Abstract
NASP (nuclear autoantigenic sperm protein) has been reported to be an H1-specific histone chaperone. However, NASP shares a high degree of sequence similarity with the N1/N2 family of proteins, whose members are H3/H4-specific histone chaperones. To resolve this paradox, we have performed a detailed and quantitative analysis of the binding specificity of human NASP. Our results confirm that NASP can interact with histone H1 and that this interaction occurs with high affinity. In addition, multiple in vitro and in vivo experiments, including native gel electrophoresis, traditional and affinity chromatography assays and surface plasmon resonance, all indicate that NASP also forms distinct, high specificity complexes with histones H3 and H4. The interaction between NASP and histones H3 and H4 is functional as NASP is active in in vitro chromatin assembly assays using histone substrates depleted of H1.
INTRODUCTION
In eukaryotes, genomic DNA is packaged into a highly ordered and regulated structure known as chromatin. The principal repeating element of chromatin is the nucleosome, which consists of ∼147 bp of DNA wrapped around the histone proteins: H2A, H2B, H3 and H4. The transit of histone proteins throughout the cell and their subsequent deposition into chromatin is facilitated by a collection of proteins known as histone chaperones.
Histone chaperones can be grouped into a large number of families based on sequence similarity. These include the nucleoplasmin, N1/N2, CAF-1, HIR, NAP1, Asf1, Rbap46/48, Rsf-1, FACT, nucleolin and Arp families of proteins. The defining characteristic of histone chaperones is their ability to bind histone proteins and they are often highly specific with regard to these interactions. For example, chaperones such as CAF-1 and Asf1 specifically interact with complexes of histones H3 and H4, while other chaperones like nucleoplasmin and nucleolin interact with complexes of histones H2A and H2B. Histone chaperones facilitate a number of events relevant to histone biology such as their storage, transport, deposition and eviction (1–4).
The N1/N2 family of histone chaperones was originally identified in Xenopus laevis (5–7). In X. laevis, N1/N2 associates with core histones H3 and H4, providing a mechanism for the storage of these histones in Xenopus oocytes. In addition, N1/N2 also participates in mediating nucleosome assembly. The N1/N2 family was thought to be restricted to cells of the animal lineage but a potential homolog, Hif1p, was recently identified in the budding yeast Saccharomyces cerevisiae as an H3/H4-specific histone chaperone that associates with a nuclear form of the Hat1p/Hat2p type B histone acetyltransferase complex (8).
A N1/N2 family member has also recently been identified in the fission yeast Schizosaccharomyces pombe (9). Sim3 was isolated in a screen looking for mutants that disrupted the transcriptional silencing of a marker gene placed in the central core of an S. pombe centromere. Intriguingly, Sim3 was found to be important for the localization of the centromere-specific histone H3 variant CENP-A at S. pombe centromeres. In addition, Sim3 was found to physically associate with both CENP-A and histone H3.
Mammalian cells contain an N1/N2 homolog termed NASP (nuclear autoantigenic sperm protein). In mammals, NASP is present in two differentially spliced isoforms. The longer form of the protein is known as testicular NASP (tNASP) and is expressed in the testis, embryonic tissues and some transformed cells. The second form is somatic NASP (sNASP) which is found in all dividing cells. NASP plays an essential role in mammals as demonstrated by the early embryonic lethality of a mouse knockout model (10). In addition, genetic studies in Caenorhabditis elegans indicate that the NASP homolog in this organism plays a key role in development and may act as a transcriptional regulator (11).
Curiously, although NASP is clearly a member of the N1/N2 family, it has been described in the literature as a linker histone-specific chaperone (10,12–15). As the binding specificity of histone chaperones is a critical aspect of their function, we have used multiple methods to explore the biochemical properties of this protein. In vitro binding experiments using purified proteins demonstrate that sNASP binds specifically to both histone H1 and histones H3 and H4. The binding of sNASP with histones H1 and H3/H4 displays altered binding kinetics that results in high affinity as indicated by sub-micromolar dissociation constants for both interactions. Coimmunoprecipitation experiments indicated that NASP retained this spectrum of interactions in vivo. In addition, the interaction of sNASP with histones H3 and H4 was functional as sNASP was active in in vitro chromatin assembly assays that use only core histones.
MATERIALS AND METHODS
Expression and purification of recombinant sNASP
The human sNASP ORF (Invitrogen) was transferred into the Escherichia coli expression vector pDEST-17, which adds an NH2-terminal 6His tag. The resulting construct was transformed into E. coli BL21 to allow for IPTG-inducible expression. One liter cultures were grown to mid-log phase and sNASP expression was induced for 2 h. The cells were harvested and the pellets were resuspended in 30 ml start buffer (20 mM sodium phosphate pH7.4, 500 mM NaCl) and lysed by sonication. Phenylmethylsulphonyl fluoride (0.5 mM) and protease inhibitor cocktail [(1:100), Sigma, St. Louis, MO] were also added to buffers. Cell lysate was aliquoted into 1.5 ml microcentrifuge tubes and centrifuged at 10 000 r.p.m. for 10 min. The supernatant was applied to a Ni2+ charged HiTrap Chelating HP column (5 ml). After washing with 20 mM imidazole and 50 mM imidazole, respectively, the recombinant sNASP was eluted with 500 mM imidazole and confirmed by western blot with anti-6His antibody. Purified sNASP was dialyzed against DN (200) [DN buffers contain 25 mM Tris (pH 7.0), 0.1 mM EDTA, 10% glycerol and the concentration of NaCl listed in parenthesis] and the protein concentration determined.
Histone isolation
Core and linker histones were isolated from chicken erythrocyte nuclei by acid extraction (0.4 N H2SO4) followed by extensive dialysis against 50 mM Tris, pH 7.0. Core histones depleted of H1 were isolated from chicken erythrocyte nuclei by hydroxyapatite chromatography as described (16). Purified bovine histone H1 was obtained from Millipore, Billerica, MA. Recombinant histones H3 and H4 were purified as described (a generous gift from Dr M. Poirier) (17).
Native gel electrophoresis
A total of 1.5 μg of recombinant sNASP was incubated with increasing amounts of purified histone H3/H4 or histone H1 in DN (300) for at least 4 h at 4°C. The 7% native gel was prepared as follow: 1.75 ml acrylamide 40% stock, 3.75 ml 3M Tris–HCl, pH 8.8, 4.35 ml ddH2O, 100 μl 10% ammonium persulfate (APS) and 20 μl TEMED. The protein complexes were resolved by electrophoresis at 120 V with native gel running buffer (0.192 M Glycine, 0.025 M Tris, pH8.3) for 4 h. The proteins in the native gel were then visualized by Coomassie Blue staining.
Chromatography-based histone-binding assays
Purified sNASP was mixed with histones and incubated overnight at 4°C in DN (150) or DN (200) buffer. After incubation, the mixtures were applied to a Ni2+ charged HiTrap Chelating HP column (1 ml). The column was then washed extensively with wash buffers (20 mM sodium phosphate pH7.4, 150 mM NaCl 100 mM imidazole and 20 mM sodium phosphate pH7.4, 500 mM NaCl, 100 mM imidazole, respectively). A total of 0.1% of NP-40 was added to the wash buffer to minimize nonspecific binding between histones and the matrix of the column. Bound proteins were eluted with buffer containing 500 mM imidazole. Flow through and bound fractions were resolved by SDS–PAGE and visualized by Coomassie Blue staining.
Alternatively, the proteins were resolved by gel filtration chromatography (Superose 6 column, GE Healthcare, Chalfont St. Giles, UK) run with DN (300) buffer with 0.1% NP-40. Fractions were collected and resolved by 18% SDS–PAGE and visualized by Coomassie Blue staining.
Biosensor analysis
Biosensor experiments were performed using a Biacore 2000 surface plasmon resonance (SPR) instrument. Histones were coupled to a CM5 sensor chip using HBS-EP (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% Tween-20) buffer at 25°C. For the preparation of amine-coupling of the histones, the CM5 sensor chip was first washed with HBS-EP buffer at a flow rate of 10 μl/min for 10 min. Each flow cell was injected with 25 μl of a 1:1 solution of NHS/EDC [N-hydroxysuccinimide/N-ethyl-N′-(3-dimethyl-amino-propyl)-carbodiimide, 75 mg/ml/11.5 mg/ml], followed by a 30–100 μl injection of either H1 or the H3/H4 tetramer at 50–100 μg/ml in PBS, pH 7.4 buffer [typically, 30 μl of the histone(s) diluted into 100 μl of 10 mM sodium acetate, pH 4.5] was injected over flow cells 2–4. No histones were injected over the activated flow cell 1 serving as a reference cell. Flow cells were blocked with a 35 μl injection of 1 M ethanolamine, pH 8.5.
SPR binding experiments measured at 50 μl/min were performed using either HBS-EP or TBS-EN (25 mM Tris–HCl, pH 7.0, 300 mM NaCl, 0.1 mM EDTA and 0.05% NP-40) buffers. Similar NASP binding kinetics were observed over the H1 or H3/H4 tetramer-coupled surfaces using either buffer system. Nonspecific binding was not observed when NASP was injected over the underivatized flow cell 1. There were no mass transport effects seen during the association phase during NASP injections. Two-fold serial dilutions of 4–5 NASP concentrations determined the binding kinetics to the histones. Each 250 μl protein/buffer injection was followed by a 500 s dissociation period. The surface was regenerated for subsequent runs with 15 μl injections of 3 M NaCl and 4 M MgCl2.
Sensorgrams were pooled, trimmed and subtracted using BIAevaluation 4.1. A buffer sensorgram was subtracted from each NASP sensorgram before data analysis [double referencing (18)]. The NASP:histone binding kinetics fit well to a simple Langmuir biomolecular reaction model defined by a single on- and off-rate constant. Sensorgrams were globally analyzed using ClampXP (19) and the binding kinetic parameters were determined from three separate experiments.
Immunoprecipitaion of NASP
sNASP-containing complexes were immunoprecipitated from HeLa cell nuclear extract with rabbit anti-NASP antibody with coupling gel from ProFound™ Mammalian Co-immunoprecipitation Kit (Pierce, Rockford, IL) (20). After immobilization of antibody with coupling gel, 100 μl of coupling gel was incubated with 800 μl of HeLa cell nuclear extract for 5–6 h, at 4°C. The gel was then washed six times with 1 ml of DN (500)/0.1% NP-40 to remove unbound protein. The bound proteins were eluted by boiling in 100 μl of 1× SDS loading dye (0.06 M Tris–HCl, pH 6.8, 2% SDS, 10% glycerol, 0.001% bromphenol blue and 5% β-mercaptoethanol). The coupling gel without conjugation with antibody was incubated with HeLa cell nuclear extract as a negative control. The bound proteins were resolved by SDS–PAGE and detected by western blot.
In vitro chromatin assembly
Assays contained pUC18 plasmid which had been relaxed by DNA topoisomerase I (Sigma). The assembly reactions were performed in buffer containing: 10 mM Tris–HCl (pH 8.0), 1.0 mM EDTA, 100 mM NaCl, 100 μg/ml BSA, 2 mM ATP and 0.6 μg purified chicken core histones. Reactions were supplemented with 2 μl of yeast cytosolic extract as indicated (8). The reactions were incubated at 37°C for 1 h and terminated with 0.2% SDS and 100 μg/ml Protease K. After phenol extraction and ethanol precipitation, DNA was analyzed on a 1.5% agarose gel and visualized with SYBR gold nucleic acid stain (Invitrogen).
RESULTS
Human sNASP displays a high level of sequence similarity with X. laevis N1/N2 with >50% amino acid identity. In addition, sNASP and N1/N2 have a shared domain structure that is also conserved in the S. cerevisiae histone chaperone Hif1p (depicted in Figure 1A). Each of these proteins contains a large central domain that is highly enriched in acidic amino acids (glutamic acid and aspartic acid). This acidic domain is flanked by tetratricopeptide repeat (TPR) repeats with one repeat NH2-terminal to the acidic domain and two repeats COOH-terminal. Given the sequence and domain conservation between these proteins, we re-examined the histone-binding specificity of sNASP to determine whether it shares the H3/H4 specificity of N1/N2 and Hif1p or whether it specifically interacts solely histone H1, as reported in the literature.
Figure 1.
Native gel electrophoresis of sNASP:histone complexes. (A) A schematic diagram representing the conservation of sequence and domain structure between sNASP, N1 and Hif1p. (B and C) A total of 1.5 μg recombinant sNASP was incubated alone (lanes 8 and 16) or with increasing amount of histone H3/H4 (0.36, 0.81, 1.26, 1.71, 2.16 and 2.61 μg, lanes 1–6), histone H1 (0.4, 0.9, 1.4, 1.9, 2.5 and 2.9 μg, lanes 9–14) or 1.5 μg of total chicken histone (lanes 7 and 15) in the presence of DN (300) with a total volume of 30 μl. After incubation, 12 μl of each mixture was analyzed by either native gel electrophoresis (B) or SDS–PAGE (C). Proteins were visualized by Coomassie Blue staining. The migration of sNASP:histone complexes is denoted by asterisks in (B).
Multiple sNASP/histone complexes identified by native gel electrophoresis
Native gel electrophoresis has been one of the primary assays used to assess the in vitro histone-binding specificity of sNASP (12). In these experiments, specific complexes were detected between sNASP and histone H1 in the form of discreet bands that were only observed following the incubation of sNASP with linker histones. Conversely, when sNASP was incubated with histones H3 and H4, it was retained in the wells of the native gel leading to the interpretation that sNASP forms nonspecific aggregates with histones H3 and H4 (12).
We have re-examined the use of native gel electrophoresis to monitor the interaction of sNASP with histones. For these assays, recombinant human sNASP (which contains an NH2-terminal 6His tag) was expressed in E. coli and purified by Ni2+ chelate chromatography. Recombinant sNASP was incubated with a mixture of histones isolated from chicken erythrocytes that included both the core and linker histones (Figure 1C, lanes 7 and 15). Native polyacrylamide gel electrophoresis indicated that sNASP formed two distinct complexes with the histones, which had reduced mobility relative to free sNASP.
To determine whether these complexes were the result of interactions between sNASP and histones H1 or H3/H4, sNASP was incubated with either histone H1 or H3/H4 complexes separately. For these experiments, chicken erythrocyte histone H1 was separated from histones H3 and H4 by Ni2+ chelating chromatography, taking advantage of the fact that these histones display varying degrees of nonspecific interaction with column resins (Figure 4B). The sNASP was mixed with increasing amounts of either histone H3/H4 complexes or histone H1, and the mixtures resolved by native gel electrophoresis. As seen in Figure 1B, sNASP formed complexes with both histones H3/H4 and histone H1 (lanes 1–6 and lanes 9–14, respectively). While both H1 and H3/H4 were able to form complexes with sNASP, the interaction between sNASP and histone H1 occurred at lower histone concentrations suggesting that the binding of sNASP to H1 occurred with higher affinity. Surprisingly, the mobility of both the sNASP:H1 and sNASP:H3/H4 complexes was similar to the mobility of the complexes observed when sNASP was incubated with the complete histone mixture. This is likely due to the relatively low resolution afforded by native gel electrophoresis that separates macromolecules based on multiple characteristics such as size, shape and charge.
Figure 4.
Detection of sNASP:H3/H4 complex formation by affinity chromatography. (A) Purified recombinant sNASP (150 μg) and chicken erythrocyte histones (200 μg) were incubated in a total volume of 1.0 ml (input) and chromatographed on a Ni2+ chelate column. The first six fractions to wash off the column after loading (wash) and the first six fractions to elute with 500 mM imidazole buffer (elute) were resolved by SDS–PAGE and visualized by Coomassie Blue staining. A 0.24% of the input and 2.4% of each fraction was loaded on the gel. The position of each protein in the gel is indicated on the left. (B) Purified chicken erythrocyte histones (300 μg) were chromatographed and visualized as above except that 0.75% of the input was run on the gel. (C) Purified sNASP (200 μg) was incubated with bovine histone H1 (500 μg) in a total volume of 1.0 ml and chromatographed on a Ni2+ chelate column. The first five fractions to wash off the column after loading (wash) and the first five fractions to elute with 500 mM imidazole buffer (elute) were resolved by SDS–PAGE and visualized by Coomassie Blue staining. A 0.24% of the input and 2.4% of each fraction was loaded on the gel.
sNASP forms a stable complex with histones H3 and H4
We employed gel filtration chromatography to obtain a more detailed and higher resolution picture of the interaction between sNASP and histones. In the absence of sNASP, the core histones eluted as a single peak with an apparent molecular weight of ∼50 kDa (Figure 2B). Histone H1 eluted in two peaks with the first peak having an apparent molecular weight of ∼150 kDa and the second peak coeluting with the core histones suggesting that histone H1 may interact with the core histones (Figure 2B). Following incubation with sNASP, if any of the histones form a complex with this chaperone, they should coelute from the gel filtration column with sNASP with a larger apparent molecular weight. As seen in Figure 2A, histones H3 and H4 perfectly coeluted with sNASP as a complex with an apparent molecular weight of ∼350 kDa. Histone H1 did not copurify with sNASP and eluted as a single peak of ∼150 KDa apparent molecular weight. Interestingly, in the presence of sNASP, histone H1 did not coelute with the core histones suggesting that the interaction between histone H1 and the core histones may have been mediated by histones H3/H4. Importantly, the observation that sNASP does not interact with histones H2A, H2B or H1 in this assay indicated that the binding of sNASP to histones is not due to nonspecific interactions based solely on electrostatic attraction.
Figure 2.
sNASP forms a stable complex with histones H3 and H4. (A) Purified recombinant sNASP (150 μg) and chicken erythrocyte histones (200 μg) were mixed (input) and resolved by gel filtration chromatography (Superose 6). Fractions (indicated by numbers at top of gels) were resolved by SDS–AGE and visualized by Coomassie Blue staining. A 4.8% of the input and each fraction were electrophoresed. The elution positions of standards are indicated at the top. (B) Purified chicken erythrocyte histones were chromatographed and visualized as above.
The co-elution of histones H3 and H4 with sNASP during gel filtration chromatography is clear evidence that sNASP forms a distinct stable complex with H3 and H4. To determine whether the complex observed during gel filtration chromatography is related to the complexes observed by native gel electrophoresis, protein fractions from across the Superose 6 sNASP:H3/H4 peak were directly resolved on a native polyacrylamide gel. As seen in Figure 3B, these fractions contain complexes that have electrophoretic mobilities identical to those seen when the sNASP/histone mixtures were directly resolved by native gels. A number of important observations can be made from this result. First, the sNASP:H3/H4-specific bands observed by native gel electrophoresis (Figure 1B) reflect the presence of bona fide, stable complexes. Second, the differences in electrophoretic mobility observed for the sNASP:H3/H4-specific bands during native gel electrophoresis are indicative of complexes with different apparent molecular weights as the complexes that give rise to these bands elute from the gel filtration column with distinct, but overlapping, profiles. The precise nature of these complexes is not clear but may be related to the observation that sNASP can exist as both a monomer and a dimer (12). Finally, the sNASP that elutes from the gel filtration column is predominantly associated with histones H3 and H4 as there is no free sNASP detected by native gel electrophoresis.
Figure 3.
Correlation between sNASP:H3/H4 complexes detected by gel filtration chromatography and native gel electrophoresis. (A) sNASP (100 μg) was incubated with total chicken erythrocyte histones (150 μg) and resolved on a Superose 6 column. Fractions containing sNASP and histones H3 and H4 (labeled at the top of the gel) were resolved by SDS–PAGE and visualized by Coomassie Blue staining. (B) The indicated fractions from the Superose 6 column were directly resolved by native gel electrophoresis and visualized with Coomassie Blue staining. Recombinant sNASP was also analyzed alone (lane 11) or following incubation with total chicken erythrocyte histones (lane 10). The migration of sNASP:histone complexes is indicated by asterisks.
sNASP specifically interacts with histones H3 and H4
A third assay was also used to determine the in vitro binding specificity of sNASP. Recombinant sNASP (which has an NH2-terminal 6His tag), was incubated with a preparation of chicken erythrocyte histones that contained both the core and linker histones. Following incubation, the sNASP/histone mixture was applied to a Ni2+ chelate column. Unbound proteins were removed by extensive washing (with buffer containing 0.1 M imidazole) and the bound proteins eluted using buffer containing 0.5 M imidazole. As expected, the 6His-tagged sNASP bound to the column and was present in the elution fractions (Figure 4A). Histones H3 and H4 were not present in the wash fractions and coeluted with sNASP. Histones H2A, H2B and H1 were all found in the wash fractions and were not detected in the elution fractions. When the histones were applied to the Ni2+ chelate column in the absence of sNASP, all of the histones, including H3 and H4 were found in the wash fractions (Figure 4B).
This result supports the previous observations of a specific interaction between sNASP and histones H3 and H4. However, neither this assay nor the gel filtration chromatography provided evidence for an interaction between sNASP and histone H1. The lack of interaction between sNASP and histone H1 in these assays may be due to a number of factors. First, these assays contained mixtures of the core and linker histones. It may be that binding of sNASP to H3/H4 is preferred and the formation of the sNASP:H3/H4 complex precludes the interaction between sNASP and histone H1. Alternatively, the use of histones derived from chicken erythrocytes may have influenced the results as there is greater sequence identity between chicken and mammals for the core histones than for the linker histones. We tested these possibilities by directly assessing the ability of sNASP to interact with purified bovine histone H1. As seen in Figure 4C, when purified bovine histone H1 was combined with 6His-tagged sNASP, in the absence of other histones, and applied to a Ni2+ chelate column, all of the histone H1 was found in the wash fractions. In addition, no detectable histone H1 coeluted from the Ni2+ chelate column with sNASP. This result suggests that the lack of interaction between sNASP and histone H1 is not due to competition or the sequence divergence of chicken histone H1. Hence, while an interaction between sNASP and histone H1 is clearly detectable by native gel electrophoresis, it is not observed using chromatography-based assays. One explanation, which is difficult to rule out, is that the nature of the nonspecific interactions that are seen between histone H1 and column resins prevent the formation of sNASP/histone H1 complexes.
sNASP interacts primarily with histone H3
To further characterize sNASP as a histone H3/H4-specific chaperone, we tested the ability of sNASP bind individually to histone H3 and H4. For these experiments, recombinant X. laevis histones H3 and H4 were each expressed and purified from E. coli and then combined with 6His-tagged sNASP. As shown in Figure 5A, following Ni2+ chelate chromatography, all of the histone H3 coeluted with sNASP. However, even though sNASP is present in excess, histone H4 was only partially retained on the column (Figure 5B). These results suggest that sNASP can bind to both H3 and H4 but the association with histone H3 may be the primary interaction.
Figure 5.
Recombinant sNASP primarily interacts with histone H3. (A) Purified sNASP (200 μg) was incubated with recombinant X. laevis histone H3 (100 μg) in a total volume of 1.0 ml and chromatographed on a Ni2+ chelate column. The first seven fractions to wash off the column after loading (wash) and the first seven fractions to elute with 500 mM imidazole buffer (elute) were resolved by SDS–PAGE and visualized by Coomassie Blue staining. A 0.24% of the input and 2.4% of each fraction was loaded on the gel. (B) Purified sNASP (200 μg) was incubated with recombinant X. laevis histone H4 (76 μg) in a total volume of 1.0 ml and chromatographed on a Ni2+ chelate column. Fractions were analyzed as described above.
Biosensor analysis of NASP binding to histones H1 and H3/H4
The native gel electrophoresis and chromatography-based assays provided evidence that sNASP can form specific complexes with both histone H1 and histones H3/H4. To provide a more quantitative analysis of these interactions, biosensor analysis using SPR was performed to determine the binding kinetics and affinities of the sNASP:histone interactions. The H1 and H3/H4 histones were randomly coupled to the sensor chip surface using amine chemistry. Triplicate sNASP samples were injected at various concentrations over the H1 and H3/H4 surfaces (Figure 6). The binding kinetics for the NASP:H1 and NASP:H3/H4 interactions were globally fit to a single site Langmuir binding reaction model with the on (kon) and off (koff) rate constants listed in Table 1.
Figure 6.
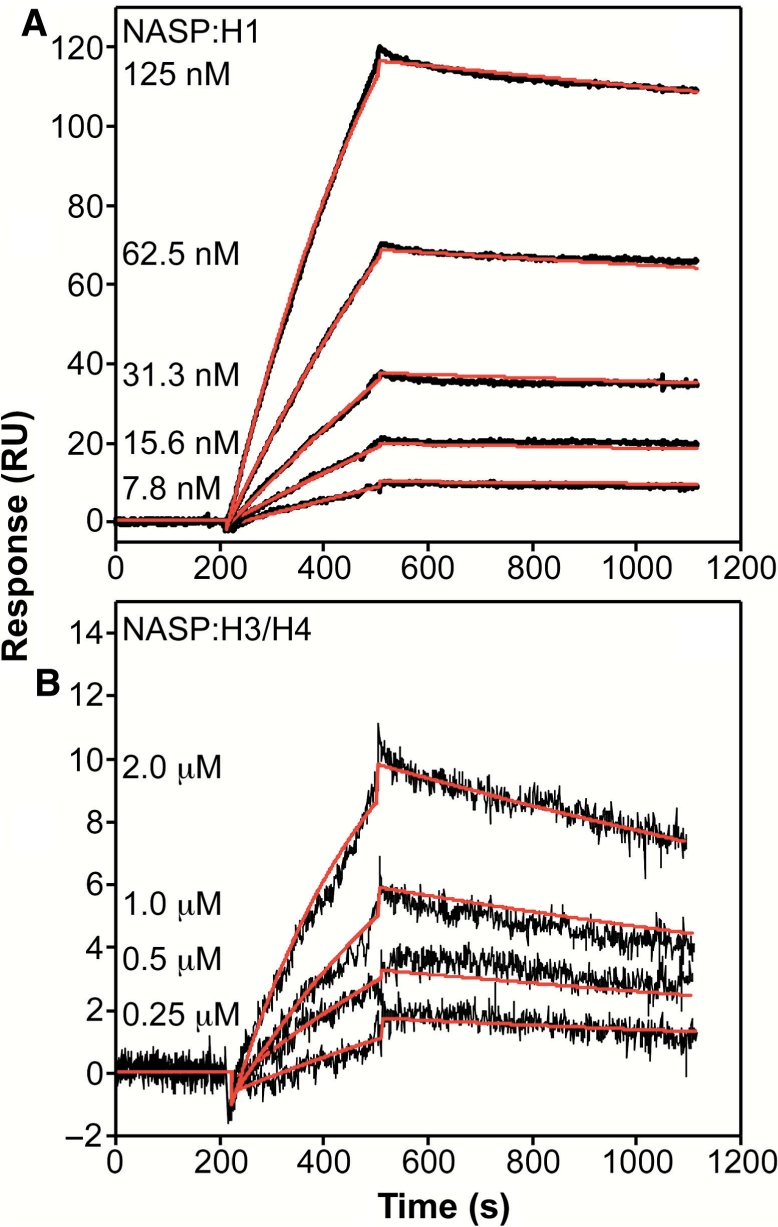
Quantitation of the interactions between sNASP and histones H1 and H3/H4 by SPR. SPR binding kinetic sensorgrams for the interactions of NASP with H1 (A) and H3/H4 (B). The black curves are the trimmed sensorgrams collected at a flow rate of 50 μl/min in HBS-EP buffer at 25°C. Global fits of the data according to the model described in the Materials and methods section are in red, and the resulting kinetic rate constants are tabulated in Table 1.
Table 1.
SPR binding kinetics and affinities for the interactions of NASP with H1 and H3/H4 tetramera
| kon (M−1 s−1) | koff (s−1) | KDb (nM) | |
|---|---|---|---|
| NASP:H1c | 1.44 (8) × 104 | 1.98 (4) × 10−4 | 13.8 (3) |
| NASP:H3/H4c | 2.92 (7) × 103 | 6.91 (4) × 10−4 | 237 (70) |
aBiosensor experiments were performed in 10 mM HEPES (pH 7.4), 150 mM NaCl, 3 mM EDTA, 0.005% Tween-20 at 25°C.
bCalculated KD = koff/kon.
cValues in parentheses represent the SD of the mean of standard errors in the final significant digit.
NASP displays altered binding kinetics and affinities during the interactions with either the H1 histone or the H3/H4 tetramer. The NASP:H1 interaction yields a kon rate of 1.44 × 104 M−1 s−1 and a koff rate of 1.98 × 10−4 s−1. The calculated equilibrium dissociation constant Kd is 13.8 nM for the NASP:H1 interaction. The NASP:H3/H4 tetramer interaction yields a kon rate of 2.92 × 103 M−1 s−1 and a koff rate of 6.91 × 10−4 s−1. The Kd is 237 nM for the NASP:H3/H4 tetramer interaction. The 17-fold weaker Kd for the NASP:H3/H4 complex in comparison to the NASP:H1 complex results from an ∼5-fold slower kon rate and an ∼3.5-fold faster koff rate. The sub-micromolar Kd values for both sNASP:H1 and sNASP:H3/H4 indicated that both complexes form with high affinity. In addition, the lower Kd value for the sNASP:H1 interaction mirrors the results obtained with native gel electrophoresis where lower concentrations of H1 were needed to form the sNASP:histone complex. Thus, the altered binding kinetics and affinities for the sNASP:H1 and sNASP:H3/H4 interactions provide a quantitative description to the qualitative gel electrophoresis and size-exclusion chromatography binding experiments.
In vivo interaction of sNASP and histones
The designation of NASP as a linker histone-binding protein was originally based on evidence from the purification of endogenous NASP from cell lysates. NASP was purified from mouse myeloma cell extracts using an anti-NASP antibody affinity column (13). Proteins bound to NASP were then eluted from the column and analyzed by reverse phase HPLC. The presence of histone H1, as well as the absence of core histones, was determined by comparison of the retention times of the NASP-associated proteins to the retention times of histone standards (13). Anti-NASP antibody columns were also used to isolate NASP from mouse testis lystes and HeLa cell lysates (14,15). In each case, histone H1 was identified as eluting from the column with NASP by western blot analysis. However, these samples were not probed for the presence of core histones.
While these experiments did not identify an in vivo interaction between NASP and histones H3/H4, other in vivo evidence supports an interaction between these proteins. Tagami and colleagues (21) used epitope tags to affinity purify complexes associated with human histones H3.1 and H3.3. While some histone chaperones, such as CAF-1 and HIRA, selectively associated with only one H3 variant, other chaperones, including Asf1 and NASP, were found complexed with both forms of H3. As histone H1 was not reported to be a component of these complexes, these results indicate that a fraction of the soluble histone H3 in mammalian cells is associated with NASP.
We have used coimmunoprecipitation to evaluate whether the interactions between NASP and histones seen in vitro are also found in vivo. For these experiments, antibodies were raised in rabbits using purified recombinant sNASP as antigen. Rabbit anti-NASP antibody was conjugated with coupling gel and incubated with HeLa cell nuclear extract. After extensive washing, the bound proteins were eluted. Western blots were then used to determine the presence of specific histones. As seen in Figure 7, the antibody immunoprecipitated both sNASP and tNASP. In addition, both histones H1 and H3 were coimmunoprecipitated with the anti-NASP antibody suggesting that NASP interacts with both linker and core histones in vivo. Hence, these results are consistent with the in vitro histone-binding data described above.
Figure 7.
sNASP interacts with both histone H1 and histone H3 in vivo. HeLa cell nuclear extract (input) was incubated with coupling gel with (lane 4) or without being conjugated to anti-NASP antibody (lane 4). Five microliters of the input (lane 1) and unbound (lane 2) fractions and 12 μl of the bound fractions were resolved by SDS–PAGE and the proteins visualized by western blots probed with antibodies recognizing the proteins indicated on the right.
sNASP participates in the deposition of core histones
It has previously been shown that sNASP can function to restore histone H1 onto chromatin that has been depleted of linker histones (12). To determine whether sNASP:H3/H4 complexes were also functional, we used an in vitro chromatin assembly assay that monitors the formation of nucleosomes through the introduction of supercoils into a relaxed circular plasmid DNA. For these assays, we used a preparation of chicken erythrocyte histones that had been purified by hydroxyapatite chromatography to remove histone H1 (data not shown). As seen in Figure 8, recombinant sNASP was unable to assemble nucleosomes using purified chicken erythrocyte core histones (compare lanes 3 and 5). As demonstrated previously, recombinant yeast Hif1p is also unable to function alone in the assembly of core histones and requires the presence of factor(s) in yeast cytosolic extracts [see lanes 4, 7–9 and ref. (8)]. Therefore, we tested whether sNASP could function in a similar manner and replace Hif1p in these assembly reactions. As seen in Figure 8 (compare lane 7 to lanes 10–12), sNASP promotes very robust chromatin assembly in the presence of core histones and a yeast cytosolic extract. These results make two important points. First, the interaction between sNASP and histones H3 and H4 is a productive one that can lead to histone deposition. Hence, sNASP can function as a core histone assembly factor. Second, the similarity in chromatin assembly activity demonstrated by sNASP and Hif1p supports the hypothesis that Hif1p is a functional ortholog of the N1/N2 family of histone chaperones in yeast.
Figure 8.
Recombinant sNASP can assemble core histones into chromatin. Chromatin assembly activity of recombinant sNASP with core histones was assayed by incubating the indicated factors with a relaxed circular plasmid. After incubation, the plasmids were extracted and resolved by 1.5% agarose gel electrophoresis, and visualized by SYBR Gold nucleic acid stain. The migration of the supercoiled (S) and relaxed (R) forms of the plasmid are indicated by arrows. Lanes 1 and 2 show the template DNA before and after relaxation, respectively.
DISCUSSION
NASP has emerged as a protein that is critical for the proper growth and development of complex eukaryotic organisms (9,11,22). As a histone chaperone, the most fundamental activity of NASP is its histone-binding specificity. The experiments presented here indicate that, in vitro, sNASP forms specific, high-affinity complexes with both histone H1 and histones H3/H4. In addition, the interaction of sNASP with histone H3/H4 complexes is functional as sNASP is able to function in the assembly of core histones onto DNA in vitro. These results are entirely consistent with the high degree of primary sequence and domain structure conservation observed between NASP and members of the N1/N2 family of histones chaperones, which have been demonstrated to interact specifically with histones H3 and H4 (5,6,8,23).
Our quantitative analysis of the binding of sNASP to histone H1 and histone H3/H4 complexes indicated that sNASP interacts with both sets of histone proteins with high affinity. The dissociation constants that we have observed, 13.8 nM and 237 nM for H1 and H3/H4, respectively, are comparable to those seen for the binding of the histone chaperone p55 to histone H4 and significantly lower than those reported for most other histone-binding proteins (typically in the 1–10 μM range) (24–30).
The effect of sNASP on in vitro plasmid supercoiling assays suggests that sNASP is not merely a histone-binding protein but that it is also a chromatin assembly factor. The requirement of an extract in these assays indicates that, under the conditions tested, sNASP is not able to generate stable nucleosomes by itself. Whether sNASP hands off histones to other factors that directly assemble nucleosomes or whether sNASP is more directly involved in mediating the histone/DNA interaction remains to be determined.
The activity of sNASP in in vitro chromatin assembly assays also sheds light on the yeast histone chaperone Hif1p. Despite a low level of primary sequence similarity, Hif1p was suggested to be a fungal ortholog of the N1/N2 family of histone chaperones based on a conserved domain structure and common histone-binding specificity (8). The ability of sNASP to functionally substitute for the yeast protein Hif1p in chromatin assays strongly supports this hypothesis that these proteins share a common function.
An interesting issue raised by our results involves the functional significance of a histone chaperone that can bind both histone H1 and histone H3/H4 complexes. One intriguing possibility suggests a potential link between the acetylation of newly synthesized histones H3 and H4 and the deposition of histone H1. Subsequent to their synthesis, histones H3 and H4 are rapidly acetylated on their NH2-terminal tails by type B histone acetyltransferases. Following deposition of these histones, these modifications are removed during chromatin maturation. While the acetylation of newly synthesized H3 and H4 is an evolutionarily conserved event, the function of this modification is not known. One potential role is suggested by the observation that preventing the deacetylation of H3 and H4 following chromatin assembly prevents association of histone H1 with chromatin (31–33). Thus, the assembly of histone H1 may be regulated by the modification state of histones H3 and H4. Combined with the finding that Hif1p is found associated with the type B histone acetyltransferase Hat1p in yeast nuclei, these observations suggest that histone chaperones such as sNASP and Hif1p may coordinate, either spatially or temporally, the assembly of histone H1 onto nucleosomes containing the proper modification state on histones H3 and H4.
FUNDING
National Institutes of Health (GM62970 to M.R.P.). Funding for open access charge: National Institutes of Health (GM62970).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Michael Poirier for the kind gift of purified recombinant histones H3 and H4. We would also like to thank Dr Paul Herman and Amy Knapp for critical reading of the article.
REFERENCES
- 1.Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr. Opin. Struct. Biol. 2003;13:6–14. doi: 10.1016/s0959-440x(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 2.Polo SE, Almouzni G. Chromatin assembly: a basic recipe with various flavours. Curr. Opin. Genet. Dev. 2006;16:104–111. doi: 10.1016/j.gde.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Jin J, Cai Y, Li B, Conaway RC, Workman JL, Conaway JW, Kusch T. In and out: histone variant exchange in chromatin. Trends Biochem. Sci. 2005;30:680–687. doi: 10.1016/j.tibs.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Kleinschmidt JA, Dingwall C, Maier G, Franke WW. Molecular characterization of a karyophilic, histone-binding protein: cDNA cloning, amino acid sequence and expression of nuclear protein N1/N2 of Xenopus laevis. EMBO J. 1986;5:3547–3552. doi: 10.1002/j.1460-2075.1986.tb04681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleinschmidt JA, Fortkamp E, Krohne G, Zentgraf H, Franke WW. Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J. Biol. Chem. 1985;260:1166–1176. [PubMed] [Google Scholar]
- 7.Kleinschmidt JA, Franke WW. Soluble acidic complexes containing histones H3 and H4 in nuclei of Xenopus laevis oocytes. Cell. 1982;29:799–809. doi: 10.1016/0092-8674(82)90442-1. [DOI] [PubMed] [Google Scholar]
- 8.Ai X, Parthun MR. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell. 2004;14:195–205. doi: 10.1016/s1097-2765(04)00184-4. [DOI] [PubMed] [Google Scholar]
- 9.Dunleavy EM, Pidoux AL, Monet M, Bonilla C, Richardson W, Hamilton GL, Ekwall K, McLaughlin PJ, Allshire RC. A NASP (N1/N2)-related protein, Sim3, binds CENP-A and is required for its deposition at fission yeast centromeres. Mol. Cell. 2007;28:1029–1044. doi: 10.1016/j.molcel.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson RT, Alekseev OM, Grossman G, Widgren EE, Thresher R, Wagner EJ, Sullivan KD, Marzluff WF, O’Rand MG. Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J. Biol. Chem. 2006;281:21526–21534. doi: 10.1074/jbc.M603816200. [DOI] [PubMed] [Google Scholar]
- 11.Grote P, Conradt B. The PLZF-like protein TRA-4 cooperates with the Gli-like transcription factor TRA-1 to promote female development in C. elegans. Dev. Cell. 2006;11:561–573. doi: 10.1016/j.devcel.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 12.Finn RM, Browne K, Hodgson KC, Ausio J. sNASP, a histone H1-specific eukaryotic chaperone dimer that facilitates chromatin assembly. Biophys. J. 2008;95:1314–1325. doi: 10.1529/biophysj.108.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson RT, Batova IN, Widgren EE, Zheng LX, Whitfield M, Marzluff WF, O’Rand MG. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J. Biol. Chem. 2000;275:30378–30386. doi: 10.1074/jbc.M003781200. [DOI] [PubMed] [Google Scholar]
- 14.Alekseev OM, Bencic DC, Richardson RT, Widgren EE, O’Rand MG. Overexpression of the Linker histone-binding protein tNASP affects progression through the cell cycle. J. Biol. Chem. 2003;278:8846–8852. doi: 10.1074/jbc.M210352200. [DOI] [PubMed] [Google Scholar]
- 15.Alekseev OM, Widgren EE, Richardson RT, O’Rand MG. Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J. Biol. Chem. 2005;280:2904–2911. doi: 10.1074/jbc.M410397200. [DOI] [PubMed] [Google Scholar]
- 16.Feng HP, Scherl DS, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–7831. doi: 10.1021/bi00081a030. [DOI] [PubMed] [Google Scholar]
- 17.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 18.Morton TA, Myszka DG, Chaiken IM. Interpreting complex binding kinetics from optical biosensors: a comparison of analysis by linearization, the integrated rate equation, and numerical integration. Anal. Biochem. 1995;227:176–185. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 19.Myszka DG, Morton TA. Clamp: a biosensor kinetic data analysis program. Trends Biochem. Sci. 1998;23:149–150. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- 20.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 22.Richardson RT, Alekseev O, Alekseev OM, O’Rand MG. Characterization of the NASP promoter in 3T3 fibroblasts and mouse spermatogenic cells. Gene. 2006;371:52–58. doi: 10.1016/j.gene.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Kleinschmidt JA, Seiter A. Identification of domains involved in nuclear uptake and histone binding of protein N1 of Xenopus laevis. EMBO J. 1988;7:1605–1614. doi: 10.1002/j.1460-2075.1988.tb02986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sims RJ, III, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuwe T, Hothorn M, Lejeune E, Rybin V, Bortfeld M, Scheffzek K, Ladurner AG. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc. Natl Acad. Sci. USA. 2008;105:8884–8889. doi: 10.1073/pnas.0712293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mateescu B, England P, Halgand F, Yaniv M, Muchardt C. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 2004;5:490–496. doi: 10.1038/sj.embor.7400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell. 2007;28:1015–1028. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Annunziato AT, Seale RL. Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 1983;258:12675–12684. [PubMed] [Google Scholar]
- 32.Perry CA, Annunziato AT. Influence of histone acetylation on the solubility, H1 content and DNase I sensitivity of newly assembled chromatin. Nucleic Acids Res. 1989;17:4275–4291. doi: 10.1093/nar/17.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry CA, Annunziato AT. Histone acetylation reduces H1-mediated nucleosome interactions during chromatin assembly. Exp. Cell Res. 1991;196:337–345. doi: 10.1016/0014-4827(91)90269-z. [DOI] [PubMed] [Google Scholar]









