Abstract
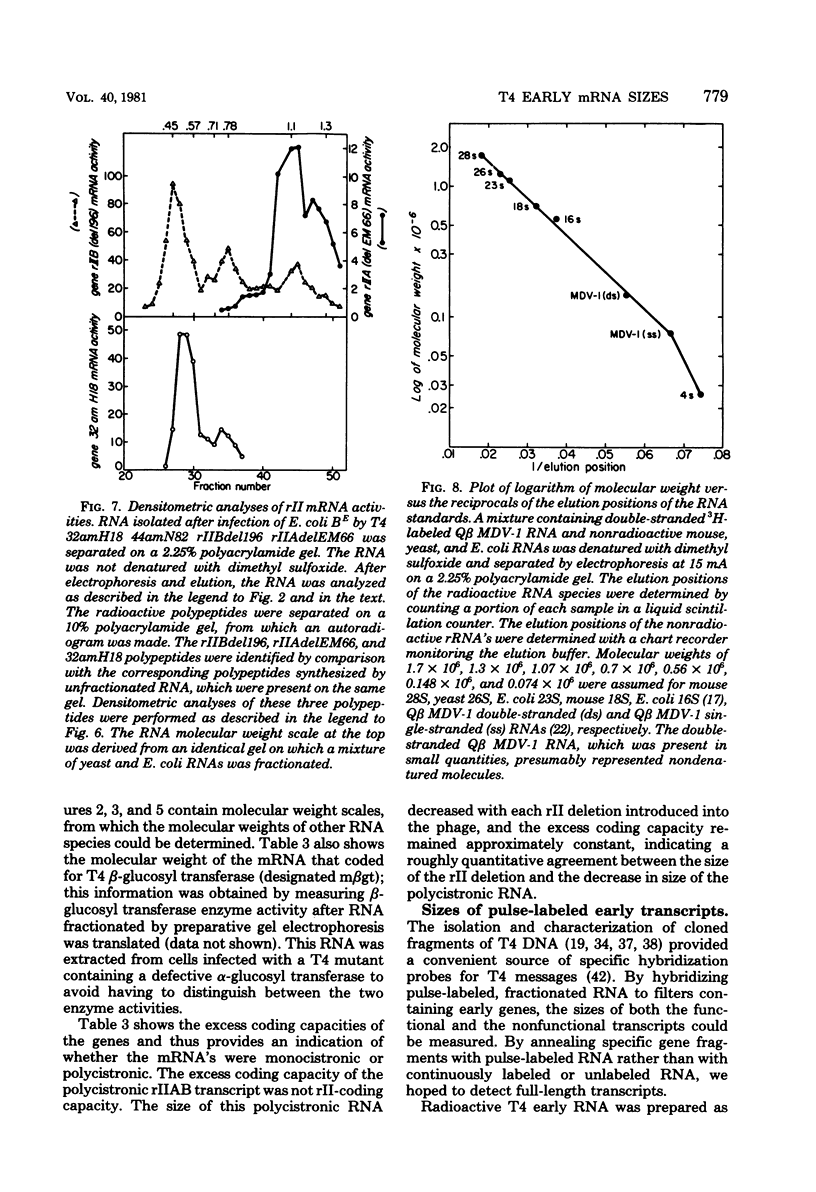
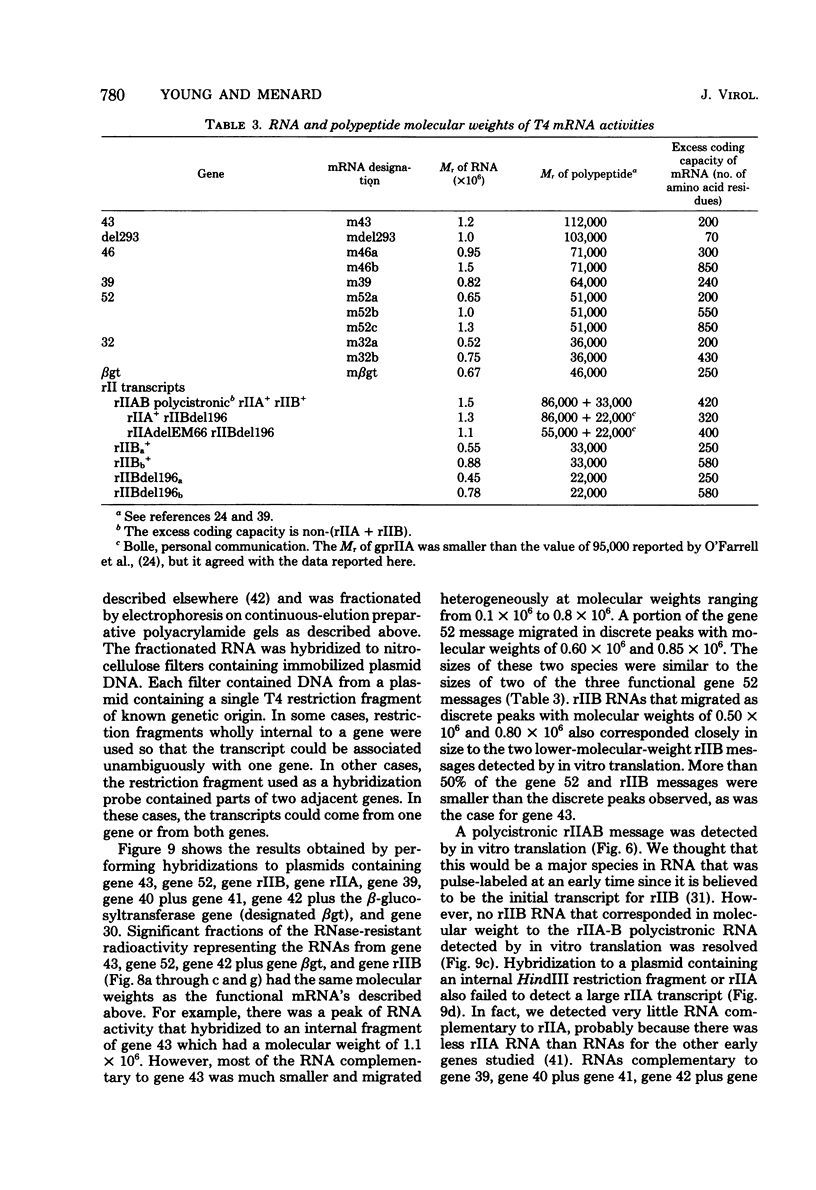
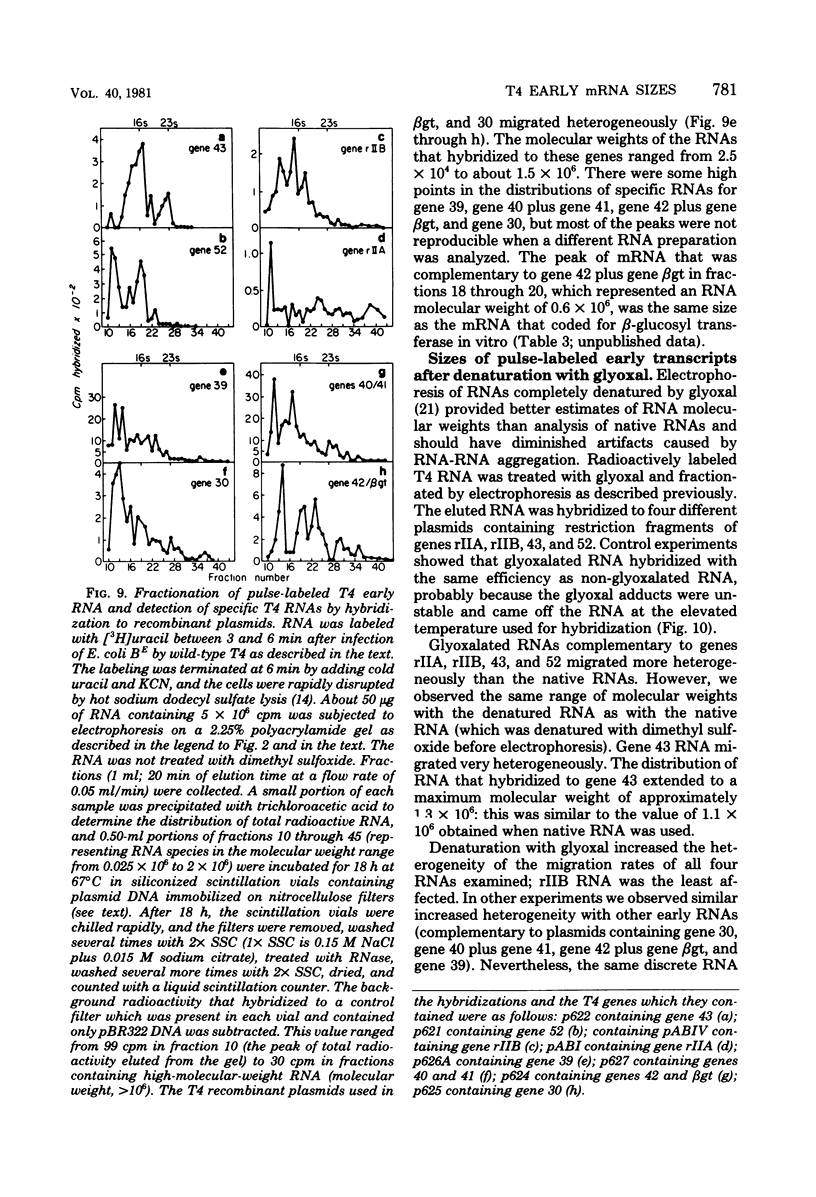
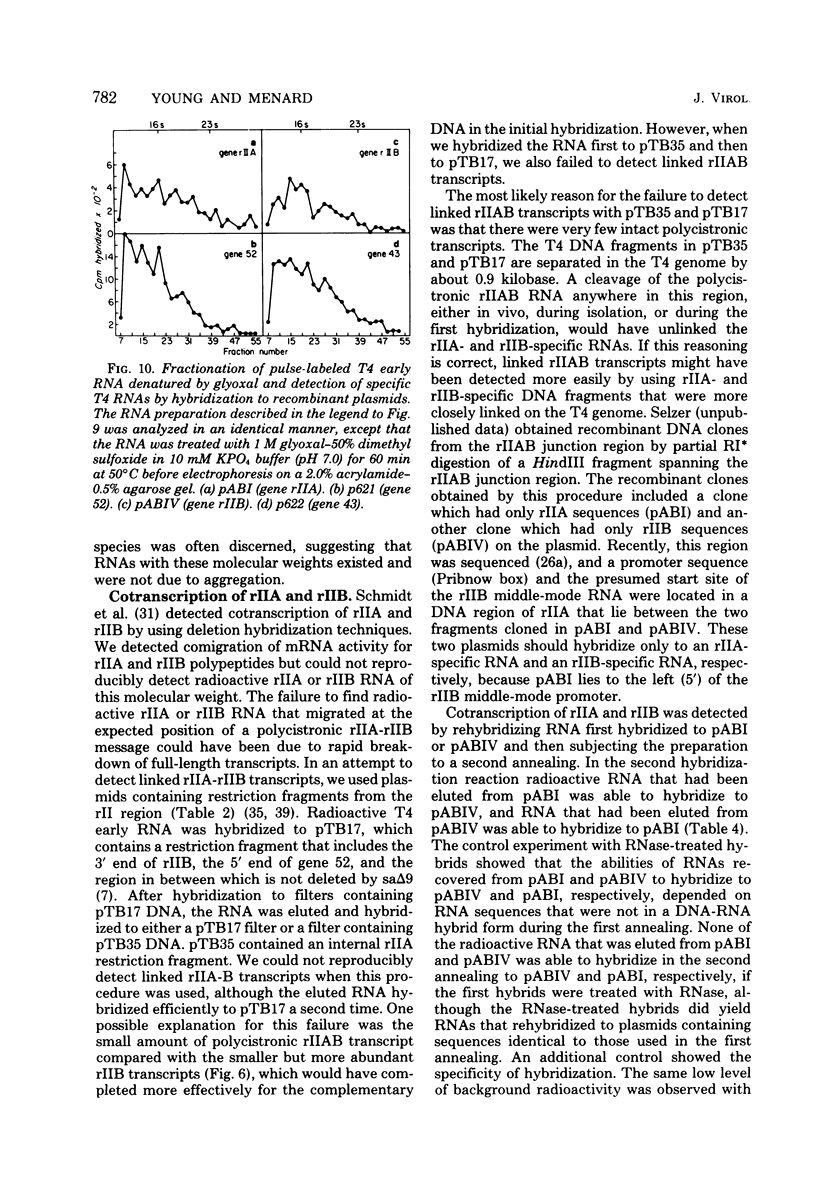
We determined the sized of specific T4 prereplicative nRNA's by preparative polyacrylamide gel electrophoresis, and we used the following two techniques to identify specific gene transcripts; cell-free protein synthesis accompanied by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to distinguish T4 polypeptides and hybridization to recombinant plasmids containing T4 DNA of known genetic composition. In our first analysis, the use of nonsense and in-phase deletion mutants allowed unambiguous identification of the functional transcripts that encoded genes 32, rIIB, and rIIA. In addition, we identified the functional transcript that encoded genes 43, 45, 30, 39, and 52, the beta-glucosyl transferase gene, and the deletion 293 region. A single peak of mRNA activity that coded for gp43, gp39, gprIIA, beta-glucosyl transferase, and the polypeptide encoded in the deletion 293 region was present; the other polypeptides were encoded in multiple mRNA species, gp46 and gp32 were encoded by two mRNA's and gp52 and gprIIB were encoded by three nRNA's. By hybridizing fractionated, pulse-labeled early RNA to cloned restriction fragments of T4 DNA, we identified the same specific transcripts for genes 43, 52, and rIIB. In addition, a lower-molecular-weight RNA (presumably degraded nRNA) was present even in pulse-labeled RNA preparations. The distribution of pulse-labeled RNAs that hybridized to gene 39, gene 30, gene rIIA, gene 40 plus gene 41, and gene 42 plus the beta-glucosyl transferase gene indicated extensive degradation. We detected cotranscription of genes rIIA and rIIB by rehybridization of RNA first annealed to an rIIB plasmid and then eluted and annealed to an rIIA plasmid. The size distributions of normal and chloramphenicol-treated RNAs that hybridized to plasmids containing T4 immediate early gene 30, gene 39, gene 40 plus gene 41, and gene 42 plus the beta-glucosyl transferase gene were not significantly different.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Black L. W., Gold L. M. Pre-replicative development of the bacteriophage T4: RNA and protein synthesis in vivo and in vitro. J Mol Biol. 1971 Sep 14;60(2):365–388. doi: 10.1016/0022-2836(71)90300-7. [DOI] [PubMed] [Google Scholar]
- Bolle A., Epstein R. H., Salser W., Geiduschek E. P. Transcription during bacteriophage T4 development: synthesis and relative stability of early and late RNA. J Mol Biol. 1968 Feb 14;31(3):325–348. doi: 10.1016/0022-2836(68)90413-0. [DOI] [PubMed] [Google Scholar]
- Brody E., Daegelen P., d'Aubenton-Carafa The role of termination factor rho in the development of bacteriophage T4 [proceedings]. Arch Int Physiol Biochim. 1978 Oct;86(4):897–898. [PubMed] [Google Scholar]
- Caruso M., Coppo A., Manzi A., Pulitzer J. F. Host--virus interactions in the control of T4 prereplicative transcription. I. tabC (rho) mutants. J Mol Biol. 1979 Dec 25;135(4):959–977. doi: 10.1016/0022-2836(79)90522-9. [DOI] [PubMed] [Google Scholar]
- Crawford L. V., Gesteland R. F. Synthesis of polyoma proteins in vitro. J Mol Biol. 1973 Mar 15;74(4):627–634. doi: 10.1016/0022-2836(73)90053-3. [DOI] [PubMed] [Google Scholar]
- Depew R. E., Snopek T. J., Cozzarelli N. R. Characterization of a new class of deletions of the D region of the bacteriophage T4 genome. Virology. 1975 Mar;64(1):144–145. doi: 10.1016/0042-6822(75)90086-0. [DOI] [PubMed] [Google Scholar]
- Dove W. The extent of rII deletions in phage T4. Genet Res. 1968 Apr;11(2):215–219. doi: 10.1017/s001667230001140x. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs are generated by site-specific cleavages. Proc Natl Acad Sci U S A. 1973 May;70(5):1559–1563. doi: 10.1073/pnas.70.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb A., Daniel V. Transcriptional control of two gene subclusters in the tRNA operon of bacteriophage T4. Nature. 1980 Jul 24;286(5771):418–420. doi: 10.1038/286418a0. [DOI] [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. A preliminary map of the major transcription units read by T7 RNA polymerase on the T7 and T3 bacteriophage chromosomes. Proc Natl Acad Sci U S A. 1974 Mar;71(3):760–764. doi: 10.1073/pnas.71.3.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso R. J., Buchanan J. M. Synthesis of early RNA in bacteriophage T4-infected Escherichia coli B. Nature. 1969 Nov 29;224(5222):882–885. doi: 10.1038/224882a0. [DOI] [PubMed] [Google Scholar]
- Hagen F. S., Young E. T. Effect of RNase III on the size of bacteriophage T7 lysozyme mRNA. J Virol. 1978 Jun;26(3):783–792. doi: 10.1128/jvi.26.3.783-792.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F. S., Young E. T. Preparative polyacrylamide gel electrophoresis of ribonucleic acid. Identification of multiple molecular species of bacteriophage T7 lysozyme messenger ribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3394–3400. doi: 10.1021/bi00713a033. [DOI] [PubMed] [Google Scholar]
- Hercules K., Sauerbier W. Two modes of in vivo transcription for genes 43 and 45 of phage T4. J Virol. 1974 Aug;14(2):341–348. doi: 10.1128/jvi.14.2.341-348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loening U. E. Molecular weights of ribosomal RNA in relation to evolution. J Mol Biol. 1968 Dec;38(3):355–365. doi: 10.1016/0022-2836(68)90391-4. [DOI] [PubMed] [Google Scholar]
- Mattson T., Richardson J., Goodin D. Mutant of bacteriophage T4D affecting expression of many early genes. Nature. 1974 Jul 5;250(461):48–50. doi: 10.1038/250048a0. [DOI] [PubMed] [Google Scholar]
- Mattson T., Van Houwe G., Bolle A., Selzer G., Epstein R. Genetic identification of cloned fragments of bacteriophage T4 DNA and complementation by some clones containing early T4 genes. Mol Gen Genet. 1977 Sep 9;154(3):319–326. doi: 10.1007/BF00571289. [DOI] [PubMed] [Google Scholar]
- Mattson T., Van Houwe G., Epstein R. H. Isolation and characterization of conditional lethal mutations in the mot gene of bacteriophage T4. J Mol Biol. 1978 Dec 15;126(3):551–570. doi: 10.1016/0022-2836(78)90058-x. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R., Spiegelman S. Complete nucleotide sequence of a replicating RNA molecule. Science. 1973 Jun 1;180(4089):916–927. doi: 10.1126/science.180.4089.916. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. Z., Gold L. M. Bacteriophage T4 gene expression. Evidence for two classes of prereplicative cistrons. J Biol Chem. 1973 Aug 10;248(15):5502–5511. [PubMed] [Google Scholar]
- O'Farrell P. Z., Gold L. M., Huang W. M. The identification of prereplicative bacteriophage T4 proteins. J Biol Chem. 1973 Aug 10;248(15):5499–5501. [PubMed] [Google Scholar]
- Pachl C. A., Yound E. T. The size and messenger RNA activity of bacteriophage T7 late transcripts synthesized in vivo. J Mol Biol. 1978 Jun 15;122(1):69–101. doi: 10.1016/0022-2836(78)90109-2. [DOI] [PubMed] [Google Scholar]
- Pachl C. A., Young E. T. Detection of polycistronic and overlapping bacteriophage T7 late transcripts by in vitro translation. Proc Natl Acad Sci U S A. 1976 Feb;73(2):312–316. doi: 10.1073/pnas.73.2.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D., Sigurdson D. C., Gold L., Singer B. S., Napoli C., Brosius J., Dull T. J., Noller H. F. rII cistrons of bacteriophage T4. DNA sequence around the intercistronic divide and positions of genetic landmarks. J Mol Biol. 1981 Jul 5;149(3):337–376. doi: 10.1016/0022-2836(81)90477-0. [DOI] [PubMed] [Google Scholar]
- Pulitzer J. F., Coppo A., Caruso M. Host--virus interactions in the control of T4 prereplicative transcription. II. Interaction between tabC (rho) mutants and T4 mot mutants. J Mol Biol. 1979 Dec 25;135(4):979–997. doi: 10.1016/0022-2836(79)90523-0. [DOI] [PubMed] [Google Scholar]
- Sakiyama S., Buchanan J. M. In vitro synthesis of deoxynucleotide kinase programmed by bacteriophage "T4-RNA. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1376–1380. doi: 10.1073/pnas.68.6.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W., Bolle A., Epstein R. Transcription during bacteriophage T4 development: a demonstration that distinct subclasses of the "early" RNA appear at different times and that some are "turned off" at late times. J Mol Biol. 1970 Apr 28;49(2):271–295. doi: 10.1016/0022-2836(70)90246-9. [DOI] [PubMed] [Google Scholar]
- Schmidt D. A., Mazaitis A. J., Kasai T., Bautz E. K. Involvement of a phage T4 sigma factor and an anti-terminator protein in the transcription of early T4 genes in vivo. Nature. 1970 Mar 14;225(5237):1012–1016. doi: 10.1038/2251012a0. [DOI] [PubMed] [Google Scholar]
- Sederoff R., Bolle A., Epstein R. H. A method for the detection of specific T4 messenger RNAs by hybridization competition. Virology. 1971 Aug;45(2):440–455. doi: 10.1016/0042-6822(71)90344-8. [DOI] [PubMed] [Google Scholar]
- Sederoff R., Bolle A., Goodman H. M., Epstein R. H. Regulation of rII and region D transcription in T4 bacteriophage: a sucrose gradient analysis. Virology. 1971 Dec;46(3):817–829. doi: 10.1016/0042-6822(71)90083-3. [DOI] [PubMed] [Google Scholar]
- Selzer G., Bolle A., Krisch B., Epstein R. Construction and properties of recombinant plasmids containing the rII genes of bacteriophage T4. Mol Gen Genet. 1978 Feb 27;159(3):301–309. doi: 10.1007/BF00268267. [DOI] [PubMed] [Google Scholar]
- Smith L. H., Sinsheimer R. L. The in vitro transcription units of bacteriophage phiX174. II. In vitro initiation sites of phiX174 transcription. J Mol Biol. 1976 Jun 5;103(4):699–710. doi: 10.1016/0022-2836(76)90204-7. [DOI] [PubMed] [Google Scholar]
- Velten J., Abelson J. The generation and analysis of clones containing bacteriophage T4 DNA fragments. J Mol Biol. 1980 Feb 25;137(2):235–248. doi: 10.1016/0022-2836(80)90327-7. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Tanyashin V. I., Murray N. E. Molecular cloning of fragments of bacteriophage T4 DNA. Mol Gen Genet. 1977 Nov 14;156(2):203–214. doi: 10.1007/BF00283493. [DOI] [PubMed] [Google Scholar]
- Witmer H. J. In vitro transcription of T4 deoxyribonucleic acid by Escherichia coli ribonucleic acid polymerase. Sequential transcription of immediate early and delayed early cistrons in the absence of the release factor, rho. J Biol Chem. 1971 Sep 10;246(17):5220–5227. [PubMed] [Google Scholar]
- Wood W. B., Revel H. R. The genome of bacteriophage T4. Bacteriol Rev. 1976 Dec;40(4):847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. T. Analysis of bacteriophage T4 chloramphenicol RNA by DNA-RNA hybridization and by cell-free protein synthesis, and the effect of Escherichia coli polarity-suppressing alleles on its synthesis. J Mol Biol. 1975 Aug 15;96(3):393–424. doi: 10.1016/0022-2836(75)90168-0. [DOI] [PubMed] [Google Scholar]
- Young E. T., Mattson T., Selzer G., Van Houwe G., Bolle A., Epstein R. Bacteriophage T4 gene transcription studied by hybridization to cloned restriction fragments. J Mol Biol. 1980 Apr 15;138(3):423–445. doi: 10.1016/s0022-2836(80)80011-8. [DOI] [PubMed] [Google Scholar]
- Young E. T., Menard R. C., Harada J. Monocistronic and polycistronic bacteriophage T4 gene 23 messages. J Virol. 1981 Dec;40(3):790–799. doi: 10.1128/jvi.40.3.790-799.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]