Abstract
The development of techniques for oncogenomic analyses such as array comparative genomic hybridization, messenger RNA expression arrays and mutational screens have come to the fore in modern cancer research. Studies utilizing these techniques are able to highlight panels of genes that are altered in cancer. However, these candidate cancer genes must then be scrutinized to reveal whether they contribute to oncogenesis or are coincidental and non-causative. We present a computational method for the prioritization of candidate (i) proto-oncogenes and (ii) tumour suppressor genes from oncogenomic experiments. We constructed computational classifiers using different combinations of sequence and functional data including sequence conservation, protein domains and interactions, and regulatory data. We found that these classifiers are able to distinguish between known cancer genes and other human genes. Furthermore, the classifiers also discriminate candidate cancer genes from a recent mutational screen from other human genes. We provide a web-based facility through which cancer biologists may access our results and we propose computational cancer gene classification as a useful method of prioritizing candidate cancer genes identified in oncogenomic studies.
INTRODUCTION
The transformation of a normal cell into a cancer cell is a multi-step process with each intermediate stage conferring a selective advantage on the cell (1). These changes result primarily from genetic alterations to the cell's DNA, although epigenetic modifications are also important contributory factors. Normal cellular homoeostasis and division are tightly controlled processes that incorporate signals from many pathways to regulate the expression of the appropriate genes. Mutations or alterations to genes involved in these processes can contribute to cellular transformation by unbalancing the natural physiological equilibrium of a cell. Indeed, cancer progression is the accumulation of a series of genetic alterations in a somatic cell.
The genetic alterations leading to cancer occur only in certain genes. Cancer-causing genes have been traditionally classified as either proto-oncogenes or tumour suppressor genes. Proto-oncogenes normally function as proliferative agents. When mutated or misregulated in cancer, they promote uncontrolled cell growth. Usually, they are phenotypically dominant requiring a pertinent mutation or chromosomal alteration of one allele to become oncogenic. Conversely, tumour suppressor genes are endowed with anti-proliferative properties and generally require inactivation of both alleles to induce cancer. In addition to proto-oncogenes and tumour suppressor genes, more recently stability genes have been proposed as a further type of cancer gene (2).
Until now, most cancer genes have been identified by positional cloning (3). However, modern cancer research has become a hybrid of molecular and bioinformatics methods. There are many molecular techniques for the analysis of tumour samples and the identification of the causative agents therein (4). Cytogenetic methods such as karyotyping, fluorescence in situ hybridization (FISH) and comparative genome hybridization (CGH), have been used to analyse large structural chromosomal changes, gains and losses of specific genes, and genome-wide gains and losses. Over the past decade, the use of cDNA microarrays to simultaneously analyse the expression of thousands of genes in tumour samples has become prevalent in cancer research. Studies have shown that gene expression data from tumours are clinically relevant in breast cancer and lymphoma prognosis (5,6) and are able to define cancer subtypes and response to therapies (7).
The use of mutational profiling of tumour genomes has yielded important results over the past few years (8). Large-scale exon resequencing of human tumours has been used to identify point mutations in candidate cancer genes in a variety of different tumours (9–14).
Indeed, more sophisticated studies exploiting data from different techniques are becoming common in cancer research (15). Recently, a number of studies have revealed the effectiveness of integrative functional genomics in cancer research, whereby information from complementary experimental data sources are combined to provide greater insight to the process of tumourigenesis (16–21). Studies have combined data from different microarray experiments (16,17), expression and copy number change data (19,20), and expression of mRNAs and microRNAs (21).
In 2004, Futreal et al. (3) published a census of human cancer genes gleaned from published literature. Subsequent additions to the initial census of 291 genes have increased the total to over 350 genes in 2006 (htpp://http://www.sanger.ac.uk/CGP). A number of criteria were used for inclusion in the census. Only genes in which cancer-causing mutations have been reported were included. Furthermore, a requirement for two independent reports of mutations in primary clinical samples was used. Genes involved in translocation or copy-number change were included. However, genes for which there was only evidence of differential expression level evidence or aberrant promoter DNA methylation in tumours were excluded.
Many issues remain to be determined in understanding oncogenesis in different tumour types, for example elucidation of candidate causative agents, distinguishing between driver and passenger alterations (22,23) and characterization of the function of cancer genes in the oncogenic process (24). Oncogenomic experiments are now providing the cancer research community with numerous candidate causative genes. However, it is then imperative to prioritize the more promising candidates from genes that are unlikely to be contributing to tumourigenesis. A number of previous computational studies have aimed at predicting cancer-associated missense mutations (25,26). Our approach is different in that we are attempting to predict genes that are likely to be involved in cancer, irrespective of the oncogenic alteration. We envisage computational cancer gene prediction as a useful method of candidate cancer gene prioritization when allied with the results of oncogenomic experiments.
We have shown before that it is possible to develop an accurate classifier for distinguishing between Cancer Gene Census genes and other human genes (27). However, it is evident from cancer biology that altered proto-oncogenes and tumour suppressor genes promote oncogenesis in different manners. Furthermore, we have also shown that differences in sequence and regulatory properties exist between these two types of cancer genes (28). These issues have prompted us to devise separate classifiers for proto-oncogenes and tumour suppressor genes. We intend to ascertain if we can accurately distinguish between the following types of human genes: (i) proto-oncogenes and other genes, (ii) tumour suppressor genes and other genes, (iii) proto-oncogenes and tumour suppressor genes and (iv) cancer genes and Mendelian disease genes. In addition, we analyse the efficacy of our classifiers on candidate cancer genes identified in a mutational screen (14) and a recent comparative oncogenomic study (29). In this study, we include in the method a large number of different properties types, and we aim to evaluate how different sets of properties perform in these tasks. In summary, the purpose of this study is (i) to develop a method to accurately distinguish oncogenes and tumour suppressor genes from the rest of genes, (ii) to evaluate the performance of different sets of properties in this task and (iii) to assess the performance of our classifiers on candidate cancer genes. The results from our classifiers are available at http://bg.upf.edu/cgprio.
METHODS
Datasets
The list of genes involved in cancer was obtained from the Cancer Gene Census (3). Using the NCBI LocusLink database (30) and the Ensembl version 37 database (31), we located the corresponding gene sequence records. T-cell receptor loci and immunoglobulin loci were not included in the list as these genes are usually implicated in cancer by translocations of other genes downstream of the promoters of the loci (3). The result was a list of 338 genes associated with human cancer (C). All other Ensembl protein-coding genes were classified as unlabelled (UNL; n = 21 787). Therefore, the unlabelled gene dataset potentially contains cancer genes that are not included in the Cancer Gene Census as yet. Cancer genes were classified as cancer dominant (CD; n = 272) or cancer recessive (CR; n = 66) according to the Cancer Gene Census (3). Genes in which both type of mutations (dominant and recessive) have been found were not used for the study. The colon cancer (CC; n = 140) and breast cancer gene (BC; n = 140) sets were obtained from Wood et al. (14), removing any genes included in the Cancer Gene Census. Dominant (DD) and recessive (DR) disease genes were manually curated from the OMIM database (32) as described elsewhere (33).
Property sets
The different types of data were used to construct six datasets, summarized in Table 1 and detailed in Supplementary Table 1. Protein conservation score (CS) was calculated as described previously (27). Protein conservation was calculated using CS, which is an estimation of the divergence that has occurred between a pair of proteins during evolution, and is independent of the length of the proteins. The Ensembl comparison proteomes used were Homo sapiens, Mus musculus, Rattus norvegicus, Canis familiaris, Bos taurus, Monodelphis domestica, Gallus gallus, Xenopus tropicalis, Takifugu rubripes, Tetraodon nigroviridis, Danio rerio, Ciona intestinalis, Anopheles gambiae, Apis mellifera, Drosophila melanogaster, Caenorhabditis elegans and Saccharomyces cerevisiae. Gene structure information was taken from the Ensembl database (31) and consisted of coding sequence length, gene length, total exon length, total intron length and total exon number. Protein domain information comprised Interpro (34) domains present in ≥30 human proteins (n = 244 domains). Overrepresentation of protein domains in the CD and CR gene sets was conducted by generating 10 000 random sets of genes and calculating z-scores for each domain. Domains with |Z-scores| of 2.5 were included. Protein–protein interaction (PPI) data were independently analysed from a previously generated human interactome (35) and consisted of total number of protein interactions per protein and number of interactions with Cancer Gene Census proteins (3). Regulatory data used were promoter conservation, number of promoter CpG islands, 3′ UTR length and number of putative microRNA targets. This information was taken from our previous study (28), which was based on alignments of human–mouse–rat–dog orthologues (36).
Table 1.
The information types in the datasets used to construct the dominant cancer gene classifiers (Onc-C) and recessive cancer gene classifiers (TSG-C) and the number of genes in each dataset
| Dataset | Information | Total | CD | CR |
|---|---|---|---|---|
| PC–GS | Protein conservation (PC), Gene structure (GS) | 22 125 | 272 | 66 |
| PC–GS–PD | PC, GS and Protein domains (PD) | 16 300 | 259 | 62 |
| PC–GS–PI | PC, GS and Protein interaction (PI) | 14 847 | 266 | 66 |
| PC–GS–RD | PC, GS and Regulatory data (RD) | 13 928 | 238 | 58 |
| PC–GS–PD–PI–RD | PC, GS, PD, PI and RD | 11 560 | 226 | 54 |
As the protein conservation and gene structure data contain some redundant information (e.g. CS in mouse and CS in rat), a feature selection step, using an adaptation of the CFS algorithm (37), was performed on this dataset. In this step, our algorithm selects a subset of variables that have a high correlation with the class (i.e. dominant or recessive cancer) and a low correlation between each other. In the protein conservation and gene structure dataset, the variables selected by the algorithm were gene length, CS in mouse, CS in yeast and total exon length for dominant cancer genes, and coding sequence length, exon number, CS in human (i.e. conservation of paralogues) and total exon length for recessive cancer genes. Feature selection was also conducted for InterPro domains to reduce the number of variables to be used for prediction. Therefore, the base for each dataset was the selected variables from the relevant protein conservation and gene structure datasets plus (i) selected InterPro domains, (ii) the protein–protein interaction data, (iii) the regulatory data and (iv) selected InterPro domains, protein–protein interaction data and regulatory data.
Prediction
Averaged positive naive Bayes
The algorithm selected for the prediction is the averaged positive naive Bayes [APNB, (38)]. This algorithm is based on the positive naive Bayes (39), which takes as input a set of positive and unlabelled instances and outputs a naive Bayes predictor (40). In order to induce the model the a priori probability of the positive class (that is, the probability that a given instance, in our case a gene, is positive, which is cancer related) is needed. This probability cannot be estimated from the dataset if no negative examples are available and, thus, we have to set it. We set the parameters of the algorithm to result in an average probability of 0.1, as it has been suggested that up to 10% of human genes could be involved in cancer (41). Setting the parameters to result in an average probability of 0.01 or 0.05 produces very similar results (data not shown). The APNB algorithm takes into account all the possible values this parameter can take, weighting each value according to a probability distribution.
In order to compute the enrichment obtained with a given classifier, we need to obtain, for the positive cases, an accurate estimation of the probability of being positive. If we simply learn the classifier from all the instances and then we use it to predict the probability of being positive for the known cancer genes, we will get an optimistically high probability (and thus an optimistically high enrichment) because we have used a these examples to train the classifier. In order to avoid this over-fitting, we have used a cross-validation scheme to estimate the probabilities of the positive cases. We convert the probabilities assigned to genes by a classifier to rank probabilities by ranking the probabilities in ascending order and dividing each rank (R) by the total number of genes (N), where the rank probability (RP) of a gene is calculated by RPi = Ri/N. In this way, the most likely candidate cancer gene has a ranking of 1 (in the absence of ties) and the least likely candidate has a ranking of 1/N (in the absence of ties).
Implementation
For prediction purposes each dataset was further divided into two sets of two distinct subsets: (i) positive dominant (containing the dominant cancer genes present in the relevant dataset; Table 1, column 4) and unlabelled dominant (containing the remainder of the genes in the dataset), and (ii) positive recessive (containing the recessive cancer genes present in the relevant dataset; Table 1, column 5) and unlabelled recessive (containing the remainder of the genes in the dataset). Recessive cancer genes are denoted as unlabelled dominant for prediction of dominant cancer genes and vice versa. A dominant and recessive prediction was conducted 10 times for each dataset with cross-validation (10-fold for dominant, 5-fold for recessive due to low sample size) (38).
Application to random gene sets
In addition, we generated 100 positive sets of randomly chosen genes from each dataset with the remainder of the genes being treated as unlabelled instances. For each of the 100 positive sets, a classifier was built and applied in an identical manner to the proto-oncogene and tumour suppressor gene classifiers. The performance of each classifier was assessed on the random positive and unlabelled datasets, and on the proto-oncogene and tumour suppressor genes.
RESULTS
Constructing the classifiers
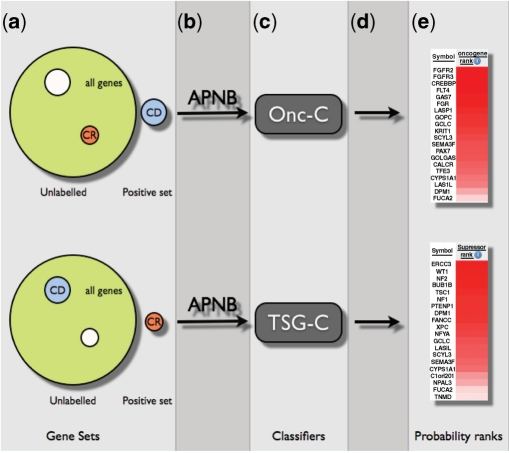
The set of properties that follow different trends in the group of proto-oncogenes and tumour suppressor genes compared to the rest of genes can be used to build a classifier that will rank all the genes with their probability of being an oncogene or a tumour suppressor gene. Since in this case there is no reliable set of negative examples available, as it is not possible to ensure that a given gene is not related to cancer, we have approached this task as a partially supervised classification problem. In order to assess how different combinations of properties are able to identify proto-oncogenes and tumour suppressor genes, we have built five different classifiers, each with a different set of properties (Table 1) to rank proto-oncogenes (Onc-C, for oncogenes classifiers) and five classifiers to rank tumour suppressor genes (TSG-C, for tumour suppressor genes classifiers) and have assessed the performance of each of them in this predictive task (Figure 1). For the purpose of proto-oncogene classification, the set of positive genes is the 272 genes annotated as cancer dominant in the Cancer Gene Census list, while the rest of human genes are considered as unlabelled. Similarly, in the case of tumour suppressor genes, the set of positive genes comprises the 66 genes annotated as cancer recessive in the Cancer Gene Census and the remainder of human genes are considered unlabelled. In total, we have constructed 10 classifiers and each of these provides us with a probability of each gene being an oncogene or a tumour suppressor gene. These probabilities have been ranked for assessment and comparison (see Methods section), and the ranked probabilities can be accessed through the following URL (http://bg.upf.edu/cgprio) and through CARGO (42).
Figure 1.
Schematic representation of the process to construct the classifiers and obtain the probability ranks. (a) The set genes labelled as CD are used as the positive set for the Onc-C and the rest as unlabelled. For the TSG-C, the set of CR genes are used as the positive set and the rest as unlabelled. (b) Next, the Averaged Positive Naïve Bayes method is applied to these sets with the corresponding property sets (PC–GS, PC–GS–PI, etc.). (c) The classifiers are obtained and applied to all genes (d) in order to obtain a probability rank for each human gene (e).
Assessing the different classifiers
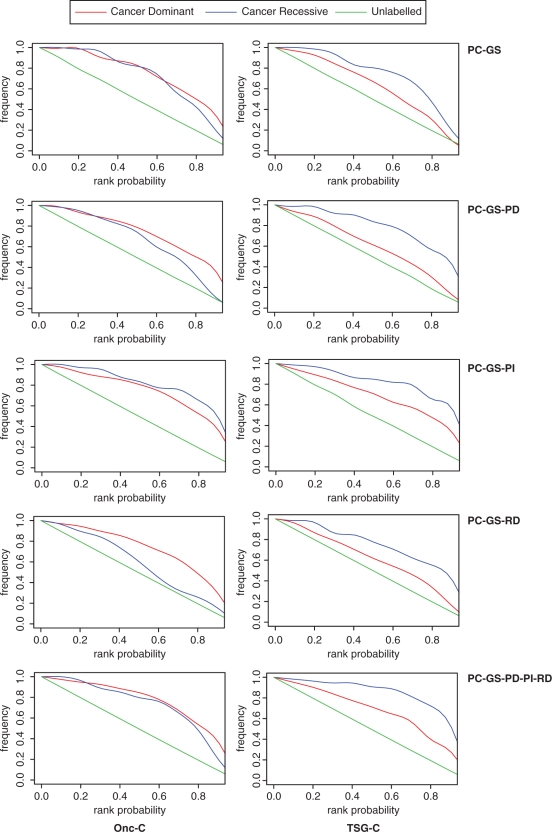
In order to assess the ability of each of the classifiers to distinguish between proto-oncogenes and the rest of genes we have compared the ranked probabilities given by the classifiers in different sets of genes (Figure 1, Table 2). The five sets of properties assign significantly higher ranked probabilities to the set of positive genes (CD or CR) than to the unlabelled genes (Figure 1). The classifiers that discriminate better between cancer (CD or CR) and unlabelled genes are the ones that include more data (PC–GS–PD–PI–RD: protein conservation—gene sequence—protein domains—protein interactions—regulatory data) (Figure 2). The mean ranked probability for oncogenes (CD) of the Onc-C is 0.75, compared to the 0.49 of the unlabelled genes (Table 2) and the mean probability of TSG-C for the recessive cancer gene classifier is 0.83 compared to 0.50 of the unlabelled genes (Table 2). However, these classifiers can give a rank probability only to those genes for which we have all the information (11 560 genes). On the other hand, the classifiers that use only protein conservation and gene sequence data (PC–GS) can rank all human genes (n = 22 125), as we have this information for all of them. This classifier, even using very simple data, can distinguish the set of CD genes fairly well (mean probability for CD is 0.73 compared to 0.50 for unlabelled and 0.71 for CR compared to 0.50 for unlabelled).
Table 2.
Mean ranked probabilities obtained by the dominant cancer gene classifier (Onc-C) and the recessive cancer gene classifier (TSG-C) for different sets of genes generated using different property sets
| Onc-C |
TSG-C |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unlabelled | CD | CR | DD | DR | BC | CC | NonCAN | Unlabelled | CD | CR | DD | DR | BC | CC | NonCAN | |
| PC–GS | 0.50 | 0.73 | 0.71 | 0.63 | 0.63 | 0.78 | 0.78 | 0.69 | 0.50 | 0.61 | 0.71 | 0.56 | 0.62 | 0.75 | 0.74 | 0.64 |
| PC–GS–PD | 0.50 | 0.72 | 0.65 | 0.57 | 0.55 | 0.73 | 0.73 | 0.63 | 0.50 | 0.59 | 0.76 | 0.53 | 0.62 | 0.76 | 0.74 | 0.63 |
| PC–GS–PI | 0.49 | 0.73 | 0.78 | 0.60 | 0.55 | 0.73 | 0.74 | 0.63 | 0.50 | 0.67 | 0.80 | 0.58 | 0.59 | 0.76 | 0.75 | 0.64 |
| PC–GS–RD | 0.50 | 0.71 | 0.58 | 0.58 | 0.46 | 0.69 | 0.76 | 0.61 | 0.50 | 0.59 | 0.73 | 0.52 | 0.58 | 0.75 | 0.73 | 0.62 |
| PC–GS–PD–PI–RD | 0.49 | 0.75 | 0.71 | 0.59 | 0.46 | 0.70 | 0.74 | 0.61 | 0.50 | 0.66 | 0.83 | 0.55 | 0.56 | 0.73 | 0.71 | 0.62 |
CD = cancer dominant, CR = cancer recessive, DD = disease dominant, DR = disease recessive, BC = breast cancer candidate genes, CC = colon cancer candidate genes, NonCAN = genes with mutations that are not candidate cancer genes.
Cancer dominant and cancer recessive coloumns are set in bold.
Figure 2.
Distribution of ranked probabilities for Cancer Gene Census dominant and recessive cancer genes and unlabelled genes (the rest of human genes) using different datasets for proto-oncogenes (left) and tumour suppressor gene (right) classifiers.
Another important question to take into account is the ability of the classifiers to distinguish between CD and CR genes. The Onc-C that uses all data (PC–GS–PD–PI–RD) cannot distinguish between CD and CR genes, as it gives high probabilities to both (mean RP of 0.75 for CD and 0.71 for CR); however, the TSG-C with the same data can distinguish the two group significantly well (mean RP of 0.83 for CR and 0.66 for CD) (Figure 2 and Table 2). Similarly, the Onc-C that uses the PC–GS–PI property set assigns high probabilities to both CD and CR genes (mean RP 0.73 and 0.78, respectively).
Finally, we also compared the probabilities assigned to Mendelian disease genes classified as dominant (DD) or recessive (DR). Some of the properties used to build the classifiers were previously found to follow specific trends in hereditary disease genes, such as conservation and sequence properties (43,44). Thus, it is important to assess if the classifiers are able to distinguish between cancer and disease genes or, alternatively, are just predicting genes that are more prone to random mutations and that may cause any disease. We observe that all classifiers assign higher probabilities to CD and CR genes than to disease genes (DD and DR). Moreover, the classifiers that include data that are more relevant (PC–GS–PD–PI–RD) seem to distinguish CD and CR genes from disease genes more successfully (Table 2, Supplementary Figure 1).
Validating the prediction methods with classifiers created with random positive sets
To further validate the performance of the methods, we constructed predictions with random positive sets of genes for each classifier (see Methods section for details). In each of these random classifiers, the positive set is a group of genes taken randomly from all human genes containing the same number of genes as the CD set or the CR set (Table 1), and the classifiers are built with the same sets of properties. For each of the 10 classifiers, we built 100 classifiers each with a different random set of positive genes and we averaged the results. With this procedure, we can examine two issues: (i) whether with these five different property sets it is possible to create a classifier that would predict any positive set of genes; thus, that it is not just a unique characteristic of cancer genes and (ii) the ability of these random classifiers to predict cancer genes.
We first analysed the ranked probabilities that each random classifier gives to the positive set. The probabilities obtained for the positive sets are not significantly different to the unlabelled set of genes (Table 3), and the SDs of the ranked probabilities of the 100 random positive sets are low (0.02–0.06), thus indicating that these sets of properties are not able to predict any set of genes. Next, we studied the ranked probabilities given by the random classifiers to the set of CD and CR. In general, these probabilities are low, but interestingly, by using some property sets the predictions are higher for CD and CR even though the classifiers were not constructed with cancer genes as positive sets. This may indicate that there is a bias in those properties with regard to the set of cancer genes. The datasets exhibiting this behaviour are those that include protein interactions and protein domains. We analysed the number of annotations for CD, CR and the rest of genes for these properties and we see that CD and CR genes have in general more annotated protein interactions and more annotated protein domains (data not shown).
Table 3.
Mean ranked probabilities and standard deviation for one hundred positive and unlabelled randomly selected datasets using the variables from the dominant cancer gene classifier (Onc-C) and the recessive cancer gene classifier (TSG-C). Means of the cancer dominant and cancer recessive sets ranked probabilities from the random classifiers are included
| Positive mean | Positive σ | Unlabelled mean | Unlabelled σ | CD mean | CR mean | |
|---|---|---|---|---|---|---|
| Onc-C | ||||||
| PC–GS | 0.49 | 0.03 | 0.50 | 0.00 | 0.51 | 0.52 |
| PC–GS–PD | 0.49 | 0.03 | 0.50 | 0.00 | 0.55 | 0.48 |
| PC–GS–PI | 0.50 | 0.02 | 0.50 | 0.00 | 0.50 | 0.51 |
| PC–GS–RD | 0.49 | 0.03 | 0.50 | 0.00 | 0.48 | 0.50 |
| PC–GS–PD–PI–RD | 0.50 | 0.03 | 0.50 | 0.00 | 0.56 | 0.53 |
| TSG-C | ||||||
| PC–GS | 0.48 | 0.06 | 0.50 | 0.00 | 0.50 | 0.50 |
| PC–GS–PD | 0.47 | 0.06 | 0.50 | 0.00 | 0.51 | 0.57 |
| PC–GS–PI | 0.48 | 0.06 | 0.50 | 0.00 | 0.53 | 0.55 |
| PC–GS–RD | 0.48 | 0.06 | 0.50 | 0.00 | 0.52 | 0.51 |
| PC–GS–PD–PI–RD | 0.49 | 0.05 | 0.50 | 0.00 | 0.54 | 0.61 |
Application of prediction to mutational screens of tumours
To investigate if our techniques are capable of identifying candidate cancer genes from other studies, we have applied our prediction methods to the results of a mutational screen of human breast and colon tumours (11,14). This study analysed transcripts representing >18 000 genes and found at least one non-silent mutation in 1718 genes in either a breast or colon tumour sample. To distinguish between driver and passenger mutations they developed a statistical technique, which culminated in the identification of a set of 140 colon cancer candidate genes and a set of 140 breast cancer candidate genes. These two sets of genes (colon, n = 113 and breast, n = 117, having removed Cancer Gene Census genes) are systematically ranked higher with all the classifiers than unlabelled genes or disease genes (Table 2). Interestingly, the sets of breast and colon cancer candidate genes are ranked even higher than cancer genes with the simplest classifier (PC–GS) indicating that this group of genes identified by Sjoblom et al. (11) have very similar conservation and sequence properties to the genes in the Cancer Gene Census (28).
In total, Wood et al. found non-silent mutations in 1718 genes. They developed a statistical technique, using an empirical Bayes analysis, to ascertain if the mutations in a candidate gene reflect a mutation rate that is greater than the passenger rate (14). Applying this procedure, they discriminated between the 280 candidate cancer genes (CAN genes) and the remainder of the candidates (non-CAN genes). In Table 2, we show that our classifiers assign lower mean rank probabilities to these non-CAN genes than to the CAN genes prioritized in the original study (Table 2 and Supplementary Table 2).
Using the predictions
In addition, we apply our method to the results of a comparative oncogenomic study which identified a gene involved in metastasis in melanoma (29). This study characterized an amplification in a mouse melanoma model syntenic to human chromosome 6p25-24, and with expression analysis identified Nedd9 as the primary candidate metastasis gene. If we examine the rank probabilities from our classifiers for human chromosomal band 6p25-24 (92 genes), we consistently see Nedd9 as one of the top candidates (Supplementary Table 3 and http://bg.upf.edu/cgprio). Furthermore, all Onc-C assign the gene a much higher ranked probability than the equivalent TSG-C (Supplementary Table 4).
DISCUSSION
The purpose of this study is to provide an aid to biologists conducting oncogenomic experiments that result in a large number of candidate cancer genes. We envisage that the necessary prioritization of some candidate genes (driver genes) over others (passengers) (22) is a task that can be facilitated by our study, in conjunction with experimental evidence. The success of integrative approaches in the study of cancer has underscored the utility of blending different interrogative techniques to produce a more thorough analysis of cancer states. Significant results have been achieved recently by the combination of comparative genomic hybridization and expression arrays (22,29,45). Furthermore, it is evident from the variety of potential oncogenic events, such as coding and non-coding point mutations, copy number variations, translocations and epigenetic alterations, that to obtain a more complete view of the underlying causes of tumourigenesis it is imperative that different types of experimental results are combined. We propose our method of computational cancer gene prediction as a different, yet complementary, approach to the prioritization of candidate cancer genes.
Our method is based on a number of salient features: (i) we have used gene and protein properties that are likely to contribute to a gene's potentiality to be oncogenic, (ii) we have attempted to use relatively unbiased data and (iii) in the case of the classifiers using protein conservation and gene structure, we can apply them in a genome-wide manner. We have previously shown that greater gene length and protein conservation are indicative of genes in the Cancer Gene Census (27). Indeed, these simple types of sequence properties are quite successful at separating cancer genes from other genes, in general, but obviously share similar patterns in this regard with Mendelian disease genes (Figure 2 and Table 2) (43). The original analysis of the Cancer Gene Census genes included a protein domain analysis, showing an over-representation of kinase and DNA-binding domains, amongst others (3). However, human proteins in total contain more than 4900 different InterPro protein domains (34), hence it was necessary to reduce this number significantly for the purpose of prediction. We used nine domains for the proto-oncogene classifier and 10 for the tumour suppressor classifier (Supplementary Table 1; see Methods section for details). These data help to distinguish between the cancer genes and disease genes, and also between dominant and recessive cancer genes (Figure 2 and Table 2).
Recently, it has been shown that Cancer Gene Census proteins participate in more PPIs than other genes (35). We have used these data to calculate two PPI variables, total number of PPIs of each human protein and total number of PPIs with Cancer Gene Census proteins, based on the premise that proteins that interact with cancer proteins are more likely to be candidate cancer proteins. Tumour suppressor genes have more PPIs than proto-oncogenes (28) and this is highlighted by the fact that both classifiers predict tumour suppressor genes better than any other type of gene (Figure 2 and Table 2).
We have also included pertinent regulatory data in this study, as we have previously described differences between cancer genes, in particular proto-oncogenes, and other genes in terms of proximal promoter conservation, number of CpG islands, 3′ UTR length and frequency of 3′ UTR putative microRNA targets (28). In particular, we felt it appropriate to include evidence of the existence of miRNA targets due to the mounting body of experimental and computational work supporting a significant role for miRNAs in tumourigenesis and metastasis (21,46–49). The addition of these data helps to significantly distinguish both proto-oncogenes and tumour suppressor genes from each other, and from disease genes (Figure 2 and Table 2). Our final dataset amalgamates all of the previous data types (PC and GS, PDs, PPIs and RD). While both the proto-oncogene and TSG-C are able to distinguish between cancer genes and both unlabelled genes and disease genes, only the tumour suppressor gene classifier differentiates between the two groups of cancer genes (Figure 2 and Table 2).
The method we describe is only useful if it can assist in the detection of previously unidentified cancer genes. We have used a set of candidate cancer genes from a mutational screen of breast and colon cancers to test our classifiers (11,14). These two sets of candidate genes, breast and colon, were included as unlabelled genes in our classifiers. In all classifiers, both the breast and colon candidate genes obtain higher average ranked probabilities than unlabelled genes, disease genes and in some cases Cancer Gene Census genes (Table 2). This is in part due to similarities between the breast and colon candidate genes Cancer Gene Census genes in gene structure and sequence conservation, as we have shown previously (28). Nevertheless, these candidate genes were found using an unbiased genome-wide screen of solid tumours in a method completely different to the discovery of most of the Cancer Gene Census genes, which were mainly detected as translocations in haematological disorders (3).
In addition, we apply our method to the results of a comparative oncogenomic study which, through array CGH and expression studies, identified the Nedd9 gene as an oncogene involved in metastasis in melanoma (29). Nedd9 is consistently ranked by our proto-oncogene classifiers as one of the top candidates in the chromosomal band in which it resides (Supplementary Table 3 and website). Other genes are ranked more highly than Nedd9 in this region by our classifiers, some of which would also be viable candidate cancer genes such as male germ cell associated kinase (a protein kinase) in different tumour types. At present our classifiers are not sufficiently sophisticated to be able to differentiate tumour-specific candidate cancer genes.
We acknowledge that even though our methods work well with the examples provided there are some limitations to the classifiers. For example, as proto-oncogenes and tumour suppressor genes share some sequence properties in general, such as higher PC, longer gene and protein sequences (27), and greater number of PPIs (28), the classifiers relying on these properties do not discriminate well between the two types of cancer genes. In addition, the use of protein domain information and to a lesser extent PPIs (and only in the case of tumour suppressor genes) appear to bias our methodology when we conduct simulations with random sets of positive genes (Table 3). In terms of PPI data, we have only incorporated the data from one study (35) and although it has been reported that human PPI maps generated from different sources only have a small overlap, it has shown to be statistically significant (50). We previously included Gene Ontology annotations (51) in a cancer gene prediction tool (27). However, as many of these are based on protein domain information included in InterPro and classifiers based on Gene Ontology annotations perform similarly to those using InterPro data (data not shown), we excluded this type of information from the present study.
Most oncogenomic experimental techniques will produce a number of false positives, and it is this very point that emphasizes the need for a combinatorial approach in cancer biology. Furthermore, with large cancer biology initiatives such as The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov) and the Cancer Genome Project (www.sanger.ac.uk/genetics/CGP) producing vast amounts of data for the foreseeable future, the need for complementary approaches for candidate cancer gene prioritization is clear. To this end, we have made all our proto-oncogene and tumour suppressor gene predictions available at http://bg.upf.edu/cgprio. Researchers are able to search for a particular gene or upload a list of genes, or search by chromosomal location. As we have included all 10 (five proto-oncogene and five tumour suppressor gene) classifiers, a researcher may choose the predictions according to the data types used to construct the classifier. In summary, it is envisaged that our classifications will be a useful prioritization aid to experimental cancer biologists in combination with experimentally derived results.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We acknowledge funding from the International Human Frontier Science Program Organization (HFSPO) and from the Spanish Ministerio de Educación y Ciencia grant number SAF2006-0459. N.L.-B. is recipient of a Ramón y Cajal contract of the Spanish Ministerio de Educación y Ciencia (MEC) and acknowledges support from Instituto Nacional de Bioinformática. Funding to pay the Open Access publication charges for this article was provided by the Ministry of the Spanish Ministerio de Educación y Ciencia grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 3.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baak JP, Janssen EA, Soreide K, Heikkilae R. Genomics and proteomics–the way forward. Ann. Oncol. 2005;16(Suppl. 2):i30–i44. doi: 10.1093/annonc/mdi728. [DOI] [PubMed] [Google Scholar]
- 5.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N. Engl. J. Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 6.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Ramaswamy S, Golub TR. DNA microarrays in clinical oncology. J. Clin. Oncol. 2002;20:1932–1941. doi: 10.1200/JCO.2002.20.7.1932. [DOI] [PubMed] [Google Scholar]
- 8.Benvenuti S, Arena S, Bardelli A. Identification of cancer genes by mutational profiling of tumor genomes. FEBS Lett. 2005;579:1884–1890. doi: 10.1016/j.febslet.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Stephens P, Hunter C, Bignell G, Edkins S, Davies H, Teague J, Stevens C, O’Meara S, Smith R, Parker A, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 10.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 11.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 12.Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 13.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 15.Liu ET, Kuznetsov VA, Miller LD. In the pursuit of complexity: systems medicine in cancer biology. Cancer Cell. 2006;9:245–247. doi: 10.1016/j.ccr.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl Acad. Sci. USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nat. Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 18.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 19.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 20.Stransky N, Vallot C, Reyal F, Bernard-Pierrot I, de Medina SG, Segraves R, de Rycke Y, Elvin P, Cassidy A, Spraggon C, et al. Regional copy number-independent deregulation of transcription in cancer. Nat. Genet. 2006;38:1386–1396. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 22.Haber DA, Settleman J. Cancer: drivers and passengers. Nature. 2007;446:145–146. doi: 10.1038/446145a. [DOI] [PubMed] [Google Scholar]
- 23.Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35:D721–D726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu P, Bader G, Wigle DA, Emili A. Computational prediction of cancer-gene function. Nat. Rev. Cancer. 2007;7:23–34. doi: 10.1038/nrc2036. [DOI] [PubMed] [Google Scholar]
- 25.Kaminker JS, Zhang Y, Watanabe C, Zhang Z. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 2007;35:W595–W598. doi: 10.1093/nar/gkm405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaminker JS, Zhang Y, Waugh A, Haverty PM, Peters B, Sebisanovic D, Stinson J, Forrest WF, Bazan JF, Seshagiri S, et al. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 2007;67:465–473. doi: 10.1158/0008-5472.CAN-06-1736. [DOI] [PubMed] [Google Scholar]
- 27.Furney SJ, Higgins DG, Ouzounis CA, Lopez-Bigas N. Structural and functional properties of genes involved in human cancer. BMC Genomics. 2006;7:3. doi: 10.1186/1471-2164-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furney SJ, Madden SF, Higgins DG, Lopez-Bigas N. Distinct patterns in the regulation and evolution of human cancer genes. In Silico Biol. 2008;8:33–46. [PubMed] [Google Scholar]
- 29.Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKusick VA. Baltimore, MD: Johns Hopkins University Press; 1998. Mendelian Inheritance in Man. A Catalog of Human Genes and Genetic Disorders. [Google Scholar]
- 33.Lopez-Bigas N, Blencowe BJ, Ouzounis CA. Highly consistent patterns for inherited human diseases at the molecular level. Bioinformatics. 2006;22:269–277. doi: 10.1093/bioinformatics/bti781. [DOI] [PubMed] [Google Scholar]
- 34.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall M, Smith L. Proceedings of the International Conference on Neural Information Processing and Intelligent Information Systems. Springer, Singapore, pp. Springer; 1997. pp. 855–858. [Google Scholar]
- 38.Calvo B, Larrañaga P, Lozano J. Learning Bayesian classifiers from positive and unlabeled examples. Pattern Recogn. Lett. 2007;28:2375–2384. [Google Scholar]
- 39.Denis F, Gilleron R, Tommasi M. The 9th International Conference on Information Processing and Management of Uncertainty in Knowledge-Based Systems, IPMU 2002. France: Annecypp; 2002. Text classification from positive and unlabeled examples. pp. 1927–1934. [Google Scholar]
- 40.Minsky M. Steps toward artificial intelligence. Proc. Inst. Radio Eng. 1961;49:8–30. [Google Scholar]
- 41.Strausberg RL, Simpson AJ, Wooster R. Sequence-based cancer genomics: progress, lessons and opportunities. Nat. Rev. Genet. 2003;4:409–418. doi: 10.1038/nrg1085. [DOI] [PubMed] [Google Scholar]
- 42.Cases I, Pisano DG, Andres E, Carro A, Fernandez JM, Gomez-Lopez G, Rodriguez JM, Vera JF, Valencia A, Rojas AM. CARGO: a web portal to integrate customized biological information. Nucleic Acids Res. 2007;35:W16–W20. doi: 10.1093/nar/gkm280. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez-Bigas N, Ouzounis CA. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 2004;32:3108–3114. doi: 10.1093/nar/gkh605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furney SJ, Alba MM, Lopez-Bigas N. Differences in the evolutionary history of disease genes affected by dominant or recessive mutations. BMC Genomics. 2006;7:165. doi: 10.1186/1471-2164-7-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 47.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comput. Biol. 2007;3:e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gusev Y, Schmittgen TD, Lerner M, Postier R, Brackett D. Computational analysis of biological functions and pathways collectively targeted by co-expressed microRNAs in cancer. BMC Bioinformatics. 2007;8(Suppl. 7):S16. doi: 10.1186/1471-2105-8-S7-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Futschik ME, Chaurasia G, Herzel H. Comparison of human protein-protein interaction maps. Bioinformatics. 2007;23:605–611. doi: 10.1093/bioinformatics/btl683. [DOI] [PubMed] [Google Scholar]
- 51.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.