Abstract
Activation of the tumour suppressor p53 on DNA damage involves post-translational modification by phosphorylation and acetylation. Phosphorylation of certain residues is critical for p53 stabilization and plays an important role in DNA-binding activity. The 14-3-3 family of proteins activates the DNA-binding affinity of p53 upon stress by binding to a site in its intrinsically disordered C-terminal domain containing a phosphorylated serine at 378. We have screened various p53 C-terminal phosphorylated peptides for binding to two different isoforms of 14-3-3, ɛ and γ. We found that phosphorylation at either S366 or T387 caused even tighter binding to 14-3-3. We made by semi-synthesis a tetrameric construct comprised of the tetramerization plus C-terminal domains of p53 that was phosphorylated on S366, S378 and T387. It bound 10 times tighter than did the monomeric counterpart to dimeric 14-3-3. We showed indirectly from binding curves and directly from fluorescence-detection analytical ultracentrifugation that 14-3-3 enhanced the binding of sequence-specific DNA to p53 by causing p53 dimers to form tetramers at lower concentrations. If the in vitro data extrapolate to in vivo, then it is an attractive hypothesis that p53 activity may be subject to control by accessory proteins lowering its tetramer–dimer dissociation constant from its normal value of 120–150 nM.
p53, as a transcription factor, exerts its tumour suppressor function by the downstream expression of various target genes including those involved in either cell-cycle arrest or apoptosis (1–4). p53 levels are low in normal unstressed cells, but rise upon stress stimuli. Various post-translational modifications of p53, such as acetylation and phosphorylation, are early events upon stress that activate p53 to the functional active form. p53 is phosphorylated at multiple sites upon DNA damage, with the pattern depending on the nature of the stress stimulus. The N-terminus of p53 is multiply phosphorylated at sites that include S6, S9, S15, S18, S20, S33, S37, S46, T55 and T81. Phosphorylation at S15 and S20 disrupts its interaction with MDM2, the negative regulator of p53, thereby leading to stabilization (5,6). Various kinases are involved in these phosphorylations, with more than one kinase for certain serine/threonine residues, suggesting the importance of fail-safe mechanisms (7–9). Cell-cycle checkpoint kinases 1 and 2 are involved in the phosphorylation of S366, S378 and T387 of the C-terminal domain (10). Despite this specific knowledge on phosphorylation events, it is not yet fully clear how post-translational modifications activate p53.
Phosphorylation of p53 at S378 and dephosphorylation at S376 create a 14-3-3-binding site and thereby increase the sequence-specific DNA-binding affinity of p53 (11). The 14-3-3 family of proteins is involved in various cellular signalling events. There are nine isoforms of 14-3-3 known in mammals; α, β, γ, δ, ɛ, η, σ, τ and ζ. The 14-3-3 family binds to phosphoserine or phosphothreonine containing motifs and regulates various cellular functions such as cell division, nuclear transport, apoptosis or differentiation (12,13). The protein 14-3-3 exists as a dimer with two binding pockets for the phosphoserine containing motifs, as shown by the crystal structure of 14-3-3 bound to a phosphoserine peptide. Two distinct 14-3-3 binding motifs, which are referred to as mode 1 [R (S/Ar)XpS(LEAM)P] (13) and mode 2 [RX(Y/F)XpS(LEAM)P] binding motifs, were identified using peptide libraries (14,15).
In the present study, we have screened the p53 C-terminal regulatory domains (p53CT) that are phosphorylated (Table 1) at various positions for binding to two different 14-3-3 isoforms, ɛ and γ, and measured the binding affinities for the various peptides using fluorescence anisotropy titration and equilibrium sedimentation experiments (AUC). We also investigated how 14-3-3 influences the sequence-specific binding of p53 to DNA using analytical ultracentrifugation (AUC) with fluorescence detection.
Table 1.
Peptides used for screening
| Peptides | Sequencea |
|---|---|
| pS314 | KRALPNNTSSSPQPKKKPLDG |
| pS315 | KRALPNNTSSSPQPKKKPLDG |
| WT | CRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
| pS366 | CRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
| pS376 | CRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
| pS378 | CRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
| pT387 | CRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
| pS392 | CRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
| pS366/pT387 | CRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
| pS378/pT387 | CRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
| pS366/pS378 | CRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
| Mode 2 binding peptide | ARLYHSLLPAA |
aPeptides were labelled using fluorescein 5′-maleimide at the cysteine introduced in the p53CT peptides. pS314, pS315, mode 2 binding peptide and all the p53TCT were labelled using fluorescein succinimidyl ester at the free amino end of the peptides. S and T indicate phosphorylated serine and threonine, respectively.
MATERIALS AND METHODS
Protein expression and purification
PET vectors encoding his6 tagged 14-3-3 ɛ and γ were transformed into Escherichia coli BL21 for overexpression. Three litres of expression cultures at 37°C and 250 r.p.m. were induced with 0.8 mM IPTG at an A600 ∼ 0.6 and allowed to express for 16 h at 20°C. The cells were harvested by centrifugation followed by suspension in cell-cracking buffer of 50 mM sodium phosphate, pH 7.5, 300 mM NaCl, 14 mM β-mercaptoethanol and lysis by sonication. After centrifugation, the soluble fraction was loaded onto a Hi-trap Ni column and eluted with a 10–250 mM imidazole gradient over 10-column volumes. The pooled fractions were digested with TEV protease overnight and then diluted with equilibration buffer followed by loading onto the Ni column. The flow-through was concentrated and further purified on a Superdex 75 26/60 preparative gel filtration column (Amersham Biosciences, UK) in 20 mM HEPES, 150 mM NaCl, 5 mM DTT, 10% glycerol, pH 7.2. The proteins were >95% pure as judged by SDS/polyacrylamide gel electrophoresis. Protein samples were flash frozen and stored in liquid nitrogen for further use. The full-length p53 carrying stabilizing mutations (16,17) was expressed and purified as previously described (18).
Electrophoretic mobility shift assay experiments
The electrophoretic mobility shift assay (EMSA) for flp53 was carried out using p21 dsDNA (4 µM) CGCGAACATGTTCGAACATGTTCGCG labelled with fluorescein at 5′-end. Phosphorylated p53 (8 µM) was incubated with various amounts of 14-3-3 for 1 h followed by addition of p21 DNA for 30 min in 20 mM HEPES, 150 mM NaCl, 5 mM DTT, 10% glycerol, pH 7.2. The reaction mixtures were then separated on 0.8% agarose gel in 1X TBE for 3 h at a rate of 5 V/cm.
Fluorescence anisotropy and analytical centrifugation
All experiments were done as described (18–20). The reported values are the means of three independent experiments. Binding data from both the fluorescence anisotropy and the equilibrium sedimentation (where applicable) matched well. We used Aviv fluorescence detection system (FDS, Aviv Biomedical, NJ, USA) for the detection of fluorescence signal from labelled protein molecules and SedVel60K-FDS fluorescence velocity cells (Spin Analytical, Inc, Durham, NH, USA) with sample volumes of 65 μl. Buffer conditions were 25 mM sodium phosphate, 150 mM NaCl, 5 mM DTT, pH 7.2, 10% glycerol, 0.2 mg/ml BSA. The samples contained full-length p53 at concentration of 12.5–100 nM. Data were analysed using SedFit software (21).
Phosphorylation of recombinant p53 and purification
The purified recombinant p53 was phosphorylated using CHKI and CHKII as guided by the supplier (New England Biolabs, UK) and as described (10). Phosphorylation was followed by mass spectrometry. After 2–3 h of reaction, the reaction mixture was loaded onto a Superdex 200 26/10 analytical gel filtration column to separate the phosphorylated p53 from the reaction cocktail and eluted in 20 mM HEPES, 150 mM NaCl, 5 mM DTT, 10% glycerol, pH 7.2. The phosphorylated p53 was used for the EMSA experiments.
Peptide synthesis and purification
Peptides were synthesized on a Pioneer peptide synthesizer (Applied Biosystems, UK) using standard Fmoc chemistry. Phosphorylated serine and threonine were incorporated into the peptides during solid phase peptide synthesis using Fmoc-Ser(PO(Obzl)OH)-OH and Fmoc-Thr(PO(Obzl)OH)-OH (Nova biochem). S362C introduced at the N-terminus of the peptide was used for either coupling to the fluorescein 5′-maleimide or for the semi-synthetic ligation. The peptides were purified on a Waters HPLC using a reverse phase C18 and C8 semipreparative column (Vydac) and a acetonitrile gradient in water as described (19).
RESULTS
Screening of C-terminal phosphopeptides for 14-3-3 binding
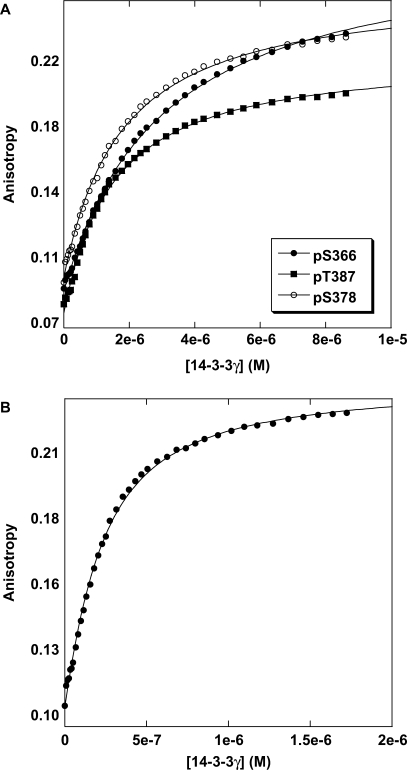
p53 does not contain any of the defined mode 1 (RSXpSXP) or mode 2 (RXXXpSXP) 14-3-3-binding motifs. However, the 14-3-3 family of proteins binds to various targets with significant deviation from the canonical sequences. We screened various p53CT phosphopeptides (362–393) for 14-3-3 binding. Table 2 lists the various monophosphorylated peptides screened with two of the 14-3-3 isoforms; γ and ɛ. Wild-type p53CT peptide did not show any detectable binding. Three peptides with phosphorylation at S366, S378 and T387 had dissociation constants of 14, 17 and 20 μM, respectively, with 14-3-3γ as measured by fluorescence anisotropy and AUC. Figure 1 shows the anisotropy curves for the three phosphopeptides pS366, pS378 and pT387 with 14-3-3γ. Two other isoforms of 14-3-3, ɛ and σ, were tested for binding. The ɛ-isoform showed similar binding affinities (SI text) as did the γ-isoform. It is reported, from immunoprecipitation and western blot assays, that 14-3-3 binds to pS378 (14). We also tested two peptides NLS (Nuclear Localization Signal, 305–322) derived from the tetramerization domain of p53 with phosphorylations at residues 314 and 315. pS314 showed weak binding, and pS315 did not show any binding at all to either of 14-3-3 isoforms.
Table 2.
Binding of mono-phosphorylated p53CT peptides (362–393) to 14-3-3
| Peptides |
Kd (µM) |
|
|---|---|---|
| 14-3-3γ | 14-3-3ɛ | |
| pT387 | 14 ± 3a | 11 ± 2a |
| pS366 | 17 ± 2a | 16 ± 2a |
| pS378 | 20 ± 2a | 18 ± 3a |
| pS392 | 105 ± 6b | 134 ± 8b |
| pS376 | 225 ± 20b | 175 ± 10b |
| pS314 | 130 ± 7b | 158 ± 8b |
| pS315 | No binding | No binding |
| WT (362–393) | Weak binding(n.q) | Weak binding (n.q) |
aMeasured using fluorescence anisotropy and AUC fitted to a simple one-state model.
bMeasured using AUC only.
Peptides are labelled at the cysteine introduced using fluorescein 5′-maleimide; n.q, not quantifiable.
Figure 1.
Binding titration profiles of 14-3-3γ with fluorescein-labelled monophosphorylated p53CT peptides. (A) 14-3-3γ binding to pS366 and pT387 peptides. (B) 14-3-3γ binding to pS378 peptide. Experiments were conducted at 20°C in 20 mM HEPES, 150 mM NaCl, 5 mM DTT, pH 7.2.
14-3-3 binds strongly to diphosphorylated peptides
In solution, 14-3-3 exists as a dimer and each 14-3-3 dimer possesses two binding pockets for phosphoserine or phosphothreonine ligands (Figure 2). The 14-3-3 binds with higher affinity to phosphopeptides containing two 14-3-3-binding sites separated by a spacer than to the single-binding motif peptide (14). Our initial experiments showed three essential phosphorylations for strong 14-3-3 binding. Hence, we synthesized three peptides carrying different combinations of the two phosphorylations: pS366/pS378, pS378/pT387 and pS366/pT387. Each peptide was tested for 14-3-3 binding (Figure 3). 14-3-3 bound to the diphosphorylated peptides much more strongly than to the monophosphorylated ones. Stoichiometric titration was carried out to find out how many 14-3-3 molecules bind to a single peptide. The sharp transition point from unbound to bound species gives the stoichiometry of binding at high concentration of the peptide when titrated with 14-3-3. One 14-3-3 dimer bound one doubly phosphorylated peptide (Figure 3C). Table 3 lists the binding data for the peptides obtained using fluorescence anisotropy measurements. A peptide with a mode 2 binding motif was synthesized to compare the binding affinities of the various peptides. As can be seen from the Table 3, the diphosphorylated peptide pS366/pT387 bound much more strongly than the consensus mode 2 peptide.
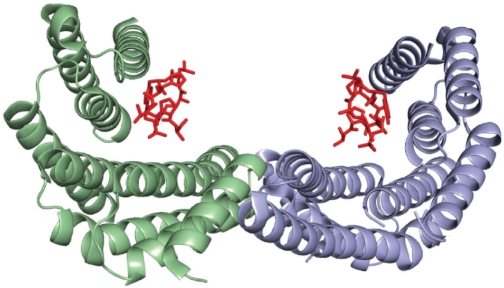
Figure 2.
Structure of the 14-3-3/mode 2 phosphopeptide complex. Each 14-3-3 monomer binds a monophosphorylated peptide (red) (24). Peptides with diphosphorylations separated by a flexible linker bind with higher affinity. This is observed in all of the p53CT diphosphorylated peptides.
Figure 3.
Binding isotherms of 14-3-3γ with fluorescein-labelled p53CT diphosphorylated peptides. (A) 14-3-3γ binding to p53CT diphosphopeptides, pS366/pT387, pS366/pS378 and pS378/pT387. (B) Binding of 14-3-3γ to the mode 1 consensus peptide. (C) Stoichiometric titration of 14-3-3γ with pS366/pT387 peptide revealed that one 14-3-3 dimer binds one diphosphorylated peptide.
Table 3.
Binding of di-phosphorylated p53CT peptides (362–393) to 14-3-3
| Peptides |
Kd (nM) |
|
|---|---|---|
| 14-3-3γ | 14-3-3ɛ | |
| Consensus | 430 ± 6a | 540 ± 10a |
| pS366/pT387 | 140 ± 7a | 180 ± 5a |
| pS366/pS378 | 480 ± 5a | 512 ± 3a |
| pS378/pT387 | 450 ± 3a | 805 ± 6a |
aMeasured using fluorescence anisotropy only.
Tetrameric p53TCT bound 10 times tighter than monomeric p53CT peptides
Since p53 is tetrameric, we looked at 14-3-3 binding to p53CT in the context of the tetramer. Five peptides (293–393) were synthesized using a chemical ligation approach: three monophosphorylated peptides (pS366, pS378 and pT387), a diphosphorylated peptide (pS366/pT387) and the wild-type peptide. Table 4 lists the binding data obtained for the tetrameric constructs. Tetrameric p53TCT pS366, pS378, pT387 had Kd values of 1.8, 1.7 and 1.2 μM, respectively, compared to 17, 14 and 20 μM, respectively, for the corresponding monomeric constructs. Figure 4 shows the binding curves for the tetrameric constructs. Wild-type p53TCT (293–393) displayed very weak binding while the diphosphorylated p53TCT only showed similar binding to the monomeric diphosphorylated ones. One reason for this apparent discrepancy may be that 14-3-3 binds to the diphosphorylated tetrameric p53 in a more complex fashion, with a mixture of bound complexes as opposed to the single complex formed between a monophosphorylated binding motif and each 14-3-3 subunit.
Table 4.
Binding of tetrameric p53TCT with mono- or di-phosphorylated subunits (293–393) to 14-3-3
| Peptides |
Kda (µM) |
|
|---|---|---|
| 14-3-3γ | 14-3-3ɛ | |
| WT TC (293–393) | Weak binding | Weak binding |
| p53TCT pS366 | 3 ± 0.5 | 6.2 ± 0.6 |
| p53TCT pT387 | 1.2 ± 0.5 | 3.5 ± 0.8 |
| p53TCT pS378 | 1.7 ± 0.3 | 1.4 ± 0.5 |
| p53TCTS366/pT387 | 0.154 ± 0.004 | 0.310 ± 0.008 |
aMeasured using fluorescence anisotropy only. Peptide concentrations used for titrations were 0.7 µM.
Figure 4.
Fluorescence anisotropy titrations of 14-3-3γ with tetrameric P53CT-phosphorylated peptides (p53TCT). (A) Binding of 14-3-3γ to fluoresceinated p53TCT pS366, pS378 and pT387 peptides. (B) Binding of 14-3-3γ to p53TCT pS366/pT387 peptide. Concentration of peptide used was 0.7 µM to ensure that p53TCT was tetrameric under experimental conditions.
14-3-3γ increases the sequence-specific DNA-binding affinity of p53
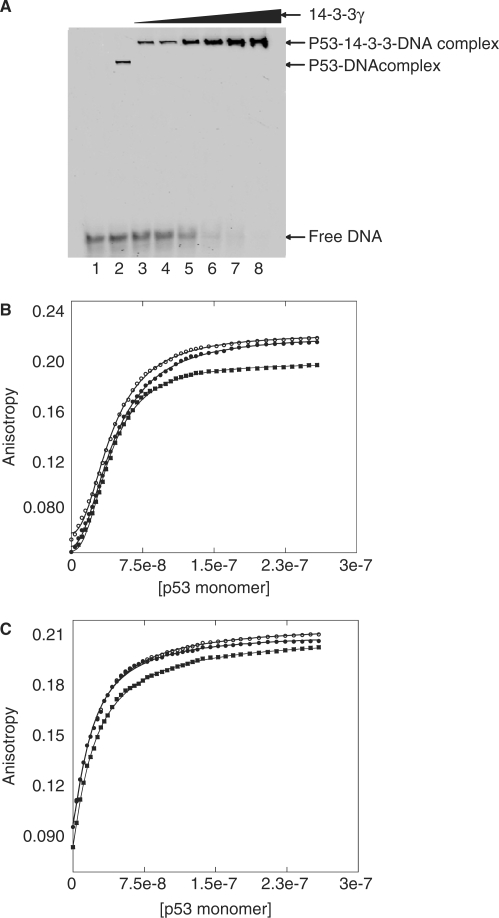
Binding of 14-3-3 enhances the sequence-specific DNA binding of p53 (11). To investigate the mechanism of enhancement, we carried out EMSA on binding to recombinant full-length p53 (flp53) that is phosphorylated using CHKI and CHKII kinases. In vitro phosphorylated p53 was incubated with increasing amounts of 14-3-3 for 1 h followed by addition of p53 recognition sequence p21 labelled at the 5′-end with fluorescein for 30 min. Increasing amount of 14-3-3 enhanced the p53/DNA complex formation (Figure 5A). Competition experiments with unlabelled p21 and mutant p21 response element showed the increase in binding is sequence specific and the effect was similar on different lengths of DNA (data not shown). Quantitative measurements by fluorescence anisotropy titrations of phosphorylated flp53 binding to fluorescein-labelled p21 response element DNA gave a Hill constant of ∼2.1 and half saturation at 45 nM. The Hill constant dropped to ∼1.1 with a Kd of 22 nM on titrating with the flp53/14-3-3 complex (Figure 5B and C). The observed binding is not the non-specific binding of DNA to the C terminus as that mode has a dissociation constant of 6–8 μM (22).
Figure 5.
DNA-binding experiments on p53. (A) EMSA experiments show specific DNA-binding enhancement in the presence of 14-3-3γ. Lane 1: only DNA (4 µM), lanes 2–8: phosphorylated p53 (8 µM), DNA (4 µM) and increasing amounts of 14-3-3γ (0, 2, 4, 8, 10, 15 and 20 µM, respectively). (B and C) Three independent binding experiments of p53 to p21 DNA labelled with fluorescein at the 5′-end in the absence and presence of 5 µM of 14-3-3, respectively. The data upon fitting to the Hill equation, [Protein]h/(Kh + [Protein]h), gave Hill constants, h, of 2.1 and 1.1, respectively. The binding curve obtained in the presence of 14-3-3 can be fitted to a simple one-state binding model [Protein]/(K + [Protein]), showing the non-cooperativity in DNA binding of p53.
Measuring the dissociation constant of full-length p53 by AUC
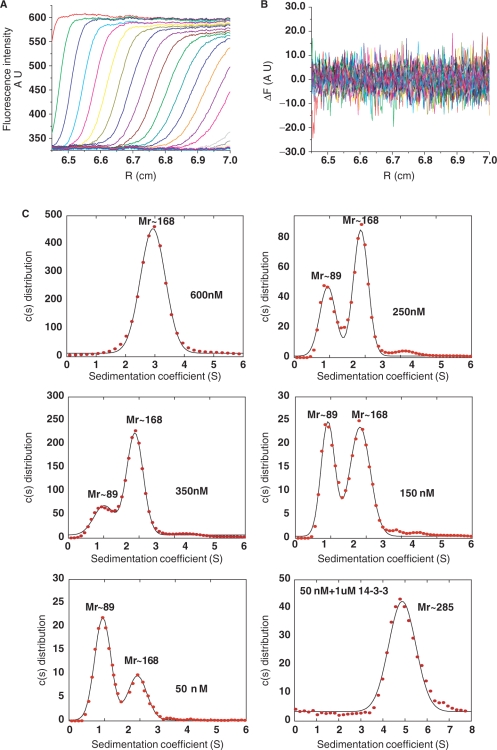
To delineate the mechanism of 14-3-3-mediated specific-DNA binding of p53, we employed an AUC method with fluorescence detection measuring sedimentation velocities. A tetracysteine (TC) motif was introduced to the C terminus of p53, which was site-specifically labelled with a FlAsH dye. The biarsenical-labelling reagent FlAsH-EDT2 becomes fluorescent when it binds to proteins containing the TC motif Cys-Cys-Pro-Gly-Cys-Cys. Introducing the dye did not affect 14-3-3 binding to p53.
p53 existed as a tetramer at as low as 600 nM (expressed in monomer concentrations). Dimers and tetramers of p53 could be distinguished at various concentrations below 500 nM. The dissociation constant for flp53 was estimated to be ∼120 nM from the integral of the area under the distributions. At 50 nM, the major species observed is the p53 dimer. When 1 µM of 14-3-3 was added to 50 nM of phosphorylated flp53, the low Mr peak disappeared and a new peak emerged at Mr ∼285 kDa, a difference of 115 kDa from the tetrameric p53, suggesting that two dimers of 14-3-3 bound a p53 tetramer (Figure 6). The stoichiometry was calculated from the differences in the apparent molecular masses obtained by the fit and hence may not reflect the true complex.
Figure 6.
AUC experiments of FlAsH tagged full-length p53. (A) Fluorescence sedimentation profiles were acquired for the flp53 at 45 000 r.p.m. A total of 400 scans in intervals of 2.5 min were acquired. For clarity, only every fifth scan is shown. (B) Residuals obtained for the fit. (C) Distributions obtained for various concentrations of FlAsH-p53; 600 nM, 350 nM, 250 nM, 150 nM, 50 nM and phosphorylated FlAsH-p53 at 50 nM in the presence of 14-3-3 using a continuous distribution model.
DISCUSSION
Two potential binding sites for 14-3-3 in p53CT domain
We screened a p53 C-terminal phosphopeptide library for potential 14-3-3 binding candidates (Table 1). Three peptides phosphorylated at either S366, S378 or T387 bound with 14–20 µM dissociation constants (Table 2). Of these, only S378 was previously shown to bind 14-3-3. The Kd values obtained for the S366 and T387 phosphorylated peptides were lower than that for S378, suggesting that these residues may also be involved in 14-3-3 recognition. S392 phosphorylation is important for regulating the oncogenic function of mutant p53 (23). The binding of the S392 phosphorylated peptide to 14-3-3 was very weak as indicated by AUC. Wild-type p53CT terminus peptide did not have detectable binding up to 200 µM.
Comparison of mono-, di- and tetrameric p53TCT peptides for 14-3-3 binding
14-3-3 is a dimeric protein, with two binding pockets for phosphopeptide motifs. A peptide with two tandem phosphoserine motifs separated by a spacer binds 14-3-3 much more strongly than the isolated motifs (14). Peptides with two phosphorylated sites, pS366/pS378, pS366/pT387 and pS378/pT387 had 50–100 times higher affinities than any of the monophosphorylated peptides, suggesting that 14-3-3 binds as a dimer to the bi-phospho motifs that are separated by a spacer (Table 3). The pS366/pT387 peptide bound much more tightly than did the other diphosphorylated peptides, suggesting that the length of the spacer may play some role in the binding events in the context of 14-3-3. Since p53CT is unstructured and flexible, it can bind 14-3-3 in an extended conformation, like many other interacting partners reported (15,24).
Since p53 functions as tetramer in binding response elements, we examined the binding affinities of 14-3-3 to a tetrameric construct of the p53CT, p53TCT. We made four p53TCT constructs with monophosphorylation at either S366, S378 or T387, and one diphosphorylation at S366 and T387. The three monophosphorylated p53TCT constructs had ∼10-fold higher affinity than did the corresponding monomeric peptides, whereas the p53TCT S366/T387 construct had a similar binding affinity to its equivalent monomeric peptide.
p53/14-3-3 complex binds DNA less cooperatively by perturbing the oligomerization equilibrium
14-3-3 increases the sequence-specific binding affinity of DNA to the core domain of p53 through its association to p53 (11), as confirmed here by EMSA experiments on phosphorylated flp53 with increasing amounts of 14-3-3. p53 exists as a dimer of dimers in solution. Two dimers of p53 bind DNA cooperatively with a Hill constant of 2 (25). p53 assembles as a tetramer on DNA. The dissociation constant of tetrameric flp53 to dimers is 120 ± 30 nM, from AUC experiments.
flp53 that was phosphorylated at various residues, particularly S366, S378 and T387 were used for DNA-binding experiments in the presence of 14-3-3. The flp53/14-3-3 complex bound DNA less cooperatively than did flp53 alone. The binding isotherms (Figure 5) fitted well to a simple one-state binding model or a cooperative Hill model of low cooperativity with h close to 1, at 1.1. As the cooperativity of binding of flp53 to response element DNA is caused by a unbound dimeric state going to a bound tetramer, we tested the effects of 14-3-3 on the tetramerization of flp53. At concentrations where flp53 is predominantly dimeric, p53 became a tetramer in the presence of 14-3-3 (Figure 6). Two dimers of 14-3-3 formed a complex with tetrameric flp53 (Figure 7). This is different from C-terminal peptides in isolation, where one diphosphorylated peptide binds one 14-3-3 dimer. In the context of the full-length tetramer with multiple phosphorylations, there is a preferential binding mode in which the phosphate-binding pockets in the two subunits of the 14-3-3 homodimer are occupied by phosphoserine/threonine residues from two different subunits of the p53 tetramer, resulting in the observed p53/14-3-3 complex.
Figure 7.
Schematic model of p53/14-3-3 interaction: p53 exists as dimer in solution. p53 binds DNA cooperatively as a dimer of dimers and assembles as a tetramer on DNA. In the presence of 14-3-3, p53 is tetramerized with two dimers of 14-3-3 forming a complex with one tetrameric p53. The p53/14-3-3 complex binds DNA less cooperatively.
The oligomerization state of p53 is modulated through its interaction with binding partner proteins
Tetramerization of p53 is important for its activity. S100 proteins are upregulated in cancer tissues and are associated with stimulation of metastasis. The S100 family of proteins regulate the oligomerization of p53. S100B and S100A bind to the tetramerizaton domain of p53 only in lower oligomerization states presumably the monomeric and the dimeric forms, in turn regulating the subcellular localization of p53 (20). Apoptosis repressor with caspase recruitment domain (ARC) is induced in many cancer tissues. ARC inactivates p53 by binding to the tetramerization domain of p53 (26). Both S100 A and ARC negatively regulate the activity of p53 by inhibiting tetramerization and affecting nuclear localization.
Another family of proteins, 14-3-3, positively regulates the function of p53 by enhancing the oligomerization of p53. The transcriptional activity of p53 is dependent on its DNA-binding ability. Though the core domains of p53 can alone bind DNA, tetrameric p53 binds DNA cooperatively with enhanced affinity. 14-3-3σ was shown to increase the transcriptional activity of p53 by facilitating the oligomerization of p53. Also, binding of 14-3-3σ increases the stability of p53 by inhibiting the MDM2-mediated ubiquitination of p53 (27).
Here, we provide direct evidence for the influence of 14-3-3 on the DNA binding of p53 by enhancing its oligomerization in vitro. Although neither the phosphorylation status of S373 and S375 (equivalent to human S376 and S378) nor 14-3-3 binding are critical for mouse p53 function, these events play an important role in activating human p53 (28). There is no direct evidence in vivo showing the activation of p53 by the modulation of its oligomeric state. However, if the dissociation constant of p53 from tetramers into dimers measured in vitro holds good for in vivo, it is expected that p53 exists predominantly in dimeric form at lower concentrations. Accordingly, it is an attractive hypothesis that the activity of p53 may be up-regulated by its interaction with accessory proteins that facilitate its tetramerization into the form that binds response elements.
FUNDING
Funding for open access charge: Medical Research Council.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr T.S. Wong for help in EMSA experiments and Dr. A. Joerger for critical reading of the article. We acknowledge Karoly Von Glos for help in peptide synthesis. We also acknowledge Structural Genomics Consortium, Oxford for the 14-3-3γ expression plasmid. S.R. is supported by a fellowship from Cambridge Commonwealth Trusts and Medical Research Council.
REFERENCES
- 1.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH. p53: death star. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 4.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 5.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 6.Unger T, Juven-Gershon T, Moallem E, Berger M, Vogt Sionov R, Lozano G, Oren M, Haupt Y. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meek DW. Multisite phosphorylation and the integration of stress signals at p53. Cell Signal. 1998;10:159–166. doi: 10.1016/s0898-6568(97)00119-8. [DOI] [PubMed] [Google Scholar]
- 8.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 9.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 10.Ou YH, Chung PH, Sun TP, Shieh SY. p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol. Biol. Cell. 2005;16:1684–1695. doi: 10.1091/mbc.E04-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat. Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 12.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 14.Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 15.Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, Gamblin SJ, Yaffe MB. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 16.Joerger AC, Allen MD, Fersht AR. Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J. Biol. Chem. 2004;279:1291–1296. doi: 10.1074/jbc.M309732200. [DOI] [PubMed] [Google Scholar]
- 17.Nikolova PV, Henckel J, Lane DP, Fersht AR. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc. Natl Acad. Sci. USA. 1998;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ang HC, Joerger AC, Mayer S, Fersht AR. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J. Biol. Chem. 2006;281:21934–21941. doi: 10.1074/jbc.M604209200. [DOI] [PubMed] [Google Scholar]
- 19.Friedler A, Veprintsev DB, Freund SM, von Glos KI, Fersht AR. Modulation of binding of DNA to the C-terminal domain of p53 by acetylation. Structure. 2005;13:629–636. doi: 10.1016/j.str.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc. Natl Acad. Sci. USA. 2005;102:4735–4740. doi: 10.1073/pnas.0501459102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinberg RL, Freund SM, Veprintsev DB, Bycroft M, Fersht AR. Regulation of DNA binding of p53 by its C-terminal domain. J. Mol. Biol. 2004;342:801–811. doi: 10.1016/j.jmb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 23.Yap DB, Hsieh JK, Zhong S, Heath V, Gusterson B, Crook T, Lu X. Ser392 phosphorylation regulates the oncogenic function of mutant p53. Cancer Res. 2004;64:4749–4754. doi: 10.1158/0008-5472.CAN-1305-2. [DOI] [PubMed] [Google Scholar]
- 24.Petosa C, Masters SC, Bankston LA, Pohl J, Wang B, Fu H, Liddington RC. 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 1998;273:16305–16310. doi: 10.1074/jbc.273.26.16305. [DOI] [PubMed] [Google Scholar]
- 25.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 26.Foo RS, Nam YJ, Ostreicher MJ, Metzl MD, Whelan RS, Peng CF, Ashton AW, Fu W, Mani K, Chin SF, et al. Regulation of p53 tetramerization and nuclear export by ARC. Proc. Natl Acad. Sci. USA. 2007;104:20826–20831. doi: 10.1073/pnas.0710017104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang HY, Wen YY, Chen CH, Lozano G, Lee MH. 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol. Cell. Biol. 2003;23:7096–7107. doi: 10.1128/MCB.23.20.7096-7107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MK, Sabapathy K. Phosphorylation at carboxyl-terminal S373 and S375 residues and 14-3-3 binding are not required for mouse p53 function. Neoplasia. 2007;9:690–698. doi: 10.1593/neo.07511. [DOI] [PMC free article] [PubMed] [Google Scholar]