plasmid to nicked, relaxed and linear forms, but does not further degrade DNA (Figure 3A), as we showed previously (9). We next examined the activity of Metnase on catenated kDNA, which is composed of interlocked DNA minicircles (25,26). As shown in Figure 3B, Metnase converts catenated kDNA into mostly linear DNA and a smaller amount of free, nicked, open circular DNA. In contrast, Topo IIα converts kDNA into fully decatenated, relaxed, covalently closed circular DNA (Figure 3C).
Figure 3.
Metnase promotes Topo-IIα-mediated decatenation of kDNA. (A) Purified recombinant Metnase nicks supercoiled plasmid to relaxed, open circular and linearized plasmid but does not degrade it. (B) Purified Metnase decatenates kDNA, forming linear DNA and a smaller amount of nicked, open circular DNA. M1, M2-markers for kDNA forms as indicated. (C) Purified Metnase enhances Topo IIα decatenation of kDNA, forming relaxed closed circular DNA. At concentrations of Metnase and Topo IIα where neither alone has an appreciable decatenation activity, they act synergistically to decatenate kDNA to nicked open circular and relaxed closed circular DNA. (D) Graph of dose–response of the enhancement of Topo IIα decatenation by Metnase. Densitometric analysis of open circular and closed circular versus well kDNA for relative kDNA decatenation from n = 3, SD too small for symbols. (E) ICRF-193 does not abrogate Metnase enhancement of Topo IIα decatenation. (F) Graph of densitometric scans of kDNA decatenation performed in the presence of ICRF-193 shown in E, n = 3, SD too small for symbols. (G) Metnase lacking nuclease activity [D483A, ref. 9)] loses most but not all of its ability to enhance Topo IIα decatenation. (H) Graph of densitometry from three D483A experiments, SD shown where larger than symbol. (I) DNase I nicks, linearizes and ultimately degrades kDNA, as opposed to Metnase activity in B.
Since Metnase both interacts with Topo IIα and possesses catenated DNA cleavage activity, we next tested whether Metnase and Topo IIα might function together in decatenating kDNA. At concentrations of Metnase and Topo IIα where neither protein had appreciable activity on kDNA, they synergistically decatenated kDNA into nicked and relaxed fully decatenated circular forms (Figure 3C and D). Averaging multiple experiments, there was more than a 6-fold increase in decatenation when Metnase was added to limiting amounts of Topo IIα (Figure 3D). The presence of ICRF-193 slightly reduced the enhancement of kDNA decatenation by Metnase, but that enhancement was still significant (Figure 3E and F). There was still more than a 4-fold enhancement of decatenation with added Metnase. However, when the nuclease activity of Metnase is deleted by mutating a required amino acid [D483A, ref. (9)] Metnase loses much but not all of its ability to enhance Topo IIα (Figure 3G and H).
To rule out the possibility that the enhanced decatenation is due to nonspecific Metnase nuclease activity, we analyzed kDNA decatenation by Topo IIα in the presence or absence of DNase I. In contrast to Metnase, DNase I did not enhance Topo IIα decatenation to circular forms, but it did produce linear DNA, which over time was degraded (Figure 3I). Indeed, the presence of DNase I inhibited Topo IIα decatenation activity.
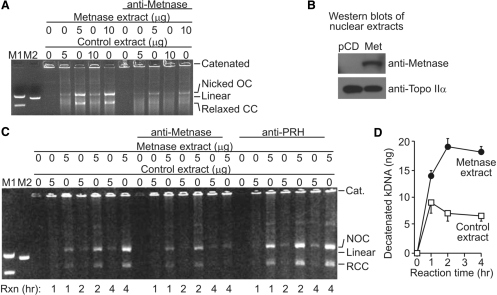
To further investigate the potentiation of Topo IIα decatenation by Metnase, we tested the effect of Metnase on kDNA decatenation with nuclear extracts from HEK-293T cells, which do not express Metnase, and a derivative stably transfected with a Metnase expression vector. As shown in Figure 4A, nuclear extracts from cells expressing Metnase were significantly more effective at decatenating kDNA than HEK-293T extracts, although the Topo IIα levels were unchanged by over-expression of Metnase (Figure 4B). This effect was Metnase specific because the decatenation activity of Metnase-containing extracts was reduced by anti-Metnase antibodies (Figure 4C). Thus, Metnase levels in nuclear extracts correlated with the ability of these extracts to decatenate kDNA. We then examined the rate of decatenation of kDNA by nuclear extracts with and without Metnase. Metnase-containing extracts decatenated kDNA at a 3-fold higher rate than HEK-293T control extracts (Figure 4C and D). As a control, decatenation with these nuclear extracts was examined in the presence of an irrelevant antibody. Anti-sera against the Prh homeobox protein used as a control did not abrogate the Metnase-mediated enhancement of decatenation (Figure 4C). These data indicate that although Topo IIα is capable of decatenating DNA in the absence of Metnase, Metnase significantly increases the reaction rate.
Figure 4.
Decatenation of kDNA by nuclear extracts is enhanced by Metnase. (A) kDNA was treated with nuclear extracts from HEK-293T cells transfected with empty vector (control extracts) or derivatives stably transfected with a Metnase expression vector (Metnase extracts). Reactions containing anti-sera against Metnase show significantly reduced decatenation. (B) Topo IIα levels were similar in the two cell lines by western analysis, while Metnase was present only in the transfected line. (C) Time course of kDNA decatenation with and without Metnase. An antibody against the homeobox gene Prh did not inhibit Metnase-mediated decatenation as did the antibody against Metnase. (D) Graph of time course of kDNA decatenation reactions catalyzed by nuclear extracts, average of three experiments.
Metnase automethylation prevents potentiation of Topo IIα-mediated decatenation
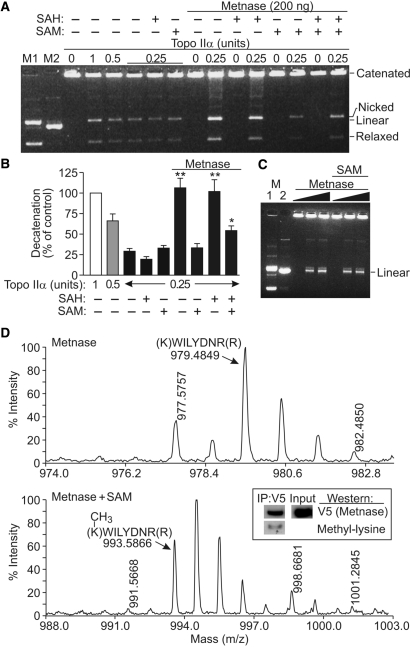
SET domain proteins were originally defined as histone methylases, but they also methylate other proteins, including themselves and transcription factors (26–28). We, therefore, investigated whether decatenation could be regulated by the histone methylase activity of Metnase. It was possible that Metnase could methylate itself or Topo IIα, modifying its synergistic effect on decatenation. S-Adenosyl methionine (SAM) is the primary cellular methyl donor, and we tested whether SAM influenced Metnase potentiation of Topo IIα-mediated decatenation. Interestingly, SAM fully abrogated the effect of Metnase on decatenation (Figure 5A and B). SAM had no effect on decatenation by Topo IIα alone, indicating that the abrogation of Metnase potentiation of Topo IIα activity is Metnase dependent. S-Adenosyl homocysteine (SAH) is a pseudo-methyl donor that binds to methyltransferase active sites but cannot donate a methyl group. SAH had no effect on the Metnase potentiation of Topo IIα decatenation activity (nor did it affect Topo IIα activity alone) (Figure 5A and B), indicating that the SAM abrogation of decatenation with Metnase depends on methyl transfer. When SAM and SAH were both present, decatenation by Metnase and Topo IIα partially reduced, indicating that SAH competes with SAM to prevent methylation. However, SAM did not alter the nuclease activity of Metnase (Figure 5C).
Figure 5.
SAM blocks the Metnase potentiation of Topo IIα decatenation. (A) kDNA decatenation assays performed as in Figure 4 in the presence or absence of the methyl donor SAM or non-donating SAM analog, SAH (2 mM each, except 1 mM each when both were present). (B) Plot of decatenation with 0.25–1 U of Topo IIα, 200 ng Metnase, 2 mM SAM or SAH based on densitometric scans of gels as in A. Shown are averages for three experiments, with all values normalized to 1 U of Topo IIα (control = 100%). Statistics calculated by t-tests. (C) SAM (2 mM) does not affect the Metnase nuclease activity toward the kDNA substrate. Metnase was present at 0, 100 or 200 ng indicated by triangles. Metnase is automethylated at residue K485. (D) MALDI tof-tof mass spectrum of Metnase alone, showing the unmodified K485 peptide at m/z of 979.4849 as predicted. Mass spectrum of Metnase incubated with SAM in methylase buffer showing methylation of the K485 peptide at m/z of 993.5866. Metnase immunoprecipitated from HEK-293 cells has metylated lysines. Methylated Metnase is present intracellularly, by western analysis.
The reduction of decatenation by SAM could reflect Metnase automethylation, or methylation of Topo IIα. Therefore, we performed MALDI Tof-Tof mass spectroscopy on Metnase and Topo IIα individually and together in the presence or absence of SAM. We found no evidence of Topo IIα methylation (data not shown), but in the presence of SAM, Metnase was automethylated at several positions. The strongest signal was monomethylation of K485, with an average of 86% of lysine residues methylated at this position (Figure 5D). Methylation of three other arginine and lysine residues was also apparent (R223, R469 and K122), but <25% of these residues were methylated (data not shown). These results suggest that Metnase automethylation regulates its ability to potentiate Topo IIα-dependent decatenation. We next asked whether methylated Metnase exists intracellularly. We found that Metnase immunoprecipitated from HEK-293 cells was recognized by anti-methyl lysine antibodies on western analysis (Figure 5D).
DISCUSSION
We showed previously that Metnase methylates histone H3 K36 and promotes DSB repair by NHEJ (5). Others have shown that Metnase has partial transposase activity and is only present in primates (6–8). We now show that Metnase interacts with Topo IIα, these proteins co-localize in vivo and Metnase enhances Topo IIα decatenation. This interaction was independent of DNA. We also found that Metnase levels regulate initiation of metaphase decatenation checkpoint arrest. Cells with increased levels of Metnase show attenuated decatenation checkpoint arrest compared to cells with lower levels of Metnase. Over-expression of Metnase compensates for ICRF-193 and VP-16 inhibition of Topo IIα, preventing engagement of this checkpoint. Based on these in vivo data, we propose that increased expression of Metnase enhances Topo IIα activity, increasing the efficiency of chromosome decatenation and inhibiting initiation of decatenation checkpoint arrest. Cancer cells also show attenuated decatenation checkpoints when Topo IIα is inhibited by ICRF-193 (11–17). It is possible that the resistance to ICRF-193 in these cells is partially mediated by robust Metnase levels.
VP-16 promotes cell cycle arrest by inhibiting Topo IIα function following its DNA cleavage step, and this DNA-damage-induced arrest is distinct from the decatenation checkpoint (11–17). However, VP-16 also triggers decatenation checkpoint arrest by inhibiting Topo IIα, although the decatenation arrest is often overshadowed by the DNA damage checkpoint. We show here that increasing Metnase levels overcomes VP-16 effects on metaphase arrest and on cell proliferation. This could result from Metnase potentiation of Topo IIα decatenation and/or DSB repair by NHEJ. The increase in proliferation with Metnase over-expression in the presence of VP-16 is greater than in the presence of ICRF-193. It is likely that this increase is due to the additive DNA repair component of Metnase on VP-16-induced DNA damage, which does not occur with ICRF-193. Alternatively, it is also possible that Metnase blocks VP-16 (and ICRF-193) access to Topo IIα. Regardless, these results suggest that Metnase is an important mediator of resistance to VP-16 used in treating human malignancies. It is also noteworthy that the Metnase gene is located at 3p26, which shows frequent abnormalities in multiple cancers including acute and chronic lymphocytic leukemia, breast cancer, hereditary prostate cancer, myeloma, myelodysplasia and non-Hodgkin's lymphoma (5). It is possible that the cancer-associated 3p26 abnormalities alter the function or expression of Metnase.
The in vivo observations implicating Metnase in promoting Topo-IIα-mediated chromosome decatenation are supported by in vitro analyses with purified proteins. Although Topo IIα alone can fully decatenate kDNA, Metnase greatly enhances this activity. The results with purified proteins are also consistent with the nuclear extract studies, which showed enhanced decatenation when Metnase levels were increased. The enhancement of decatenation by Metnase is not a result of nonspecific nuclease activity because DNase I did not promote Topo IIα decatenation activity. The nuclease activity of Metnase is important for its enhancement of decatenation, as deleting that activity (9) markedly decreases its Topo IIα enhancement. However, there is still some enhancement remaining, and Metnase can still enhance Topo IIα decatenation in buffers and at levels where there is little nuclease function seen. Thus, Metnase may also function as a structural co-factor to increase Topo IIα activity. This model is consistent with the known mechanism of Topo IIα, as DNA cleavage, strand passage and religation are catalyzed by Topo IIα in a coordinated set of covalent reactions (10).
Metnase may promote Topo IIα activity in a manner analogous to extra-cellular signal-related kinase (ERK2). ERK2 promotes Topo IIα activity, and this was initially thought to depend on phosphorylation. ERK2 does indeed phosphorylate Topo IIα in vitro. However, this phosphorylation is not required to enhance Topo IIα activity as kinase-inactive ERK2 also stimulates Topo IIα (29). ERK2, therefore, functions as a structural co-factor for Topo IIα activity in vitro. This is also similar to HMGB1 interacting with Topo IIα to enhance its DNA binding and its decatenation activity in vitro (30). This mechanism is in contrast to BRCA1, which may activate Topo IIα by ubiquitylation (31).
Besides Metnase, only BRCA1 has been demonstrated to regulate the in vivo decatenation checkpoint response. Topo IIα exists in a large complex termed the toposome, and components of this complex may regulate Topo IIα function (32,33). Metnase promotes both decatenation and NHEJ, and its varied activities could be regulated by its presence or absence in the toposome. For example, Metnase recruitment to DNA damage sites could deplete it from the toposome, and this could reduce Topo IIα activity, slowing decatenation and cell cycle progression, thereby increasing time for repair before mitosis. We recently reported that Metnase interaction with the DNA repair component Pso4 is induced by DNA damage and is important for the ability of Metnase to enhance NHEJ (34). It is possible that Metnase is sequestered with Pso4 after DNA damage, which might preclude it from Topo IIα interaction, and thus slowing decatenation.
There is another example of an SET domain protein regulated by automethylation. G9a negatively regulates its histone methylation activity by automethylation (27,28). Metnase methylates histone H3 in vitro (5), and we show here that Metnase is strongly automethylated on K485 and weakly automethylated at three other residues. Metnase does not appear to methylate Topo IIα, but SAM blocks Metnase potentiation of Topo IIα decatenation. If SAM binding to the Metnase methyltransferase active site inhibited the Metnase–Topo IIα interaction, then SAH would be expected to produce the same effect, but this was not observed. Metnase potentiation of Topo IIα decatenation is inhibited by SAM, and this requires the donation of a methyl group to Metnase. Also, lysine-methylated Metnase species exist intracellularly, as such species could be immunoprecipitated. This represents a novel function for an SET domain protein and suggests a new regulatory mode for this important class of protein post-translational modification.
Metnase is widely expressed in human tissues and cell lines (5), but three cell lines have been identified that do not express Metnase, all of which are transformed by SV40 T-antigen, including the HEK-293T cells used here (also the BPH and GM05849 lines). It is possible that T-antigen represses Metnase expression, or that it functionally replaces Metnase in some manner, selecting for cells that do not express it.
The data here have general significance for several reasons. First, the precise biochemical process whereby replicated chromosomes are untangled is not well understood, but it is clear that Metnase enhances this process, perhaps at an early, rate-limiting step of decatenation. Second, the automethylation of Metnase is a unique feedback regulatory mechanism to slow down decatenation. It also shows that SET domain protein automethylation can affect other functions besides histone methyltransferase activity, as seen with G9a (27,28). Third, the ability of cells expressing Metnase to continue to proliferate in the presence of lethal concentrations of the clinically relevant Topo IIα inhibitor VP-16 (Figure 2D) suggests a combination chemotherapy strategy in which an inhibitor of Metnase would potentiate the lethal effects of topoisomerase inhibitors on human cancer. Finally, since transposase activity would be deleterious to humans, the benefit of a transposase domain protein in humans has not been defined (6,9,35). However, these data demonstrate such a benefit, and perhaps shed light on how Metnase was selected for in primates.
FUNDING
National Institutes of Health (R01 CA100862 to J.A.N.); APRC supplement to CA100862 (to J.A.N. and R.H.); National Institutes of Health (R01 CA102283 to R.H., R01 HL075783 to R.H.); the Leukemia and Lymphoma Society (SCOR 7388-06 to R.H.). Funding for open access charge: NIH RO1 CA102283.
Conflict of interest statement. None declared.
Footnotes
The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint First Authors.
REFERENCES
- 1.Brillet B, Bigot Y, Auge-Gouillou C. Assembly of the Tc1 and mariner transposition initiation complexes depends on the origins of their transposase DNA binding domains. Genetica. 2007;130:105–120. doi: 10.1007/s10709-006-0025-2. [DOI] [PubMed] [Google Scholar]
- 2.Miskey C, Izsvak Z, Kawakami K, Ivics Z. DNA transposons in vertebrate functional genomics. Cell Mol. Life Sci. 2005;62:629–641. doi: 10.1007/s00018-004-4232-7. [DOI] [PubMed] [Google Scholar]
- 3.Dupuy AJ, Jenkins NA, Copeland NG. Sleeping beauty: a novel cancer gene discovery tool. Hum. Mol. Genet. 2006;15(Spec No 1):R75–R79. doi: 10.1093/hmg/ddl061. [DOI] [PubMed] [Google Scholar]
- 4.Plasterk RH, Izsvak Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 5.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc. Natl Acad. Sci. USA. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl Acad. Sci. USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol. Cell Biol. 2007;27:1125–1132. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miskey C, Papp B, Mates L, Sinzelle L, Keller H, Izsvak Z, Ivics Z. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell Biol. 2007;27:4589–4600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roman Y, Oshige M, Lee YJ, Goodwin K, Georgiadis MM, Hromas RA, Lee SH. Biochemical characterization of a SET and transposase fusion protein, Metnase: its DNA binding and DNA cleavage activity. Biochemistry. 2007;46:11369–11376. doi: 10.1021/bi7005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 11.Downes CS, Clarke DJ, Mullinger AM, Gimenez-Abian JF, Creighton AM, Johnson RT. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- 12.Clarke DJ, Vas AC, Andrews CA, Diaz-Martinez LA, Gimenez-Abian JF. Topoisomerase II checkpoints: universal mechanisms that regulate mitosis. Cell Cycle. 2006;5:1925–1928. doi: 10.4161/cc.5.17.3200. [DOI] [PubMed] [Google Scholar]
- 13.Toyoda Y, Yanagida M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol. Biol. Cell. 2006;17:2287–2302. doi: 10.1091/mbc.E05-11-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikhailov A, Shinohara M, Rieder CL. Topoisomerase II and histone deacetylase inhibitors delay the G2/M transition by triggering the p38 MAPK checkpoint pathway. J. Cell Biol. 2004;166:517–526. doi: 10.1083/jcb.200405167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skoufias DA, Lacroix FB, Andreassen PR, Wilson L, Margolis RL. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol. Cell. 2004;15:977–990. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Damelin M, Bestor TH. The decatenation checkpoint. Br. J. Cancer. 2007;96:201–205. doi: 10.1038/sj.bjc.6603537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deming PB, Cistulli CA, Zhao H, Graves PR, Piwnica-Worms H, Paules RS, Downes CS, Kaufmann WK. The human decatenation checkpoint. Proc. Natl Acad. Sci. USA. 2001;98:12044–12049. doi: 10.1073/pnas.221430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews CA, Vas AC, Meier B, Gimenez-Abian JF, Diaz-Martinez LA, Green J, Erickson SL, Vanderwaal KE, Hsu WS, Clarke DJ. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/securin. Genes Dev. 2006;20:1162–1174. doi: 10.1101/gad.1367206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doherty SC, McKeown SR, McKelvey-Martin V, Downes CS, Atala A, Yoo JJ, Simpson DA, Kaufmann WK. Cell cycle checkpoint function in bladder cancer. J. Natl Cancer Inst. 2003;95:1859–1868. doi: 10.1093/jnci/djg120. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa T, Hayashita Y, Maeno K, Masuda A, Sugito N, Osada H, Yanagisawa K, Ebi H, Shimokata K, Takahashi T. Identification of decatenation G2 checkpoint impairment independently of DNA damage G2 checkpoint in human lung cancer cell lines. Cancer Res. 2004;64:4826–4832. doi: 10.1158/0008-5472.CAN-04-0871. [DOI] [PubMed] [Google Scholar]
- 21.Deming PB, Flores KG, Downes CS, Paules RS, Kaufmann WK. ATR enforces the topoisomerase II-dependent G2 checkpoint through inhibition of Plk1 kinase. J. Biol. Chem. 2002;277:36832–36838. doi: 10.1074/jbc.M206109200. [DOI] [PubMed] [Google Scholar]
- 22.Ishida R, Miki T, Narita T, Yui R, Sato M, Utsumi KR, Tanabe K, Andoh T. Inhibition of intracellular topoisomerase II by antitumor bis(2,6-dioxopiperazine) derivatives: mode of cell growth inhibition distinct from that of cleavable complex-forming type inhibitors. Cancer Res. 1991;51:4909–4916. [PubMed] [Google Scholar]
- 23.Lukes J, Guilbride DL, Votypka J, Zikova A, Benne R, Englund PT. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell. 2002;1:495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapiro TA. Kinetoplast DNA maxicircles: networks within networks. Proc. Natl Acad. Sci. USA. 1993;90:7809–7813. doi: 10.1073/pnas.90.16.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hromas R, Van Ness B. Nuclear factors bind to regulatory regions of the mouse kappa immunoglobulin gene. Nucleic Acids Res. 1986;14:4837–4848. doi: 10.1093/nar/14.12.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 27.Chin HG, Esteve PO, Pradhan M, Benner J, Patnaik D, Carey MF, Pradhan S. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res. 2007;35:7313–7323. doi: 10.1093/nar/gkm726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro PS, Whalen AM, Tolwinski NS, Wilsbacher J, Froelich-Ammon SJ, Garcia M, Osheroff N, Ahn NG. Extracellular signal-regulated kinase activates topoisomerase IIa through a mechanism independent of phosphorylation. Mol. Cell Biol. 1999;19:3551–3560. doi: 10.1128/mcb.19.5.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stros M, Bacíková A, Polanská E, Stokrová J, Strauss F. HMGB1 interacts with human topoisomerase IIalpha and stimulates its catalytic activity. Nucleic Acids Res. 2007;35:5001–5013. doi: 10.1093/nar/gkm525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou Z, Minter-Dykhouse K, Chen J. BRCA1 participates in DNA decatenation. Nat. Struct. Mol Biol. 2005;12:589–593. doi: 10.1038/nsmb953. [DOI] [PubMed] [Google Scholar]
- 32.Borowiec JA. The toposome: a new twist on topoisomerase IIalpha. Cell Cycle. 2004;3:627–628. doi: 10.4161/cc.3.5.822. [DOI] [PubMed] [Google Scholar]
- 33.Lee CG, Hague LK, Li H, Donnelly R. Identification of toposome, a novel multisubunit complex containing topoisomerase IIalpha. Cell Cycle. 2004;3:638–647. [PubMed] [Google Scholar]
- 34.Beck BD, Park SJ, Lee YJ, Roman Y, Hromas RA, Lee SH. Human PSO4 is a Metnase (SETMAR) binding partner that regulates Metnase function in DNA repair. J. Biol. Chem. 2008;283:9023–9030. doi: 10.1074/jbc.M800150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]