Abstract
Rigid spin-labeled nucleoside Ç, an analog of deoxycytidine that base-pairs with deoxyguanosine, was incorporated into DNA oligomers by chemical synthesis. Thermal denaturation experiments and circular dichroism (CD) measurements showed that Ç has a negligible effect on DNA duplex stability and conformation. Nucleoside Ç was incorporated into several positions within single-stranded DNA oligomers that can adopt two hairpin conformations of similar energy, each of which contains a four-base loop. The relative mobility of nucleotides in the alternating C/G hairpin loops, 5′-d(GCGC) and 5′-d(CGCG), was determined by electron paramagnetic resonance (EPR) spectroscopy. The most mobile nucleotide in the loop is the second one from the 5′-end, followed by the third, first and fourth nucleotides, consistent with previous NMR studies of DNA hairpin loops of different sequences. The EPR hairpin data were also corroborated by fluorescence spectroscopy using oligomers containing reduced Ç (Çf), which is fluorescent. Furthermore, EPR spectra of duplex DNAs that contained Ç at the end of the helix showed features that indicated dipolar coupling between two spins. These data are consistent with end-to-end duplex stacking in solution, which was only observed when G was paired to Ç, but not when Ç was paired with A, C or T.
INTRODUCTION
Electron paramagnetic resonance (EPR) spectroscopy has been used extensively to study structure and dynamics of proteins (1–4) and to a lesser extent of nucleic acids (5–8). EPR studies require unpaired electrons and thus, studies of biopolymers rely on incorporation of free radicals or paramagnetic metal ions. Nitroxides are stable organic free radicals that have been extensively used as labels for EPR spectroscopy. In recent years, there have been advances in the spin-labeling of nucleic acids, both through conjugation of nitroxide spin labels to the sugar moieties (9,10), the phosphodiester (9,11,12) or nucleotide bases (13–18), providing nucleic acid samples for EPR studies.
Most of the spin labels that have been described to date are linked to the nucleic acid with a tether that has some degree of flexibility. Thus, these spin labels have movement independent of the nucleic acid to which they are attached. The mobility varies, depending on the structure of the tether. For example, flexible tethers (14) result in a relatively large degree of freedom for the spin label. Therefore, the dominant feature of EPR spectra of DNAs containing mobile spin labels is a very fast moving component resulting from rapid movement of the spin label itself. Spin labels that are tethered to the biopolymer with semi-flexible (7,10) or semi-rigid tethers have less motion, such as 2,2,5,5-tetramethyl-pyrrolin-1-yloxyl-3-acetylene (TPA) connected by an acetylenic tether to the 5-position of pyrimidines, originally synthesized and incorporated into DNA by Hopkins and coworkers (13,15) and later used for spin-labeling of both DNA and RNA (17,18). However, TPA spin labels have some motion due to rotation of the spin label around the single bonds flanking the acetylene tether (19).
Mobility of the spin label can affect the study of nucleic acid structure and dynamics. Even semi-flexible or semi-rigid labels that have been shown to report on the motion of the nucleotide to which they are attached have some inherent mobility that complicates analysis of their motions (10,20–22). Consequently, the motion of the label needs to be taken into account when studying movements (dynamics) of the nucleotide to which the spin label is attached. Furthermore, when using the spin labels for distance measurements, the residual motion of the spin label will yield a larger distance distribution. Therefore, it is of interest to prepare rigid spin labels, where the nitroxide does not move independent of the nucleic acid to which it is attached.
The first rigid spin label for nucleic acids (Q) was described by Hopkins and coworkers (23). The nitroxide was immobilized by fusing the five-membered ring of the nitroxide to a nucleoside base and enabled the study of DNA internal dynamics by EPR spectroscopy (24–27). However, the preparation of this novel C-nucleoside requires over 20 synthetic steps. In addition, its base-pairing partner is a nonnatural nucleotide containing 2-aminopurine as a base. Therefore, we decided to design and synthesize new rigid spin label (Ç), which is an N-nucleoside and base-pairs with natural nucleotide G(1) (28).

In this article, we describe the details of the incorporation of Ç into DNA oligonucleotides, characterization of the spin-labeled oligomers and the use of Ç-labeled oligomers to study DNA folding/conformation. Nitroxides are known to be partially reduced using the standard reaction conditions for chemical synthesis of nucleic acids (29). Reduction of Ç renders the nucleotide fluorescent (28,30), which enabled us to readily monitor its reduction and optimize the reaction conditions used for oligomer syntheses, in addition to using fluorescence spectroscopy to study oligomer folding. The incorporation of the spin label was verified by mass spectrometry and enzymatic digestion. Circular dichroism (CD) measurements and thermal denaturation studies indicate that Ç has negligible effect on stability and conformation of DNA duplexes. We also show that Ç is a sensitive EPR probe for studying nucleic acid folding. Furthermore, incorporation of Ç at the end of duplexes enabled observation of end-to-end duplex stacking in solution by EPR spectroscopy.
MATERIALS AND METHODS
General
Water was purified on a MILLI-Q water purification system. Analytical thin layer chromatography (TLC) was performed on glass plates (Merck Kieselgel 60 F254). DNA oligomers were synthesized on an ASM 800 DNA synthesizer from Biosset, Novosibirsk, Russia. All commercial phosphoramidites and columns were purchased from ChemGenes, Wilmington, MA, USA. Solvents and reagents were purchased from ChemGenes, SigmaAldrich, St. Louis, MO, USA and Applied Biosystems, Foster City, CA, USA. Unmodified DNAs were purchased from TAG Copenhagen, Copenhagen, Denmark. UV-VIS spectra were recorded on a PerkinElmer Lambda 25 UV/VIS spectrometer. Enzymatically digested DNAs were run on a Beckman Coulter Gold HPLC system using Beckman Coulter Ultrasphere C18 4.6 × 250 mm analytical column with UV detection at 254 nm. Solvent gradients for analytical RP-HPLC were run at 1.0 ml/min using following gradient program: solvent A, 50 mM Et3NOAc (pH 7.0); solvent B, 100% CH3CN; 4% B isocratic for 4 min, 10 min linear gradient from 4% B to 20% B, 10 min linear gradient to 50% B, 5 min linear gradient to 80% B, isocratic for 1 min, then a 5-min gradient to initial conditions (4% B). CD spectra were recorded on a JASCO J-810 spectropolarimeter at 25°C with path length of 1 mm (Hellma, Essen, Germany), 10 scans, scanned from 500 to 200 nm with response of 1 s, data pitch of 0.1 nm and band width of 1.0 nm. Continuous wave (CW) EPR spectra were recorded on a MiniScope MS200 (Magnettech, Berlin, Germany) X-band spectrometer using 100 kHz modulation frequency, 1.0 G modulation amplitude and 2.0 mW microwave power. Molecular weight (MW) of DNA was determined by MALDI-Tof analysis and mass spectra recorded on a Bruker Autoflex III [50–100 μM of 3-hydroxy picolinic acid (3HPA) containing 5–10 μM ammonium citrate/acetate in H2O, concentration of oligomers was ca. 10–20 μM].
Reaction conditions for chemical synthesis of DNA oligomers
The nitroxide in Ç was substantially reduced using these standard conditions and, therefore, we systematically determined what reaction conditions were compatible with the nitroxide as described in more detail in Results and Discussion. In our optimized condition, t-butylhydroperoxide [19.4% tert-butyl hydroperoxide/toluene (v/v)] was used for oxidation instead of iodine and 3% dichloroacetic acid (DCA) in CH2Cl2 (v/v) was used for deprotection of the trityl groups (reaction time 90 s). When using small amounts of spin-labeled phosphoramidite, we used 1,2-dichloroethane for preparation of the spin-labeled phosphoramidite solution and the reaction time for this coupling step was increased from 70 to 380 s. A summary of the standard and modified reaction conditions for DNA synthesis is listed in Table S1.
DNA synthesis and purification
Unmodified and spin-labeled DNAs were synthesized by a trityl-off synthesis on 1.0 μmol scale (1000 Å CPG columns) using phosphoramidites with standard protecting groups. For spin-labeled DNA, the spin-labeled phosphoramidite (28) was site-specifically incorporated into the DNA by automated chemical synthesis. The DNA was deprotected in concentrated ammonia solution at 55°C for 8 h and purified by 23% denaturing polyacrylamide gel electrophoresis (DPAGE). The oligonucleotides were visualized by UV shadowing and the bands were excised from the gel. The gel slices containing the DNA were crushed and soaked in TEN buffer (250 mM NaCl, 10 mM Tris, 1 mM Na2EDTA, pH 7.5). The DNA elution solutions were filtered through 0.45 μm polyethersulfone membrane (disposable filter device from Whatman, Maidstone, Kent, UK) and desalted using Sep-Pak cartridge (Waters Corporation, Milford, MA, USA) according to manufacturer's instructions. After removing the solvent in vacuo, the DNA was dissolved in de-ionized and sterilized water (100 μl). Concentrations of oligonucleotides were calculated from Beer's law based on measurements of absorbance at 260 nm. Extinction coefficients were determined by using the UV WinLab oligonucleotide calculator (V2.85.04, PerkinElmer). The MWs of spin-labeled DNA were calculated using the ‘Mongo Oligo Mass Calculator,’ v2.06 (http://library.med.utah.edu/masspec/mongo.htm). The MWs of spin-labeled oligonucleotides were determined via MALDI-Tof MS analysis (in linear mode, external calibration): 5′-d(GACCTCGÇATCGTG): 4442.4 (calcd. 4442.1); 5′-d(GTGÇGCAC): 2613.5 (calcd. 2611.7); 5′-d(GTGCGÇAC): 2613.1 (calcd. 2611.7); 5′-d(CGCGAATTÇGCG): 3849.9 (calcd. 3848.2); 5′-d(ÇACCTCGCATCGTG): 4402.9 (calcd. 4401.6); 5′-d(GATGÇGCGCGCGCGACTGAC): 6347.2 (calcd. 6345.8); 5′-d(GATGCGÇGCGCGCGACTGAC): 6346.9 (calcd. 6345.8); 5′-d(GATGCGCGÇGCGCGACTGAC): 6345.1 (calcd. 6345.8); 5′-d(GATGCGCGCGÇGCGACTGAC): 6350.2 (calcd. 6345.8); 5′-d(GATGCGCGCGCGÇGACTGAC): 6347.4 (calcd. 6345.8); 5′-d(GTÇAGTCGCGCGCGCGCATC): 6295.5 (calcd. 6296.8); 5′-d(GTÇAGTGCGCGCGCGCGATC): 6336.0 (calcd. 6336.8); 5′-d(GATCGCGCGÇGCGCACTGAC): 6305.7 (calcd. 6305.8); 5′-d(GATCGCGÇGCGCGCACTGAC): 6306.1 (calcd. 6305.8)
UV-monitored thermal denaturation
The DNA duplexes (ca. 2–3 nmol) were prepared by mixing a spin-labeled strand with 1.25 Eq of the complementary strand in PNE buffer (10 mM Na2HPO4, 100 mM NaCl, 0.1 mM Na2EDTA, pH 7.0; 100 μl) and using the following annealing protocol: 90°C for 2 min, 60°C for 5 min, 50°C for 5 min, 40°C for 5 min, 22°C for 15 min. The samples were diluted to 1 mL with PNE buffer and degassed with argon. The DNA duplexes were slowly denatured by heating from 20°C to 90°C (0.5°C/min) and the absorbance at 260 nm recorded at 0.5°C intervals.
Enzymatic digestion of DNA and HPLC analysis
A solution of the unmodified DNA 8-mer 5′-d(GTGCGCAC) (8.3 nmol, 0.6 OD) in Tris buffer (50 mM Tris, 10 mM MgCl2, pH8.0; 40 μl) was treated with 0.1 U of snake venom phosphodiesterase I and 4 U of calf intestinal alkaline phosphatase and incubated at 37°C for 50 h. Spin-labeled DNA 8-mer 5′-d(GTGCGÇAC) (8.3 nmol, 0.6 OD) was digested using the same protocol, however, 3 U of nuclease P1 from Penicillium citrinum were added to effect cleavage of the phosphodiester bonds adjacent to Ç. Samples were analyzed by analytical RP-HPLC (Figure S2).
CD measurements
Spin-labeled DNA single strand 5′-d(GACCTCGÇATCGTG) (2.5 nmol) and its complementary strand 5′-d(CACGATGCGAGGTC) (2.5 nmol) were dissolved in PNE buffer (100 μl), annealed and diluted to 200 μl with PNE buffer. CD spectra of unmodified 14-mer duplex possessed negative and positive molar ellipticities at ca. 250 and 280 nm, characteristic of a right-handed B-DNA (Figure S3).
EPR measurements
The DNA oligomer 5′-d(GACCTCGÇATCGTG) (2 nmol) was dissolved in PNE buffer (10 μl, final conc. 200 μM). For spin-labeled DNA duplex, the DNA oligomer 5′-d(GACCTCGÇATCGTG) (2 nmol) was annealed to its complementary strand 5′-d(CACGATGCGAGGTC) (2.4 nmol) in PNE buffer (final conc. 200 μM of duplex). Samples (10 μl) were placed in a quartz capillary for EPR measurements. EPR spectra were recorded at 20°C.
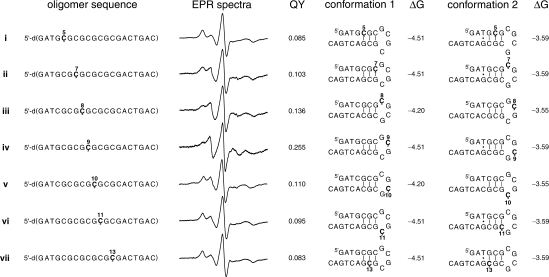
In the folding study of oligomers i–ix (Figure 2), each DNA (0.5 nmol) was dissolved in PNE buffer (10 μl, final conc. 50 μM) and directly measured. All EPR spectra of i–ix were recorded at 20°C.
Figure 2.
Oligomers used for DNA folding studies and their X-band EPR spectra. Also listed are the DNA conformations that are predicted by the mfold software along with their Gibbs free energy (ΔG, kJ/mol) at 20°C and 0.1 M Na+. For sequences i–vii, the number above Ç indicates its position from 5′-end. Fluorescence quantum yields (QY) were obtained after reduction of the spin-labeled oligomers.
Steady-state fluorescence and quantum yield determination
All DNA samples were measured at 5 × 10−6 M in 400 μl of PNE buffer at 20°C using an excitation wavelength of 365.5 nm. Fluorescence spectra were averaged over three scans. Quantum yields were determined using anthracene as a standard for all DNA samples in PNE buffer (Figure 2). Quantum yields of all DNAs were determined using the following equation: ΦF(x) = (As/Ax) · (Fx/Fs) · (nx/ns)2 · ΦF(s), where S is the standard (anthracene), X is the unknown (sample being measured), A is the absorbance at the excitation wavelength (365.5 nm), F is the area under the emission curve, n is the refractive index of the solvent (H2O = 1.3439, EtOH = 1.3610) and Φ is the quantum yield.
RESULTS AND DISCUSSION
Incorporation of Ç into DNA by chemical synthesis
Spin labels have been site-specifically incorporated into DNA through synthesis of phosphoramidites of spin-labeled nucleosides and subsequent chemical synthesis of oligonucleotide (13,15,23,28). Nucleoside Ç was converted to spin-labeled phosphoramidite (2) by tritylation and phosphitylation (28) and used for synthesis of spin-labeled DNA. The initial attempts to synthesize spin-labeled DNA using standard conditions resulted in a fluorescent side-product, which significantly reduced the yield of the spin-labeled DNA. We have previously shown that Ç can be reduced with DTT (28) and Na2S (30) to yield the corresponding hydroxylamine and amine (Çf), respectively, both of which are highly fluorescent. Several lines of evidence suggest that the nitroxide was reduced to the corresponding amine, rather than the hydroxyl amine. For example, our experience shows that hydroxyl amines are readily oxidized to the nitroxide upon normal handling of samples on the bench. The fluorescent byproduct from synthesis of spin labeled DNA did not show such oxidation and could not even be oxidized back to the nitroxide in the presence of either Na2WO4/H2O2 or copper wire/O2. Because of the fluorescence, we could readily study the nitroxide decomposition that occurred during DNA synthesis and thereby allowed optimization of the reaction conditions for chemical synthesis of DNAs containing nitroxides (Table S1).
We first studied stability of Ç in the presence of the iodine oxidation solution, because reduction of nitroxides with iodine has been reported (18,29). After incubating Ç in the iodine oxidation solution for a short time (ca. 1 min) a fluorescent product was observed by TLC analysis. After 90 min, the nitroxide was completely reduced to the corresponding amine. Another commonly used oxidizing agent for oligonucleotide synthesis is tert-butyl hydroperoxide in toluene, employed for the synthesis of RNA using the ACE-chemistry (31). No decomposition of Ç was observed upon incubation in the tert-butylhydroperoxide solution, even after 24 h.
We also tested stability of Ç in the 3% TCA/CH2Cl2 solution used to remove the trityl groups during oligomer synthesis. After 10 min at 22°C, 5–10% of Ç had been reduced to the corresponding amine and was fully reduced after 24 h. Reduction of nitroxides with TCA through a disproportionation mechanism has previously been reported (18,32,33). Therefore, we tested the weaker acid DCA, which has also been used in oligomer synthesis (34). Indeed, reduction of Ç in 3% DCA/CH2Cl2 was much slower; after 10 min at 22°C, no decomposition was observed and only ca. 50% of Ç was reduced after 24 h. Thus, we used DCA instead of TCA for synthesis of spin-labeled DNA. For all other reagents, such as the acetic anhydride/THF solution and the N-methylimidazole/pyridine/THF used in the capping step (or a 50:50 mixture of the two); the activator (0.25 M ethylthiotetrazole/acetonitrile) used in the coupling step, and the concentrated ammonia solution used in the final deprotection step, no reduction of Ç was observed.
When using small amounts of spin-labeled phosphoramidite, we found it advantageous to use 1,2-dichloroethane as a solvent instead of acetonitrile, presumably because it is less hygroscopic than acetonitrile. Since changing the solvent can affect the coupling reaction, we used 31P-NMR to study the coupling reaction of thymine phosporamidite in deuterated 1,2-dichloroethane (0.05 M). Reaction with 5-ethylthiotetrazole gives a peak around 125 p.p.m., presumably corresponding to formation of phosphoramidite/tetrazole intermediate. Reaction of thymine phosporamidite with 5-ethylthiotetrazole in 1,2-dichloroethane was somewhat slower than in acetonitrile, because ca. 5% of the phosporamidite was still unreacted after 95 s but was fully reacted after 180 s. Therefore, we used 1,2-dichloroethane only for preparation of the spin-labeled phosphoramidite solution and the reaction time for this coupling step was increased from 70 to 380 s.
Synthesis and purification of spin-labeled DNAs
The spin-labeled DNA was prepared by automated chemical synthesis using the optimized reaction conditions and purified by DPAGE. During purification, we still observed ca. 5–10% reduction of spin-labeled DNA, as quantified for the 14-mer 5′-d(GACCTCGÇATCGTG), presumably due to the acid treatment. However, this by-product was readily removed by DPAGE purification.
EPR analysis of DPAGE-purified DNA samples showed that spin-labeled DNAs contained some ‘free-spin’ impurity, which refers to a contaminant that displayed narrow lines in the EPR spectrum and indicates a fast isotropic motion of a small molecule (Figure S1). This impurity was efficiently removed by ethanol precipitation. After purification, the spin-labeled DNAs were analyzed by MALDI-Tof MS (see Materials and methods section).
Enzymatic digestion and HPLC analysis
Incorporation of spin label was further verified by enzymatic digestion of the spin-labeled oligomer 5′-d(GTGCGÇAC). We found that digestion of modified oligomer using standard conditions with phosphodiesterase I and alkaline phosphatase resulted in an incomplete digestion. However, adding nuclease P1 from P. citrinum resulted in complete digestion. HPLC analysis of an enzymatically digested unmodified oligomer revealed four peaks, corresponding to natural nucleosides dC, dG, dT and dA (Figure S2A). Similar analysis of the modified oligomer showed, in addition to the natural nucleosides, a new strongly retained peak corresponding to nucleoside Ç (Figure S2B), which was verified by co-injection with an equimolar amount of an authentic sample of Ç (Figure S2C).
Thermal denaturation studies and CD measurements
To determine if Ç affects the stability of DNA duplexes, we measured the melting temperatures (TMs) of unmodified and spin-labeled DNAs (Table 1). Thermal stability of DNA duplexes containing the structurally related phenoxazine (tCO) and phenothiazine (tC) has been studied by Wilhelmsson and coworkers (35,36). Incorporation of tCO and tC resulted in an increase or decrease in TMs (−3°C < ΔTMs < +7°C), depending on the identity of the flanking bases. The TM of spin-labeled DNA 14-mer duplex VII was only one degree lower than the umodified duplex VI (Table 1). Indeed, a similar decrease in TM was observed when G and A were the bases flanking tCO [5′-d(-GtCOA-)] and tC [5′-d(-GtCA-)]. Consistent with this result, the self-complementary doubly spin-labeled DNA 8-mer duplex II has the same flanking sequence [5′-d(-GÇA-)] and a ca. 3°C lower TM. Another doubly spin-labeled DNA 8-mer duplex III had a ca. 3.2°C lower TM than that of unmodified I, while the TMs of tCO/tC-containing oligomers with the same flanking sequence [5′-d(-GÇG-)] were 0–3°C higher. However, molecular modeling indicates that there is a steric clash between the methyl groups of the spin labels in duplex III, which could explain lower than expected TM. In contrast, the TM of double spin-labeled DNA duplex V was ca. 6°C higher than the corresponding unmodified duplex IV, which is a similar result as obtained for a 10-mer containing tCO and tC in the same flanking sequence [5′-d(-TtCOG-)] and [5′-d(-TtCG-)]. A slight increase in TM (2–3°C) was observed when Ç was incorporated at the 5′-end of a duplex (XI, VIII). Combined, our results indicate that Ç has a minor effect on duplex stability. Furthermore, our data suggest that the TMs of spin-labeled duplexes of a particular sequence containing Ç can be predicted based on extensive thermal denaturation studies on DNAs containing the structurally similar analogs tCO and tC (35,36). Thus, any variations in TM originate in the phenoxazine moiety, rather than in the spin label addition, except when spin labels are in close proximity.
Table 1.
TMs of DNA duplexes I–XI containing Ça
| Duplex | Sequence | TMb | ΔTMc |
|---|---|---|---|
| I | 5′-d(GTGCGCAC) | 49.5 ± 0.5 | |
| 3′-d(CACGCGTG) | |||
| II | 5′-d(GTGCGÇAC) | 46.6 ± 0.6 | −2.9 |
| 3′-d(CAÇGCGTG) | |||
| III | 5′-d(GTGÇGCAC) | 46.3 ± 0.5 | −3.2 |
| 3′-d(CACGÇGTG) | |||
| IV | 5′-d(CGCGAATTCGCG) | 57.4 ± 0.5 | |
| 3′-d(GCGCTTAAGCGC) | |||
| V | 5′-d(CGCGAATTÇGCG) | 63.1 ± 0.5 | +5.7 |
| 3′-d(GCGÇTTAAGCGC) | |||
| VI | 5′-d(GACCTCGCATCGTG) | 62.8 ± 0.3 | |
| 3′-d(CTGGAGCGTAGCAC) | |||
| VII | 5′-d(GACCTCGÇATCGTG) | 61.7 ± 0.6 | −1.1 |
| 3′-d(CTGGAGCGTAGCAC) | |||
| VIII | 5′-d(CACCTCGCATCGTG) | 65.4 ± 0.3 | |
| 3′-d(GTGGAGCGTAGCAC) | |||
| IX | 5′-d(ÇACCTCGCATCGTG) | 67.3 ± 0.2 | +1.9 |
| 3′-d(GTGGAGCGTAGCAC) | |||
| X | 5′-d(ACCTCGCATCGTG) | 60.7 ± 0.3 | |
| 3′-d(TGGAGCGTAGCAC) | |||
| XI | 5′-d(ÇACCTCGCATCGTG) | 65.3 ± 0.4 | +4.6 |
| 3′-d(TGGAGCGTAGCAC) |
a2 μM duplex in 10 mM sodium phosphate, 100 mM NaCl, 0.1 mM Na2EDTA, pH 7.0.
bTMs of duplexes (°C).
cΔTM is the difference in the TM values between a duplex having Ç or C in a particular sequence.
To determine if the spin label affects the DNA conformation, a CD spectra of two duplexes containing identical sequences (VII and VI), except that one contained Ç instead of C, were recorded and observed to be nearly identical (Figure S3). Both possessed negative and positive molar ellipticities at ca. 250 and 280 nm, respectively, characteristic of right-handed B-DNA double helix (Figure S3). Thus, CD and thermal denaturation studies indicate that the spin-labeled nucleoside Ç does not significantly alter the conformation and stability of DNA duplexes.
Characterization of spin-labeled oligomers by EPR spectroscopy
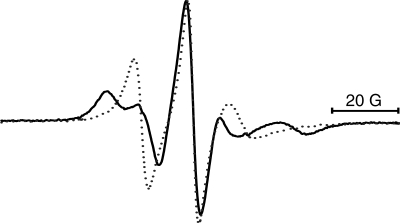
The EPR spectra of spin-labeled oligomer 5′-d(GACCTCGÇATCGTG) and spin-labeled duplex VII (Table 1) are very different (Figure 1). The EPR spectrum of the single strand shows three broad lines. Upon annealing the complementary strand, the spectrum becomes much broader; specifically, the high- and low-field components split into two, indicative of motion in the slow-motion regime. This is consistent with immobilization of the spin label within the duplex as the rotational correlation time estimated from the EPR spectra is similar to that expected for a cylinder with the dimensions of this 14-mer duplex (28). Thus, the rigid spin label is an excellent probe of base-pairing and thereby of nucleic acid secondary structure formation.
Figure 1.
Overlay of EPR spectra of spin-labeled, single-stranded 14-mer DNA 5′-d(GACCTCGÇATCGTG) (dotted line) and spin-labeled duplex DNA 5′-d(GACCTCGÇATCGTG)·5′-d(CACGATGCGAGGTC) (VII) (bold line).
Folding of DNA by EPR spectroscopy
For application of Ç to study DNA folding, we prepared spin-labeled oligomers that could fold into more than one hairpin conformation (Figure 2). Oligomers i–vii represent one series, where the spin label was systematically walked through the stem and the hairpin loop using two similar sequences. Five of those oligomers contained the sequence 5′-d(GATGCGCGCGCGCGACTGAC) (i, ii, iv, vi and vii) and the other two contained the related sequence 5′-d(GATCGCGCGCGCGCACTGAC) (iii and v), where the CG registry in the middle of the sequence has been shifted. These oligomers, because of the alternating 5′-d(CG)-sequence, can form both self-dimers and hairpins. Since the hairpin-dimer equilibrium is concentration dependent, EPR spectra of oligomer iv was recorded at different concentrations (Figure S4). The data showed that self-dimer formation dominated (>90%) at 2 mM concentration, while ca. 40% of the oligomer is present as a self-dimer at 50 μM concentration, where the EPR data in Figure 2 were collected. However, information about nucleotide mobility in the hairpin conformation could be readily extracted from the EPR spectra.
Inspection of the EPR spectra of oligomers i–vii reveals that there are noticeable differences between some of the spectra. In particular, the EPR spectrum of oligomer iv is very different from all the others. In fact, the dominant feature of the EPR spectrum of oligomer iv is similar to a single strand (Figure 1). The smaller peaks that are furthest away from the central peak are features seen in duplex spectra and originate from the self-dimer. In contrast to oligomer iv, the spectra of other oligomers have a stronger duplex-like component. Thus, the EPR data indicate that Ç in position 9 is located in a single-stranded region while the Çs in the other positions are predominantly base-paired or have restricted motion due to stacking. Indeed, the lowest energy DNA conformation of oligomer iv, predicted by the mfold software (37), places Ç9 in the second position of the four-base hairpin loop, where it would be expected to display increased mobility relative to the other Çs (Figure 2). For sequence iv, there are actually two conformations that are predicted to have a similar free energy, both of which place Ç at the tip of the hairpin loop, either in the second or third position from the 5′-end.
Comparison of the EPR spectra for iii and v, both of which contain the same sequence, provides further insights into the mobility of individual nucleotides in the hairpin loop. In particular, the spin label in iii is clearly more mobile than in v, even if both sequences have two conformations that place the spin label either adjacent to the helix or at the tip of the hairpin loop, one from the 5′-end and the other from the 3′-end. Judging from the data for oligomer v, the positions that are at the tip of the hairpin loop are the most mobile and, therefore, the data for oligomers iii and v indicates that the most mobile position in the loop is the second, rather than the third position from the 5′-end. This result is corroborated by NMR studies of DNA hairpin loops, although the NMR structures have not been solved for alternating G and C loops. For example, the previously reported NMR structures of the four-base DNA hairpin loops [5′-d(-GTTA-)] (38) or [5′-d(-TTTU-)] (39) clearly show that the nucleotide second from the 5′-end is the most mobile. The other three nucleotides have a strong potential to stack on 5′- and 3′-end of the stems or the neighboring bases.
The EPR spectra that display the lowest mobility in the i–vii series are that of oligomers i and vii, which is not surprising given the fact that both of the lowest energy folds for these oligomers place the spin label in a duplex. In contrast, the spin label in oligomers ii and vi, both of which contain the same sequence, is placed in a duplex region in one conformation and in the hairpin loop in the other. Both ii and vi display higher mobility by EPR than oligomers i and vii. Oligomers ii and vi also enable the determination of the relative mobility of the nucleotides that are adjacent to the stem, on the 5′- and the 3′-end of the loop. The EPR spectrum of ii shows more mobility, even if the major fold places the label in the duplex region, indicating that the nucleotide at the 5′-end of the loop [5′-d(-ÇGCG-)] is more mobile than the one on the 3′-end [5′-d(-GCGÇ-)]. These data are also supported by NMR structures of DNA hairpin loops that show that the nucleotide on the 3′-end of the loop is stacked between the helix and the adjacent nucleotide in the loop (38,39).
The nitroxide in Ç can be reduced under mild conditions to the corresponding amine (Çf), which is highly fluorescent (30). Fluorescent nucleotides that can form base-pairs with natural nucleotides and are sensitive to their microenvirionment have been used to study nucleic acid folding and local conformational changes (40,41). Therefore, we treated the spin-labeled oligomers i–vii with sodium sulfide (30) and determined their fluorescence quantum yields (Figure 2). Comparison of the EPR- and fluorescence data reveals a direct correlation between the quantum yields and the mobility determined by EPR spectroscopy; the higher the mobility by EPR, the higher the quantum yield. For example, oligomers i and vii, for which the labels are placed in a duplex region in the hairpin conformations, have EPR spectra that display the low mobility of a label locked within a duplex and also the lowest quantum yields. In fact, the quantum yields for a duplex, prepared from oligomer iv and an unmodified complementary sequence, was determined to be almost identical (0.079). The other extreme is oligomer iv, which has the highest quantum yield and the highest mobility. Thus, the fluorescence data are fully consistent with the EPR results and the structural interpretation based on previous NMR studies, discussed above.
Mismatches at the 5′-end and helical stacking by EPR spectroscopy
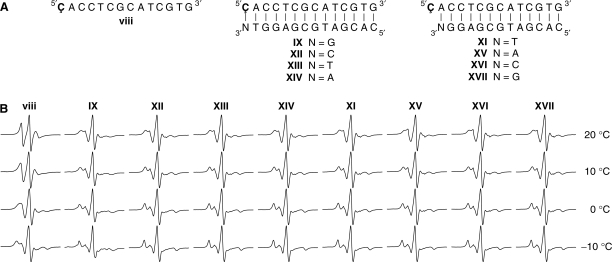
There is currently interest in the detection of single-base mismatches, because many diseases can be traced to genetic disorders based on single nucleotide polymorphisms (42,43). We tried unsuccessfully to use Ç in combination with EPR spectroscopy to readily detect and distinguish between single-base mismatches within a duplex region (data not shown). We argued that it would be easier to detect mismatches if the probe was placed at the end of a duplex region, which would provide more conformational flexibility for the label. Therefore, Ç was incorporated into the oligomers as shown in Figure 3, where Ç was present in a single strand or paired to G, C, T or A at the end of a duplex. The EPR spectra of these spin-labeled oligomers at several different temperatures are shown in Figure 3B. As expected, the EPR spectra of DNA duplexesIX, XII–XIV, fully base-paired or mismatched, were much broader than spectra of single-stranded spin-labeled 14-mer viii at all temperatures. However, the spectra of IX, XII–XIV all look similar, indicating that spin-labeled nucleoside could not only base-pair with G, but also with C, T and A. This result was somewhat unexpected but suggests that stacking is a major determinant of the spin label mobility at the end of duplexes, rather than hydrogen bonding to the opposite base.
Figure 3.
(A). Sequences of single-stranded DNA 14-mer (x) and duplexes (IX and XI–XVIII). (B). X-band EPR spectra of the oligomers in PNE buffer at 20, 10, 0 and −10°C.
To directly determine the propensity of Ç to stack on the end of duplexes, DNA duplexes XI, XV–XVII containing a Ç overhang at the 5′-end were prepared (Figure 3A). These EPR spectra look very similar to that of duplexes IX, XII–XIV, indicating that Ç stacks readily at the 5′-end and does not have substantial movement independent of the duplex itself (Figure 3B). Kool and coworkers (44,45) reported that adding a single dangling natural nucleoside (A,C,T and G) or nucleotides containing nonpolar aromatic bases such as benzene, naphthalene, phenanthrene and pyrene at the end of duplexes increased their TMs. In general, the nonpolar analogs stacked more strongly than the more polar natural bases. The extended ring-system of Ç confers added hydrophobicity and it is, therefore, not surprising that Ç stacks well on top of the duplex. Further evidence for efficient stacking of Ç at the end of the duplex comes from thermal denaturation studies; TMs of duplex XI (Table 1) containing a Ç overhang at the 5′-end is 4.6°C higher than the unmodified 13-mer duplex X.
The EPR spectra of IX (Figure 3B), which contains a Ç·G base-pair at the end of the duplex, displays additional features at low temperatures that are consistent with dipolar coupling between two nitroxides. This spectrum is shown enlarged in Figure 4A with arrows pointing to the dipolar features. The dipolar coupling is not observed for duplex VII at low temperatures (data not shown), showing that it is related to placement of the label at the end of the duplex. One probable explanation of this dipolar feature is duplex stacking, which would bring the nitroxides close to each other. To determine if this dipolar coupling arose from duplex stacking, we titrated an unlabeled duplex of the same sequence into the spin-labeled sample of oligomer IX, which should diminish the dipolar coupling due to dilution of the sample (Figure 4). This was indeed observed to be the case; after adding 10 equivalents of the unlabeled duplex, the dipolar features were no longer observed (Figure 4D). Interestingly, the same effect is not seen for duplexes XII–XIV, which suggests that the stacking only takes place when a Watson–Crick base-pair is located at the end of the duplex. End-to-end stacking of DNA duplexes has been observed in crystals (46), in liquid crystals (47) and frozen solution at 40 K (18,48), but according to our knowledge, has until now never been detected in solution.
Figure 4.
Concentration dependence on dipolar coupling. (A). EPR spectrum of oligomer IX in PNE buffer at −10°C. Arrows show additional features in the spectrum that are consistent dipolar coupling between two nitroxides resulting from helical stacking in solution. Unlabeled duplex of the same sequence was added to this sample; 1, 4 and 10 equivalents, shown in (B, C and D, respectively).
CONCLUSION
The rigid, spin-labeled nucleoside Ç, which is an analog of dC and base-pairs to dG, has been incorporated into DNA and shown to have a negligible effect on duplex stability and conformation as determined by CD and thermal denaturation studies. The EPR spectra of oligomers containing Ç in a single strand are very different from that observed for duplexes. Therefore, Ç holds promise for studying single-strand to duplex mixtures/equilibria, such as found in bistable nucleic acids (49). We have used EPR to systematically study the mobility of individual nucleotides in DNA oligomers that can fold up into two different hairpin loops. Variations in the EPR spectra were used to qualitatively explain the different mobilities of different positions and are consistent with previous NMR-based structural studies of a DNA hairpins. Fluorescence spectroscopy of reduced spin-labeled oligomers provided further support for the hairpin EPR data. A direct correlation was observed between the mobility determined by EPR spectroscopy and the quantum yields; the higher the mobility, the higher the quantum yields. EPR spectroscopy and thermal denaturation studies show that when Ç is placed at the end of a DNA duplex as a single overhanging nucleotide, it stacks efficiently on the end of the duplex. Surprisingly, we have also detected end-to-end duplex stacking in solution by placing a Ç·G pair at the end of a DNA helix. Combined, these results lay the groundwork for further studies of nucleic acid structure and conformation using Ç in combination with EPR spectroscopy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Icelandic Research Fund (060028023); the National Institutes of Health (GM65944, GM62360 and NIEHS P30ES07033); the Eimskip Fund of the University of Iceland (doctoral fellowship to P.C.). Funding for open access charge: Science Institute, University of Iceland.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Sigurdsson and Robinson research groups for helpful discussions.
REFERENCES
- 1.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 2.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 3.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem. Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 4.Steinhoff HJ. Inter- and intra-molecular distances determined by EPR spectroscopy and site-directed spin-labeling reveal protein-protein and protein-oligonucleotide interaction. Biol. Chem. 2004;385:913–920. doi: 10.1515/BC.2004.119. [DOI] [PubMed] [Google Scholar]
- 5.Prisner T, Rohrer M, MacMillan F. Pulsed EPR spectroscopy: biological applications. Annu. Rev. Phys. Chem. 2001;52:279–313. doi: 10.1146/annurev.physchem.52.1.279. [DOI] [PubMed] [Google Scholar]
- 6.Qin PZ, Dieckmann T. Application of NMR and EPR methods to the study of RNA. Curr. Opin. Struct. Biol. 2004;14:350–359. doi: 10.1016/j.sbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Kim NK, Murali A, DeRose VJ. A distance ruler for RNA using EPR and site-directed spin-labeling. Chem. Biol. 2004;11:939–948. doi: 10.1016/j.chembiol.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Cai Q, Kusnetzow AK, Hubbell WL, Haworth IS, Gacho GPC, Van Eps N, Hideg K, Chambers EJ, Qin PZ. Site-directed spin-labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Res. 2006;34:4722–4730. doi: 10.1093/nar/gkl546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagahara S, Murakami A, Makino K. Spin-labeled oligonucleotides site specifically labeled at the internucleotide linkage—separation of stereoisomeric probes and EPR spectroscopical detection of hybrid formation in solution. Nucleos. Nucleot. 1992;11:889–901. [Google Scholar]
- 10.Edwards TE, Okonogi TM, Robinson BH, Sigurdsson ST. Site-specific incorporation of nitroxide spin labels into internal sites of the TAR RNA; structure-dependent dynamics of RNA by EPR spectroscopy. J. Am. Chem. Soc. 2001;123:1527–1528. doi: 10.1021/ja005649i. [DOI] [PubMed] [Google Scholar]
- 11.Fidanza JA, Ozaki H, Mclaughlin LW. Site-specific labeling of DNA-sequences containing phosphorothioate diesters. J. Am. Chem. Soc. 1992;114:5509–5517. [Google Scholar]
- 12.Qin PZ, Butcher SE, Feigon J, Hubbell WL. Quantitative analysis of the isolated GAAA tetraloop/receptor interaction in solution: a site-directed spin labeling study. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- 13.Spaltenstein A, Robinson BH, Hopkins PB. A rigid and nonperturbing probe for duplex DNA motion. J. Am. Chem. Soc. 1988;110:1299–1301. [Google Scholar]
- 14.Keyes RS, Bobst EV, Cao YY, Bobst AM. Overall and internal dynamics of DNA as monitored by five-atom-tethered spin labels. Biophys. J. 1997;72:282–290. doi: 10.1016/S0006-3495(97)78667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischhaber PL, Reese AW, Nguyen T, Kirchner JJ, Hustedt EJ, Robinson BH, Hopkins PB. Synthesis of duplex DNA containing a spin labeled analog of 2′-deoxycytidine. Nucleos. Nucleot. 1997;16:365–377. [Google Scholar]
- 16.Qin PZ, Hideg K, Feigon J, Hubbell WL. Monitoring RNA base structure and dynamics using site-directed spin labeling. Biochemistry. 2003;42:6772–6783. doi: 10.1021/bi027222p. [DOI] [PubMed] [Google Scholar]
- 17.Schiemann O, Piton N, Mu YG, Stock G, Engels JW, Prisner TF. A PELDOR-based nanometer distance ruler for oligonucleotides. J. Am. Chem. Soc. 2004;126:5722–5729. doi: 10.1021/ja0393877. [DOI] [PubMed] [Google Scholar]
- 18.Piton N, Mu YG, Stock G, Prisner TF, Schiemann O, Engels JW. Base-specific spin-labeling of RNA for structure determination. Nucleic Acids Res. 2007;35:3128–3143. doi: 10.1093/nar/gkm169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchner JJ, Hustedt EJ, Robinson BH, Hopkins PB. DNA dynamics from a spin probe—dependence of probe motion on tether length. Tetrahedron Lett. 1990;31:593–596. [Google Scholar]
- 20.Spaltenstein A, Robinson BH, Hopkins PB. DNA structural data from a dynamics probe—the dynamic signatures of single-stranded, hairpin-looped and duplex forms of DNA are distinguishable. J. Am. Chem. Soc. 1989;111:2303–2305. [Google Scholar]
- 21.Spaltenstein A, Robinson BH, Hopkins PB. Sequence-dependent and structure-dependent DNA-base dynamics—synthesis, structure and dynamics of site and sequence specifically spin-labeled DNA. Biochemistry. 1989;28:9484–9495. doi: 10.1021/bi00450a036. [DOI] [PubMed] [Google Scholar]
- 22.Edwards TE, Sigurdsson ST. Electron paramagnetic resonance dynamic signatures of TAR RNA–small molecule complexes provide insight into RNA structure and recognition. Biochemistry. 2002;41:14843–14847. doi: 10.1021/bi026299a. [DOI] [PubMed] [Google Scholar]
- 23.Miller TR, Alley SC, Reese AW, Solomon MS, Mccallister WV, Mailer C, Robinson BH, Hopkins PB. A probe for sequence-dependent nucleic acid Dynamics. J. Am. Chem. Soc. 1995;117:9377–9378. [Google Scholar]
- 24.Okonogi T, Reese AW, Alley SC, Hopkins PB, Robinson BH. Flexibility of duplex DNA on the submicrosecond timescale. Biophys. J. 1999;77:3256–3276. doi: 10.1016/S0006-3495(99)77157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okonogi TM, Alley SC, Reese AW, Hopkins PB, Robinson BH. Sequence-dependent dynamics in duplex DNA. Biophys. J. 2000;78:2560–2571. doi: 10.1016/S0006-3495(00)76800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okonogi TM, Alley SC, Reese AW, Hopkins PB, Robinson BH. Sequence-dependent dynamics of duplex DNA: the applicability of a dinucleotide model. Biophys. J. 2002;83:3446–3459. doi: 10.1016/S0006-3495(02)75344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okonogi TM, Alley SC, Harwood EA, Hopkins PB, Robinson BH. Phosphate backbone neutralization increases duplex DNA flexibility: a model for protein binding. Proc. Natl Acad. Sci. USA. 2002;99:4156–4160. doi: 10.1073/pnas.072067799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barhate N, Cekan P, Massey AP, Sigurdsson ST. A nucleoside that contains a rigid nitroxide spin label: a fluorophore in disguise. Angew. Chem. Int. Ed. Engl. 2007;46:2655–2658. doi: 10.1002/anie.200603993. [DOI] [PubMed] [Google Scholar]
- 29.Gannett PM, Darian E, Powell J, Johnson EM, Mundoma C, Greenbaum NL, Ramsey CM, Dalal NS, Budil DE. Probing triplex formation by EPR spectroscopy using a newly synthesized spin label for oligonucleotides. Nucleic Acids Res. 2002;30:5328–5337. doi: 10.1093/nar/gkf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cekan P, Sigurdsson ST. Single base interrogation by a fluorescent nucleotide: each of the four DNA bases identified by fluorescence spectroscopy. Chem. Commun. 2008;29:3393–3395. doi: 10.1039/b801833b. [DOI] [PubMed] [Google Scholar]
- 31.Scaringe SA. RNA oligonucleotide synthesis via 5′-silyl-2′-orthoester chemistry. Methods. 2001;23:206–217. doi: 10.1006/meth.2000.1132. [DOI] [PubMed] [Google Scholar]
- 32.Abakumov GA, Tikhonov VD. Reaction of stable radical 2,2,6,6-tetramethylpiperidone-4-oxyl-1 with acids. Bull. Acad. Sci. USSR Ch+ 1969:796–801. [Google Scholar]
- 33.Rozantsev EG, Sholle VD. Synthesis and reactions of stable nitroxyl radicals II. Reactions. Synth. Int. J. Methods. 1971:401–414. [Google Scholar]
- 34.Adams SP, Kavka KS, Wykes EJ, Holder SB, Galluppi GR. Hindered dialkylamino nucleoside phosphite reagents in the synthesis of 2 DNA 51-mers. J. Am. Chem. Soc. 1983;105:661–663. [Google Scholar]
- 35.Engman KC, Sandin P, Osborne S, Brown T, Billeter M, Lincoln P, Norden B, Albinsson B, Wilhelmsson LM. DNA adopts normal B-form upon incorporation of highly fluorescent DNA base analogue tC: NMR structure and UV-Vis spectroscopy characterization. Nucleic Acids Res. 2004;32:5087–5095. doi: 10.1093/nar/gkh844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandin P, Borjesson K, Li H, Martensson J, Brown T, Wilhelmsson LM, Albinsson B. Characterization and use of an unprecedentedly bright and structurally non-perturbing fluorescent DNA base analogue. Nucleic Acids Res. 2008;36:157–167. doi: 10.1093/nar/gkm1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.vanDongen MJP, Mooren MMW, Willems EFA, vanderMarel GA, vanBoom JH, Wijmenga SS, Hilbers CW. Structural features of the DNA hairpin d(ATCCTA-GTTA-TAGGAT): formation of a G-A base pair in the loop. Nucleic Acids Res. 1997;25:1537–1547. doi: 10.1093/nar/25.8.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh M, Kumar NV, Varshney U, Chary KVR. Structural characterisation of a uracil containing hairpin DNA by NMR and molecular dynamics. Nucleic Acids Res. 1999;27:3938–3944. doi: 10.1093/nar/27.19.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rist MJ, Marino JP. Fluorescent nucleotide base analogs as probes of nucleic acid structure, dynamics and interactions. Curr. Org. Chem. 2002;6:775–793. [Google Scholar]
- 41.Tinsley RA, Walter NG. Pyrrolo-C as a fluorescent probe for monitoring RNA secondary structure formation. RNA. 2006;12:522–529. doi: 10.1261/rna.2165806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olivier M. From SNPs to function: the effect of sequence variation on gene expression. Focus on ‘A survey of genetic and epigenetic variation affecting human gene expression’. Physiol. Genomics. 2004;16:182–183. doi: 10.1152/physiolgenomics.00194.2003. [DOI] [PubMed] [Google Scholar]
- 43.Suh Y, Vijg J. SNP discovery in associating genetic variation with human disease phenotypes. Mutat. Res.-Fundam. Mol. Mech. Mutagen. 2005;573:41–53. doi: 10.1016/j.mrfmmm.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Guckian KM, Schweitzer BA, Ren RXF, Sheils CJ, Paris PL, Tahmassebi DC, Kool ET. Experimental measurement of aromatic stacking affinities in the context of duplex DNA. J. Am. Chem. Soc. 1996;118:8182–8183. doi: 10.1021/ja961733f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guckian KM, Schweitzer BA, Ren RXF, Sheils CJ, Tahmassebi DC, Kool ET. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinemann U, Alings C, Bansal M. Double helix conformation, groove dimensions and ligand-binding potential of a G/C stretch in B-DNA. EMBO J. 1992;11:1931–1939. doi: 10.1002/j.1460-2075.1992.tb05246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakata M, Zanchetta G, Chapman BD, Jones CD, Cross JO, Pindak R, Bellini T, Clark NA. End-to-end stacking and liquid crystal condensation of 6-to 20-base pair DNA duplexes. Science. 2007;318:1276–1279. doi: 10.1126/science.1143826. [DOI] [PubMed] [Google Scholar]
- 48.Bowman MK, Maryasov AG, Kim N, DeRose VJ. Visualization of distance distribution from pulsed double electron-electron resonance data. Appl. Magn. Reson. 2004;26:23–39. [Google Scholar]
- 49.Kreutz C, Kahlig H, Konrat R, Micura R. Ribose 2′-F labeling: a simple tool for the characterization of RNA secondary structure equilibria by F-19 NMR spectroscopy. J. Am. Chem. Soc. 2005;127:11558–11559. doi: 10.1021/ja052844u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.