Abstract
MicroRNAs (miRNAs, miRs) are genomically encoded small ∼22 nt RNA molecules that have been shown to mediate translational repression of target mRNAs involved in cellular proliferation, differentiation and death. Despite intensive studies on their physiological and pathological functions, the molecular mechanism of how miRNA gene transcription is regulated remains largely unknown. Microarray profiling revealed 21 miRNAs clustered on chromosome 12, including miR-433 and miR-127, that were co-upregulated in small heterodimer partner (SHP, NR0B2) SHP knockouts (SHP–/–) liver. Gene cloning revealed that the 3′-coding region of pri-miR-433 served as the promoter region of pri-miR-127. Estrogen related receptor (ERRγ, NR3B3) robustly activated miR-433 and miR-127 promoter reporters through ERRE, which was transrepressed by SHP. The strong elevation of miR-433 and miR-127 in Hepa-1 cells correlated with the down-regulation of SHP and up-regulation of ERRγ. Ectopic expression of ERRγ induced miR-433 and miR-127 expression, which was repressed by SHP coexpression. In contrast, knockdown ERRγ decreased miR-433 and miR-127 expression. In addition, the ERRγ agonist GSK4716 induced miR-433 and miR-127 expression both in vitro and in vivo, respectively. In summary, the coupled miR-433 and miR-127 genes were transcribed from independent promoters regulated by nuclear receptors ERRγ/SHP in a compact space by using overlapping genomic regions.
INTRODUCTION
MicroRNAs have emerged recently as a new class of non-protein-coding RNA molecules that have been shown to participate in the regulation of many cellular processes and changes of their expression are frequently observed in human diseases (1–3). Since the discovery of miRNAs, studies have been focused on the function of miRNAs (4,5) and their post-transcriptional regulation of mRNA targets (6,7). A major gap exists regarding how the expression of miRNA genes are regulated by transcription factors including nuclear receptors. This area of study is particularly challenging because little is known about miRNA gene structure and their transcriptional initiation sites.
Small heterodimer partner (SHP, NROB2) is a unique orphan nuclear receptor and transcriptional repressor that plays important roles in several metabolic diseases and in liver carcinogenesis (8–11). Due to lack of DNA-binding domain, SHP is unable to bind directly to its target gene promoter, but exerts its transrepressive effect through physical interaction with its regulatory partners (12), including estrogen related receptor (ERRγ, NR3B3) (8,13). To determine the potential regulatory function of SHP in miRNA gene expression, we conducted miRNA expression profiling using SHP–/– mice and cloned the primary transcripts encoding miR-433 and miR-127 gene cluster. We for the first time revealed that pri-miR-433 and pri-miR127 were derived from two separate but overlapping genes (manuscript under review). In this study, we characterized the molecular mechanism that controls miR-433 and miR-127 gene transcription. Our study showed that the transcription of miR-433 and miR-127 genes was tightly regulated by ERRγ and SHP through independent promoters which used overlapping genomic coding regions. Our results revealed a novel mechanism by which the coupled miR-433 and miR-127 genes were regulated by nuclear receptors in a compact genomic space.
MATERIALS AND METHODS
Total RNA isolation and miRNA microarray analysis
Total RNA with miRNA was isolated from livers of 2-month-old male mice (n = 3) using mirVana™ miRNA Isolation Kit (Ambion, Austin, TX, USA). The RNA quality control was performed using Bioanalyzer 2100. SHP knockouts (SHP–/–) and wild-type (WT) mice on a pure C57/BL6 background were analysed. The mice were with normal chow diet under feeding conditions.
MiRNA microarray including labelling, hybridization, image scanning and initial data analysis was carried out by LC sciences (http://www.LCsciences.com, Houston, TX). All protocols were deposited at ArrayExpress. LC-miRHumanMouseRat-9.1-070207-MRA-1030 array was used which was deposited in MIAMExpress. In brief, arrays are made based μParaflo microfluidic technology (Atactic Technologies, Houston, TX, USA). On the μParaflo microfluidic chip, each detection probe consists of a chemically modified nucleotide coding segment complementary to target microRNA (from miRBase, http://microrna.sanger.ac.uk/sequences/) or other RNA (control or customer defined sequences) and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. The detection probes are made by in situ synthesis using photogenerated reagent (PGR) chemistry (Array Protocol: LC Mir-Array-Prtl-060518). Small RNAs (<300 nt) are 3′-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag is then ligated to the poly(A) tail for later fluorescent dye staining; two different tags are used for the two RNA samples in dual-sample experiments (Labeling Protocol: LC Mir-Label Prtl-060518). Hybridization is performed using a micro-circulation pump (Atactic Technologies). The hybridization conditions are 100 μl 6 × SSPE buffer (0.90 M NaCl, 60 mM Na2HPO4, 6 mM EDTA, pH 6.8) containing 25% formamide, 34°C and overnight (Hybridization Protocol: LC Mir-Hyb Prtl-060518). Hybridization images are collected using a laser scanner (GenePix 4000B, Molecular Device, Sunnyvale, CA, USA). Scan resolution is set at 10 μ and PTM is set between 350 and 700 V (Scanning Protocol: LC Mir-Scan Prtl-060518). Data are analysed by first subtracting the background and then normalization. The background is determined using a regression-based background mapping method. The regression is performed on 5–25% of the lowest intensity data points excluding blank spots. Raw data matrix is then subtracted by the background matrix. Normalization is carried out using a LOWESS (Locally weighted Regression) method on the background-subtracted data (Normalization Protocol: LC Mir-Norm Prtl-060518). The data were deposited to the ArrayExpress database and the accession number is E-MEXP-1721.
Real-time RT–PCR quantification of miRNAs
Real-time reverse transcription polymerase chain reaction (RT–PCR) quantification of miRNA expression was carried out using mirVana™ quantitative real-time PCR (qRT-PCR) miRNA Detection Kit (Ambion, Austin, TX, USA) according to manufacturer's protocol. Briefly, cDNAs were synthesized from total RNA using gene-specific primers. Reverse transcription reactions contained 25 ng RNA samples, 1 μl of RT primer, 2 μl of RT buffer and 0.4 μl of ArrayScript Enzyme Mix. The 10 μl reactions were incubated for 30 min at 37°C, 15 min at 95°C, then held at 4°C. Real-time PCR was performed using an Applied Biosystems 7500 sequence detection system. The 25 μl PCR included 10 μl RT product, 12.5 μl of SYBR green mix (Applied Biosystems, Foster City, CA, USA) and 0.5 U Super Taq (Ambion) and 0.5 μl of mirVana PCR primers. Reactions were incubated in a 96-well optical plate at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The threshold cycle (Ct) was determined using default threshold settings. The Ct value is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. All experiments were done in triplicates and repeated three times. 5S small RNA was used as an internal control to normalize microRNA input in the real-time RT–PCR assay. Data were then analysed using the equation 2−ΔΔ Ct, where ΔΔCt = (Ct, Target–Ct, 5S) Target sample – (Ct, Target–Ct, 5S) Calculator (14).
In vitro and in vivo treatment of ERRγ agonist
ERRγ agonist GSK4716 (Cat #: C0926) was obtained from Sigma. In vitro cell culture experiments with GSK4716, Hepa-1 cells were treated with different doses of GSK4716 (5, 10 and 20 μm) or ethanol (vehicle) for 18 h (first time) and 8 h (second time). For in vivo i.p. injection in mice, GSK4716 was made up as a 20% solution in Cavitron (15). The mice were injected twice with 100 mg/kg of GSK4716, and the livers were collected for miR-433 and miR-127 gene expression analysis.
Transient transfection
The promoters of pri-miR-433 and pri-miR-127 were cloned into pGL3 basic vector, respectively. Hela cells were maintained in Dulbecco's Modified Eagle's Medium in the presence of 10% fetal bovine serum. Eight additional deletion constructs of pri-miR-127 and pri-miR-433 promoter reporters were generated using eight upstream primers (see Supplementary Materials). All deletion constructions were verified by sequencing. SHP and ERRγ expression vectors are available in our laboratory. For luciferase assays, cells were plated in 24-well plates 1 day before transfection and transfection was carried out using Fugene HD (Roche, Indianapolis, IN, USA). Total DNA in each transfection was adjusted by adding appropriate amounts of pcDNA3 empty vector. Approximately 48 h after transfection, cells were harvested and luciferase activities were measured and normalized against β-galactosidase activities as an internal control. The transfection experiments were carried out independently three times with similar efficiency and one representative result is shown.
Chromatin immunoprecipitation (ChIP) assays
The ChIP assays were performed using the ChIP Assay Kit (Upstate Biotechnology, Lake Placid, NY, USA). Hepa-1 cells were cultured until 70–80% confluence. Chromatin was cross-linked with 1% formaldehyde at 37°C for 10 min. Cells were washed with cold PBS twice and disrupted in SDS lysis buffer containing the protein inhibitor cocktail. Chromatin was sonicated to shear DNA to an average length between 200 and 1000 bp as verified by agarose gel. The sonicated cell supernatants were diluted 10-fold in ChIP dilution buffer containing the protein inhibitor cocktail and an aliquot of the solution was reserved for input control. Ten micrograms of ERRγ antibody (Aviva Systems Biology, San Diego, CA, USA) was added and the chromatin solution was gently rotated overnight on ice. The protein A agarose slurry was added to the antibody-bound chromatin solution and incubated at 4°C for 1 h with constant rotation. The agarose beads were collected by centrifugation, washed and the antibody-bound chromatin was released from the agarose beads. Finally, the DNA was purified by phenol/chloroform extraction and ethanol precipitation. The ERRE region was detected using primers (see Supplementary Materials) in PCR reactions. A 10 kb region downstream from the ERRE was selected and detected using primers listed in the Supplementary Material.
Electrophoretic mobility shift assay (EMSA)
EMSA was done using the LightShift Chemiluminescent EMSA Kit from Pierce Biotechnology (Rockford, IL, USA). In vitro translated ERRγ was incubated with the biotin-labelled duplex oligonucleotide probes (see Supplementary Materials). The binding reactions contained biotin-labelled DNA were incubated with the in vitro translated proteins in 1× binding buffer and 1 μg/μl Poly (dIċdC) for 20 min at room temperature. Competitor oligonucleotides were included at a 10- to 100-fold molar excess as indicated. DNA–protein complexes were resolved by electrophoresis in 6% native acrylamide gels and visualized by chemiluminescence.
RNA interference (RNAi)
ShRNA constructs against Mus musculus ERRγ were purchased from Origene Company (Rockville, MD, USA). Hepa-1 cells were cultured until 70–80% confluence. ShRNA constructs were transfected into the cells using transfection reagent Fugene HD (Roche) according to the manufacturer's instructions. The level of ERRγ expression was determined using real-time PCR.
Adenoviral transduction
Recombinant adenovirus vectors for expression of mSHP and the GFP control were prepared as described (8). Viral supernatants were generated by standard methods at Baylor College of Medicine BCM virus Core facility. Hepa-1 and Huh7 cells were cultured to 70–80% confluence. They were plated at 2 × 106/10 cm dish and infected the next day with viral supernatant at different multiplicities of infection for 2 h. Virus containing media were removed and cells were continuously cultured.
RESULTS
MiRNAs microarray profiling in SHP–/– mice
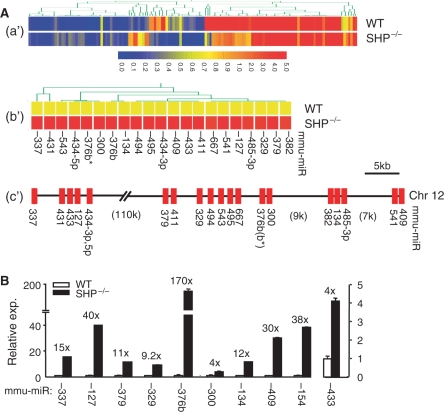
As an initial step to establish a correlation between SHP and miRNA gene expression, we conducted miRNA expression profiling by miRNA-microarray analysis using RNAs isolated from livers of WT and SHP–/– mice (Figure S1). Gene profiling and hierarchical clustering identified a total of 143 miRNAs that were differentially expressed (P < 0.01) (Figure 1A-a′ and Table S1). Among the 143 differentially expressed miRNAs, 82 were up-regulated and 61 were down-regulated in SHP–/– liver relative to the WT control. Out of the 82 up-regulated miRNAs, a surprising co-localization of 21 miRNAs on a single chromosome (Chr 12), that were arranged in a cluster, was revealed by comparative analysis (Figure 1A-b′ and -c′). The down-regulated miRNAs were located among different chromosomes. The coordinated up-regulation of miRNAs clustered on chromosome 12 in SHP–/– mice raised the possibility of a common repressive mechanism by SHP on the expression of these miRNAs. The expression levels of selected up-regulated miRNAs were further validated by qPCR using primer–probe sets that are commercially available (Figure 1B). The fold increases of miRNA expression in SHP–/– mice were generally higher by qPCR analysis as compared with microarray analysis, consistent with qPCR being a more sensitivity method for quantitating gene expression.
Figure 1.
Analysis of miRNA expression in the livers of WT and SHP–/– mice. (A-a′) Hierarchical clustering of miRNA expression profiling by microarray analysis. (A-b′) Hierarchical clustering of the 21 upregulated miRNAs in SHP–/– mice as compared with WT mice. (A-c′) The 21 up-regulated miRNAs in SHP–/– mice formed a cluster on the chromosome 12. (B) Real-time PCR verification of the miRNA expression in WT and SHP–/– mice.
Cloning of miRNA primary transcripts to determine the transcriptional initiation sites (TIS) is the first step towards elucidating the regulation of miRNA gene expression. Using database mining method and EST extension, we first cloned the primary transcripts of miR-433 and miR-127 by 5′ and 3′ RACE and observed that the expression of both transcripts was strongly up-regulated in livers of SHP–/– mice as compared with WT mice (Song, G.S. and Wang, L., unpublished data). In addition, the basal expression of pri-miR-127 appeared to be higher than that of pri-miR-433 in SHP–/– mice, which was consistent with the expression pattern of mature miR-433 and miR-127 (Figure 1B).
Cloning miR-433 and miR-127 promoters and identifying nuclear receptor-binding elements
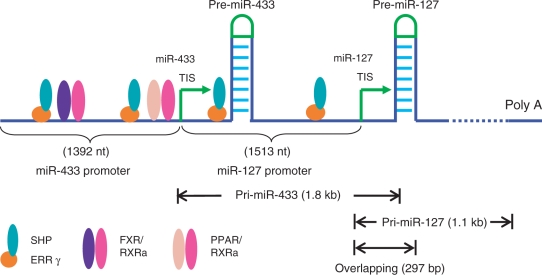
Despite similarities in their precursor organization, miR-433 and miR-127 exhibited an increased, but different, fold induction of expression in SHP–/– mice (Figure 1B). This suggests that they share a common inhibited mechanism by SHP, but are independently regulated at the transcriptional level. It is postulated that the region between the pre-miR-433 TIS and the pri-miR-127 TIS that contains part of pri-miR-433 may function as the promoter for pri-miR-127. Thus, the promoter of pri-miR-127 is located within the genomic region that encodes pri-miR-433 (Figure 2).
Figure 2.
Schematic of the overlapping miR-433 and miR-127 gene structure and the location of nuclear receptor-binding sites in the miR-433 and miR-127 promoters.
We cloned the region ∼1.4–1.5 kb upstream from TIS of pri-miR-433 and pri-miR-127 into luciferase reporters, respectively. Sequence analysis of the miR-433 and miR-127 promoters with the MatInspector program predicted two potential binding sites (−647/−656 nt and −1271/−1280 nt from TIS of miR-433; −375/−384 nt and −894/−903 nt from TIS of miR-127) for the ERRγ (Table 1). Putative binding sites for farnesoid X receptor (FXR) and peroxisome proliferator-activated receptor (PPAR) were also identified in the miR-433 promoter (Table 2).
Table 1.
Predicted ERR binding sites on the miR-433 and miR-127 gene promoters
| miR-433 Promoter (from miR-433-TIS) | miR-127 Promoter (from miR-127-TIS) |
| ERRE1: TGAAGGTgA (−647 to −656) | ERRE1: ACAAGGTCA (−375 to −384) |
| ERRE2: cCAAGGTCA (−1271 to −1280) | ERRE2: TcACCTTCA (−894 to −903) |
The predicted hits were mainly filtered out based on the criteria of Core Similarity 1 and a Matrix similarity of more than 0.8. For ERRE sites, mismatches with the consensus sequence are indicated by lower case letters.
Table 2.
Predicted binding sites for NRs on the miR-433 gene promoter
| TF name | Optimized matrix threshold | Strand | Core similarity | Matrix similarity | Sequence (capitals: core sequence) |
|---|---|---|---|---|---|
| Farnesoid X - activated receptor (RXR/FXR dimer), IR1 sites | 0.8 | (+) | 1 | 0.884 | AGGTcagtgtccc |
| Farnesoid X - activated receptor (RXR/FXR dimer), IR1 sites | 0.8 | (+) | 0.875 | 0.8 | GGGTcactggcat |
| PPAR/RXR heterodimers, DR1 sites | 0.76 | (−) | 0.753 | 0.764 | cccagccagggaacAGGCaaggc |
The NR-binding sites on the promoter regions of the two primary transcripts were predicted using software MatInspector. The predicted hits were mainly filtered out based on the criteria of Core Similarity 1 and a Matrix similarity of more than 0.8.
Common regulation of miR-433 and miR-127 promoters by nuclear receptors ERRγ/SHP
To explore the mechanisms of nuclear receptor regulation of miR-433 and miR-127 expression, transient transfection assays were performed. Among the nuclear receptors tested, ERRγ (Eγ), but not LXR (L), TR (T), ER (E), CAR (C), PPARα (Pα), PPARβ (Pβ) and ERRα (Eα), robustly stimulated the transactivation of both the miR-433 and miR-127 promoter reporters (Figure 3A). Ectopic expression of FXR/RXRα (FR), PPARα/RXRα (PαR) and PPARγ/RXRα (PγR) heterodimers also moderately activated the miR-433Luc, but not miR-127Luc. As expected, the response of the miR-433Luc and miR-127Luc to ERRγ was dose-dependent and was strongly repressed by SHP coexpression (Figure 3B), consistent with ERRγ being a target for potent SHP repression (8,13).
Figure 3.
Promoter analysis of the miR-433 and miR-127 luciferase reporters. (A) Transient transfection assays to determine nuclear receptor regulation of miR-433 and miR-127 promoter (Pro.) transactivation. The promoters of pri-miR-433 and pri-miR-127 were cloned into a pGL3-basic vector, respectively. Hela cells were transfected with the miR-433Luc or miR-127Luc in the presence of various nuclear receptor expression plasmids (200 ng). Luciferase (Luc.) activities (act.) were determined, which were normalized by β-gal activities. Con, control (pcDNA3); R, RXRa; L, LXRa; F, FXR; T, TRα; E, ERα; C, CAR; Pα, PPARα; Pβ, PPARβ; Pγ, PPARγ; Eα, ERRα; Eγ, ERRγ. (B) Dose-dependent transactivation of miR-433 and miR-127 promoter (Pro.) reporters by ERRγ (50, 100 and 200 ng), which were inhibited by SHP (50, 100 and 200 ng). (C) Deletion inactivation of the putative ERRE sites in the miR-433 (E1 and E2) and miR-127 (E1′ and E2′) promoter reporters. Each deletion reporter was transfected with the ERRγ expression plasmid (white bar, 20 ng; black bar, 40 ng) and luciferase activities were determined and normalized.
The ERR elements were designated as E1 and E2 for the miR-433Luc, E1′ and E2′ for the miR-127Luc (Figure 3C). Inactivation of E2 by deletion the entire ERRE site markedly decreased miR-433 (-1088) Luc activity induced by low (empty bar) and high (filled bar) doses of ERRγ (left). The effect of deleting both E1/E2 (-622 and -328Luc) appeared to be no different than that of deleting E2 alone (-1088Luc), whereas the activity of the smallest -92Luc was further decreased. Serial deletion of E2′ and its surrounding region reduced miR-127 (-990, -713 and -508) Luc activity induced by low doses but not by high doses of ERRγ (right). Further deletion of E1′ significantly decreased -287Luc transactivation. Thus, E2 in the miR-433 promoter and E1′/E2′ in the miR-127 promoter appeared to be potent ERRE sites for ERRγ activation. It is noted that deletion both of E1/E2 or E1′/E2′ did not completely block transactivation of either promoter to the basal level, suggesting additional mechanisms for the regulation of the miR-433 and miR-127 promoters.
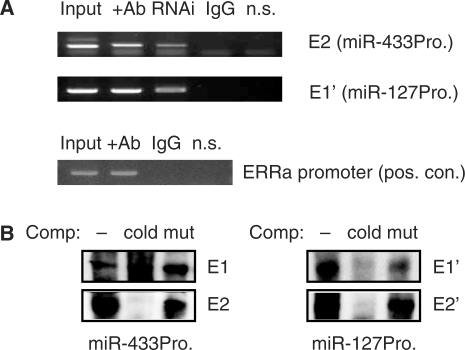
ChIP analyses showed that ERRγ was physically associated with the endogenous miR-433 and miR-127 promoters in Hepa-1 cells by strong Co-IP at E2 and E1′ sites, respectively (Figure 4A). In contrast, a non-specific primer set located ∼11 kb downstream of miR-127 promoter did not produce PCR product. In addition, RNAi knockdown ERRγ in the cells markedly reduced ERRγ binding to the promoters. It has been reported that ERRγ can activate ERRα promoter through an ERRE (16). Thus, ChIP assays were performed on ERRα promoter, which served as a positive control. As shown in Figure 4A (lower panel), ERRγ was Co-IP on ERRα promoter using ERRγ antibodies. Taken together, the results support our conclusion that ERRγ can bind to the endogenous miR-433 and miR-127 promoters in Hepa-1 cells.
Figure 4.
(A) ChIP analysis of ERRγ co-immunoprecipitation (Co-IP) on the miR-433 and miR-127 promoter region containing putative ERRE sites. The endogenous promoters in Hepa-1 cells were analysed. RNAi, ERRγ knockdown; non-specific (n.s.), using primers ∼11 kb downstream of miR-127 promoter which served as negative control. The ERRE in ERRα promoter served as positive control (pos. con.). The n.s. primers were located ∼10 kb upstream of the ERRα promoter. Nmuli cells were used. (B) Gel shift analysis to determine specific ERRγ protein binding to the putative ERRE sites in the miR-433 and miR-127 promoters. Comp, competitor; mut, mutant probe; cold, cold probe (10×).
EMSA showed that the ERRγ protein efficiently bound to E2, E1′ and E2′, but not E1 (Figure 4B). Thus, E1 appeared to be a non-specific ERRE. These results identified ERRγ and SHP as potent transcriptional regulators of miR-433 and miR-127 promoter activity.
Regulation of miR-433 and miR-127 expression by ERRγ/SHP
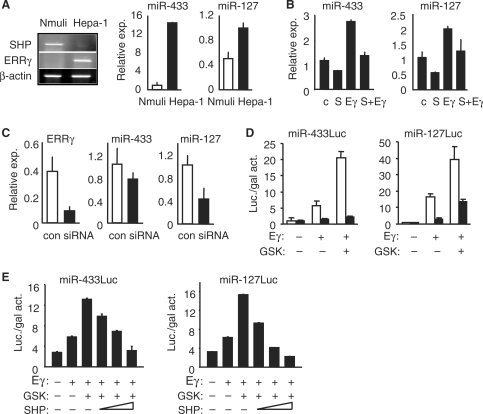
To test the direct regulatory effects of ERRγ and SHP on miR-433 and miR-127 expression, mouse hepatoma Hepa-1 cells, which have lost SHP expression (11), were used. Increased expression of ERRγ (Figure 5A, left) and ERRβ (Figure S2A), but not ERRα, correlated with the up-regulation of miR-433 and miR-127, as compared with normal hepatocyte Nmuli cells (Figure 5A, right). Despite increased expression of ERRβ in Hepa-1 cells, luciferase activity with the miR-433 and miR-127 promoter constructs was rather weak, thus we consider it as non-responsive (Figure S2B). Thus, ERRβ is an unlikely regulator of miR-433 and miR-127 expression. Ectopic overexpression of ERRγ in Hepa-1 cells significantly induced both miR-433 and miR-127 expression, which was repressed by coexpression of SHP (Figure 5B). Knocking down ERRγ in Hepa-1 cells by siRNA (Figure S2C) resulted in a corresponding reduction of miR-127 expression, although the effect on miR-433 was somewhat less efficient (Figure 5C). The data suggest that the transcription of the miR-433 gene may also be regulated by other nuclear receptors, as indicated by the transfection results (Figure 3A).
Figure 5.
SHP/ERRγ regulation of miR-433 and miR-127 expression. (A) Left: semi-quantitative RT-PCR analysis of SHP and ERRγ mRNA levels in normal mouse hepatocyte Nmuli cells and Hepa-1 hepatoma cells. Right: real-time PCR analysis of miR-433 and miR-127 expression in Nmuli and Hepa-1 cells. (B) Real-time PCR analysis of miR-433 and miR-127 expression in Hepa-1 cells that overexpressed with ERRγ (Eγ) and SHP (S). Control (c.). (C) Real-time PCR analysis of miR-433 and miR-127 expression in Hepa-1 cells that were transfected with siRNA-ERRγ. (D) Transient transfection assays to determine the effect of GSK4716 (1 μM) on ERRγ (10 ng) transactivation of miR-433Luc (white bar, -1392Luc; black bar, -92Luc) and miR-127Luc (white, -1513Luc; black, -287Luc). (E) Transient transfection assays to determine the effect of SHP (200, 100 and 50 ng) on ERRγ (40 ng) and GSK4716 (2.5 μM) transactivation of miR-433Luc (-1513Luc) and miR-127Luc (-1392Luc).
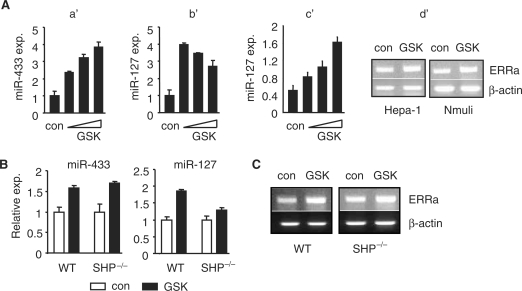
GSK4716 has been reported to be a selective ERRγ agonist which enhances ERRγ activity (15). Indeed, the transactivation of ERRγ on miR-433Luc and miR-127Luc containing ERRE was markedly enhanced by GSK4716 (Figure 5D), whereas no or weak activation on -92miR-433Luc and -287miR-127Luc was observed, respectively. Co-transfection of SHP dose-dependently inhibited the effect of GSK4716 on ERRγ activity (Figure 5E). Consistently, activation of ERRγ by GSK4716 induced miR-433 expression in a dose-dependent manner (Figure 6A, a′). The lack of dose response for the miR-127 suggested that miR-127 gene may be more sensitive to GSK4716 and that the lowest dose of GSK4716 that we used may have induced a maximal response (b′). Indeed, a dose-responsive activation of miR-127 was observed when a low dose range of GSK4716 was applied (c′). To confirm the effect of GSK4716, we analysed ERRγ target gene ERRα mRNA expression (16). GSK4716 treatment induced ERRα expression in both the Hepa-1 and Nmuli cells (Figure 6A, d′). When administrated in vivo in mice, GSK4716 induced miR-433 expression to a similar extent in both the WT and SHP–/– mice (Figure 6B, left). Interestingly, the induction of miR-127 by GSK4716 was more dramatic in WT than in SHP–/– (right). It is possible that the expression of ERRγ mRNA (not shown) and miR-127 (Figure 1B) was already highly up-reguated in SHP–/– mice, and that GSK may not be able to further enhance ERRγ activity. In addition, the expression of ERRα was induced by GSK4716 in both the WT and SHP–/– mice (Figure 6C), further confirm the effect of GSK4716.
Figure 6.
(A) Real-time PCR analysis of miR-433 and miR-127 expression in Hepa-1 cells that were treated with the ERRγ agonist GSK4716 (a′ and b′: 5, 10 and 20 μM; c′: 1, 2.5 and 5 μM). con, solvent (ethanol). d′: semi-quantitative PCR analysis of ERRα expression in Hepa-1 and Nmuli cells treated with GSK4716 (2.5 μM). (B) Real-time PCR analysis of miR-433 and miR-127 expression in the livers of WT and SHP–/– mice in vivo administrated GSK4716. (C) Semi-quantitative PCR analysis of ERRα expression in the livers of WT and SHP–/– mice in vivo administrated GSK4716.
DISCUSSION
Recently, emerging studies have established molecular links between miRNAs and nuclear receptors (17–20). However, essentially all these studies have focused on miRNA regulation of post-transcription of nuclear receptors. Although in one report it was shown that the expression of several miRNAs was altered in nuclear receptor PPARα knockout mice, the transcriptional mechanism of how PPARα regulates these miRNAs remains unknown (20). Our study is the first report which characterized the detailed molecular basis for SHP/ERRγ controlling miR-433 and miR-127 expression.
It is noted that the clustered 21 miRNAs on chromosome 12 were all up-regulated in SHP–/– mice, suggesting that they were directly or indirectly suppressed by SHP. Because we have not cloned all the primary transcripts and promoters for these up-regulated miRNAs, it is currently unknown if these clustered miRNAs are regulated through SHP/ERRγ dependent mechanism or through SHP dependent but ERRγ independent mechanism. It would be of great interest to investigate in future studies.
A recent report suggests that the human miR-127 is expressed as part of a miRNA cluster including miR-433 and is processed from a large transcript including all miRNAs (21). Our data show that the mouse miR-127 and miR-433 are generated from two distinct but overlapping primary transcripts controlled by independent promoters. Our new preliminary data suggest that a similar regulatory mechanism exists in other mammalian species including humans (Song, G.S. and Wang, L., unpublished data). Therefore, miR-433 and miR-127 expression regulation that we identified in mice may represent a conversed mechanism in a variety of mammalian species.
One interesting question is whether the elements upstream of miR-433 may also regulate miR-127. Eukaryotic gene promoters can have regulatory elements in proximal region or several kilobases away from the transcriptional start site. In general, the proximal promoter contains primary regulatory elements and the distal promoter may contain additional regulatory elements, often with a weaker influence than the proximal promoter. It is possible that the ERREs upstream of the miR-433 TIS start site may also regulate miR-127. Because the sites are over 2 kb away from the TIS of pri-miR127, they may represent weak regulatory elements. Therefore, miR-127 is believed to be primarily regulated by the proximal ERREs between miR-433 TIS and miR-127 TIS. Nevertheless, the results, that the two pri-miRNAs produced from the miR-433-127 locus are transcribed from two separate overlapping genes and the basal expression of pri-miR-127 is much higher than pri-miR-433, strongly suggest that miR-127 and miR-433 are independently and differentially regulated.
Another question is whether the ERREs in the promoters of miR-433 or miR-127 may also contribute to the increased expression of the flanking miR-431 and miR-343-3p/5p. The question is expected to be addressed upon determining TIS and TTS of both miRs and the cloning of full-length pri-miR-431 and pri-miR-343.
SHP has been recently shown to function as a tumour suppressor in liver cancer (10,11). It remains to be determined if miR-127 regulates BCL-6 in HCC, as it does in other human cancer cells (21). A direct association between SHP, miR-127 and HCC progression is currently under investigation.
In summary, our identification of the common regulation of miR-433 and miR-127 gene transcription by the nuclear receptors SHP and ERRγ and potential differential regulation of miR-433 by other nuclear receptors support the notion of a complex and interactive regulation of miRNA gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Liver Foundation/American Association for the Study of Liver Diseases (Liver Scholar Award); American Diabetes Association (Junior Faculty Award); American Heart Association (BGIA Award); NIH NIDDK (DK080440 to L.W.). Funding to open access charge: NIH DK080440.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Curt Hagedorn for critical reading of the manuscript. We are particularly grateful to Dr Timothy Willson for providing the GSK4716 information.
REFERENCES
- 1.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 2.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 3.Chang TC, Mendell JT. MicroRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2007;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 7.Sandmann T, Cohen SM. Identification of Novel Drosophila melanogaster MicroRNAs. PLoS One. 2007;2:e1265. doi: 10.1371/journal.pone.0001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Liu J, Saha P, Huang JS, Chan L, Spiegelman B, Moore DD. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 9.Huang JS, Iqbal J, Saha P, Liu J, Chan L, Hussain M, Moore DD, Wang L. Molecular characterization of the role of orphan receptor SHP in development of fatty liver. Hepatology. 2007;46:147–157. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- 10.He N, Park KT, Zhang YX, Huang JS, Lu S, Wang L. Epigenetic inhibition of nuclear receptor SHP is associated with and regulates hepatocellular carcinoma growth. Gastroenterology. 2008;134:793–802. doi: 10.1053/j.gastro.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Zhang YX, Xu P, Park KT, Kim YH, Moore DD, Wang L. Orphan receptor SHP suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48:289–298. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bavner A, Sanyal S, Gustafsson JA, Treuter E. Transcriptional corepression by SHP: molecular mechanisms and physiological consequences. Trends Endocrinol. Metab. 2005;16:478–488. doi: 10.1016/j.tem.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Sanyal S, Kim JY, Kim HJ, Takeda J, Lee YK, Moore DD, Choi HS. Differential regulation of the orphan nuclear receptor small heterodimer partner (SHP) gene promoter by orphan nuclear receptor ERR isoforms. J. Biol. Chem. 2002;277:1739–1748. doi: 10.1074/jbc.M106140200. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Zuercher WJ, Gaillard S, Orband-Miller LA, Chao EY, Shearer BG, Jones DG, Miller AB, Collins JL, McDonnell DP, Willson TM. Identification and structure-activity relationship of phenolic acyl hydrazones as selective agonists for the estrogen-related orphan nuclear receptors ERRbeta and ERRgamma. J. Med. Chem. 2005;48:3107–3109. doi: 10.1021/jm050161j. [DOI] [PubMed] [Google Scholar]
- 16.Zhang ZP, Teng CT. Interplay between estrogen-related receptor alpha (ERRα) and gamma (ERRγ) on the regulation of ERRα gene expression. Mol. Cell Endocrinol. 2007;264:128–141. doi: 10.1016/j.mce.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takagi S, Nakajima M, Mohri T, Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J. Biol. Chem. 2008;283:9674–9680. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 18.Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, Wang W, Liu R, George J, Ng HH, Perera RJ, et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;26:17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- 19.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol. Endocrinol. 2007;21:1132–1147. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 20.Shah YM, Morimura K, Yang Q, Tanabe T, Takagi M, Gonzalez FJ. Peroxisome proliferator-activated receptor alpha regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Mol. Cell Biol. 2007;27:4238–4247. doi: 10.1128/MCB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with down-regulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.