Abstract
In flowering plants, the male gametophyte, the pollen, develops in the anther. Complex patterns of gene expression in both the gametophytic and sporophytic tissues of the anther regulate this process. The gene expression profiles of the microspore/pollen and the sporophytic tapetum are of particular interest. In this study, a microarray technique combined with laser microdissection (44K LM-microarray) was developed and used to characterize separately the transcriptomes of the microspore/pollen and tapetum in rice. Expression profiles of 11 known tapetum specific-genes were consistent with previous reports. Based on their spatial and temporal expression patterns, 140 genes which had been previously defined as anther specific were further classified as male gametophyte specific (71 genes, 51%), tapetum-specific (seven genes, 5%) or expressed in both male gametophyte and tapetum (62 genes, 44%). These results indicate that the 44K LM-microarray is a reliable tool to analyze the gene expression profiles of two important cell types in the anther, the microspore/pollen and tapetum.
Keywords: Anther, Laser microdissection, Microarray, Oryza sativa, L. Microspore/pollen, Tapetum
Introduction
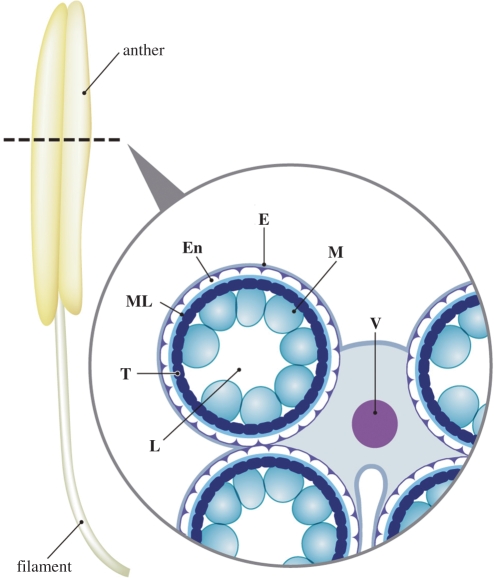
In flowering plants, the male gametophyte, the pollen, develops in the anther. The free pollen cells are released in the anther locule, whose wall consists of the tapetum, middle layer, endothecium and epidermis (Fig. 1). Development of pollen cells in the locule is synchronized with functional anther development (Goldberg et al. 1993, Scott et al. 2004). After differentiation of the male germline, pollen mother cells undergo meiosis to form tetrads of haploid microspores in the anther locule. These microspores then mature into pollen grains through cell division and formation of the complex pollen wall. Through an asymmetric mitosis, the uninuclear microspore develops into bicellular pollen comprising a larger vegetative cell and a smaller generative cell (McCormick 1993, McCormick 2004). In plant species with tricellular pollen (including Arabidopsis and rice), the generative cell then undergoes a second mitosis to form two sperm cells. During pollen maturation, the tapetum acts as a nutritive tissue, providing materials for pollen wall formation, and subsequently disintegrates in the later stages of pollen development (Goldberg et al. 1993, Scott et al. 2004).
Fig. 1.
Scheme of a cross-section of a rice anther containing immature microspores. L, locule; T, tapetum; ML, middle layer; En, endothecium; E, epidermis; M, microspores; V, vascular bundle.
The important biological events described above are regulated by complex patterns of gene expression in both the gametophytic and sporophytic tissues during anther and male gametophyte development (McCormick 2004, Scott et al. 2004). Gene expression in the anther has been studied intensively in important crops and model plants by using conventional cDNA cloning, promoter analysis and microarrays (Koltunow et al. 1990, Scott et al. 1991, Tsuchiya et al. 1994, Hihara et al. 1996, Rubinelli et al. 1998, Jeon et al. 1999, Endo et al. 2002, Amagai et al. 2003, Masuko et al. 2006). In particular, genes specifically expressed in the tapetum have attracted the attention of phytologists because the tapetum exhibits high metabolic activity (Scott et al. 2004) and is related to both male sterility (Wilson et al. 2001, Sorensen et al. 2003, Yui et al. 2003, Ariizumi et al. 2004, Yang et al. 2007) and sporophytic self-incompatibility (Takayama et al. 2000, Watanabe et al. 2001), indicating a critical role for the tapetum in pollen development and maturation.
Anther transcriptomes analyzed by microarrays offer important information for understanding the genetic regulation of anther and pollen development. Of particular scientific interest are the gene expression profiles of two important cell types within the anther: the microspore/pollen and tapetum. However, most of the gene expression analyses conducted to date have employed whole anther tissue taken at different developmental stages (Endo et al. 2002, Mandaokar et al. 2003, Endo et al. 2004, Lan et al. 2004, Yamaguchi et al. 2004, Jung et al. 2005, Wang et al. 2005, Ma et al. 2006) or mature pollen grains (Honys and Twell 2003), and have not distinguished the cell types within the anther. Microarray technology is able to explore a large number of genes whose transcript levels change between states or development stages. However, sufficient specific tissue must be available in order to isolate enough mRNA to conduct the hybridization, typically 1–2 μg (Duggan et al. 1999, Richmond and Somerville 2000). Furthermore, it can be technically difficult to isolate single cell types from plant organs. Thus, most microarray studies have been conducted using RNAs extracted from a mixture of tissues and/or cell types (Girke et al. 2000). Together these technical challenges have made it impossible to analyze a large number of gene expression profiles in precisely separated cell types with high resolution. Honys and Twell (2004) established stage-separated spore isolation procedures for Arabidopsis, and performed transcriptome analyses of male gametophyte development. In brief, inflorescences were ground in mannitol, and the mixture filtered through nylon mesh. By centrifugation of the filtered microspores in a Percoll step gradient, uninucleate microspores, bicellular pollen and immature tricellular pollen were discriminated and isolated. The haploid male gametophyte-specific transcriptome revealed large-scale repression of various genes and an increase in the proportion of male gametophyte-specific transcripts during pollen maturation (Honys and Twell 2004). Although comparison of the successful male gametophyte-specific transcriptome with the tapetum-specific transcriptome would undoubtedly be informative, it has not been possible to date because of the technical challenge of precise isolation of tapetal cells, which are attached to the middle layer of the anther wall.
Laser microdissection (LM) is a powerful tool for isolating specific cell types from sectioned specimens of heterogeneous tissues, including plant tissues (Asano et al. 2002, Kerk et al. 2003, Nakazono et al. 2003, Day et al. 2005, Nelson et al. 2006, Ohtsu et al. 2007b). A tissue section that contains the cell type of interest is placed on a microscope stage, a laser beam is targeted on the cells of interest and then the target cells are separated from the rest of the tissue. In general, there are two types of LM methodology: laser capture and laser cutting (Ohtsu et al. 2007b). Laser capture reliably targets cells for collection but sometimes also collects surrounding cells that remain attached to the target cells. On the other hand, laser cutting minimizes the collection of non-target cells, but is subject to interference by various factors such as static electricity when the target cells are very small. However, a newly available LM system (Veritas Laser Microdissection System, Molecular Devices) can compensate for the disadvantages of laser capture and laser cutting by combining the two methods, thereby providing LM with higher precision and efficiency. Recently, many LM-microarray analyses have been conducted in plants, demonstrating wide applicability of the technology. Successfully targeted plant cell types include embryos (Casson et al. 2005, Spencer et al. 2007), coleoptile epidermis and vascular tissues (Nakazono et al. 2003), shoot apical meristems (Ohtsu et al. 2007a, Zhang et al. 2007), root pericycles (Woll et al. 2005, Dembinsky et al. 2007), silique replums (Cai and Lashbrook 2006) and stamen abscission zones (Cai and Lashbrook 2008).
In this report, and the following articles in the same issue (Hobo et al. 2008, Hirano et al. 2008), the LM-microarray approach has been used to characterize the two separated transcriptomes of the microspore/pollen and tapetum in rice. By using LM technology, the tapetal cell layers were collected with minimal contamination by other anther wall tissues or microspore/pollen. With minimum input of labeled cDNAs from the precisely separated rice microspore/pollen and tapetal cells, a 44K microarray analysis was achieved. We were able to validate the LM-microarray developed in this study by comparing our data with the results of a 4,304 cDNA array (4K array) previously published by Endo et al. (2004), which identified 156 genes specifically expressed in rice anthers, some of which were confirmed by RNA in situ hybridization.
Results and Discussion
Isolation of male gametophyte- and tapetum-specific RNAs
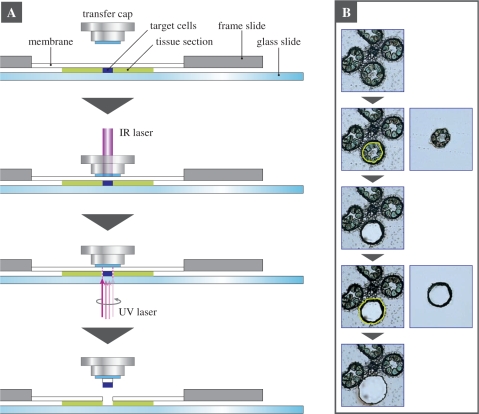
In order to perform the LM-microarray of microspore/pollen and tapetum in rice, we prepared cross-sections of Oryza sativa cv. Nipponbare anthers at various developmental stages. In rice, anther development has been precisely classified into five stages (from meiosis to tricellular pollen), according to glume length and cell numbers in the male gametophytes. These are meiosis (MEI), tetrad (TET), uninuclear microspore (UN), bicellular pollen (BC) and tricellular pollen (TC) (stages described as An4 to An8 in Itoh et al. 2005). We isolated microspore/pollen from each of the five stages. Because degradation of the tapetum begins at the BC stage, tapetum cells could only be isolated from MEI, TET and UN anthers. Using the Veritas Laser Microdissection System (Molecular Devices; Fig. 2A), the microspore/pollen or tapetal cells from each developmental stage were isolated from the anther sections. In the case of MEI, TET and UN anthers, the microspores were isolated first, followed by the tapetum; Fig. 2B.
Fig. 2.
Laser microdissection (LM) of pollen/microspores and tapetum cells from a cross-section of a rice anther. (A) A schematic procedure for isolating plant target cells by laser microdissection using the Veritas Laser Microdissection System (Molecular Devices), which can use both laser capture and laser cutting. In brief, a transfer cap is placed on a tissue section, which contains the target cells, mounted on a membrane of a frame slide. To anchor and capture the target cells to the transfer cap, a focused low energy infrared (IR) laser beam is targeted on the cells of interest through the transfer cap, causing the cap to fuse to the target cells. The area around the target cells is cut by a UV laser beam. The transfer cap is removed from the tissue section. The laser-microdissected target cells that fused to the transfer cap are separated from the rest of the tissue. (B) Isolation of the pollen/microspore and tapetum cells by the two-step LM. In the case of the TET anther, the microspores were isolated by the first cutting, and the tapetum cells were isolated by the second cutting.
Total RNAs were isolated from the cells dissected from 250–400 sections, corresponding to anthers of four or five spikelets, and were subjected to the 44K rice oligoarrays with 44,000 features of 60mer oligonucleotides (Agilent Technologies), corresponding to the full-length cDNAs characterized by the Rice Annotation Project (RAP). For each microarray experiment, an average 12.6 ng (minimum 2.2 ng) of total RNAs were used for Cy3 labeling, for complementary RNA (cRNA) synthesis and amplification, according to the Agilent protocol with slight modification. Labeled cRNAs of average 660 ng (minimum 277 ng) were then used to perform the one-color microarray analysis. The microarray experiment was repeated at least three times in each stage/cell type using RNAs isolated independently. The significance of this study is the isolation of microspore/pollen- and tapetum-specific RNAs in sufficient quantity and quality for microarray analysis, making it possible to compare the male gametophyte and tapetum transcriptomes (see Hobo et al. 2008, Hirano et al. 2008 in this issue).
Gene expression profiles of reported genes in the 44K LM microarray
We focused on validating the 44K LM-microarray by re-characterizing genes previously reported as tapetum specific (see below for detail) and/or reported in a 4K microarray analysis of the developmental anther stages in rice (Endo et al. 2004). The expression profile data from the 4K microarray can be accessed at the following web address: http://www.jstage.jst.go.jp/article/ggs/79/4/79_213/_article.
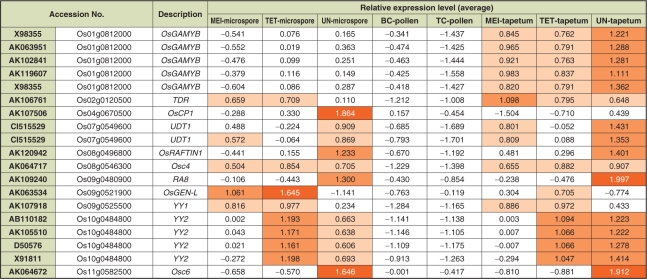
Of the 156 anther-specific genes on the 4K array identified by Endo et al. (2004), the expression profiles of Osc4, Osc6, YY1, OsRAFTIN1 and TDR have already been characterized in detail (Tsuchiya et al. 1992, Tsuchiya et al. 1994, Hihara et al. 1996, Wang et al. 2003, Li et al. 2006). These five genes are expressed mainly in the tapetum, as confirmed by RNA in situ hybridization and/or promoter analysis using a glucronidase (GUS) gene. In addition to these, we identified six previously reported tapetum-expressed genes, YY2, RA8, OsGAMYB, OsCP1, UDT1 and OsGEN-L (Hihara et al. 1996, Jeon et al. 1999, Kaneko et al. 2004, Lee et al. 2004, Jung et al. 2005, Moritoh et al. 2005). Expression profiles of these 11 known genes were re-analyzed using the 44K LM-microarray developed here (Fig. 3).
Fig. 3.
Gene expression profiles of reported genes in the 44K LM-microarray. Expression level of 19 probes corresponding to 11 selected genes, which were characterized by previous reports. The same gene accession numbers were annotated to independent probes in the case of OsGAMYB (X98355) and UDT1 (CI515529). Relative values of 0.5–1.0, 1.0–1.5 and 1.5–2.0 were labeled by light orange, orange and dark orange, respectively.
The probes for OsGAMYB (X98355, AK063951, AK102841 and AK119607) clearly demonstrated that this gene was highly expressed in the MEI, TET and UN tapetum, but not in the male gametes of all stages (Fig. 3), consistent with previous reports (Kaneko et al. 2004, Tsuji et al. 2006). Such distinct gene expression profiles between microspores/pollen and the tapetum indicate that the LM technology isolated each cell type accurately, and there were few contaminating tapetum RNAs detectable in the microspore cells which were isolated first from the anther sections (Fig. 2B). It is noteworthy that genes similar to β-ketoacyl-ACP synthase (AK067275), transaldolase family protein (AK058325), acyl carrier protein III (AK058903), phytochelatin synthetase-like protein 2 (AK070472) and GCN5-related N-acetyltransferase domain-containing protein (AK068410) also showed similar tapetum-specific expression profiles to OsGAMYB in the 44K LM array (data not shown). Among these, tapetum-specific localization of transcripts for AK058903 and AK070472 has been confirmed by RNA in situ analysis (Endo et al. 2004).
Osc6, RA8 and OsRAFTIN1 were predominantly expressed in the UN tapetum, and lower expression was also observed in the microspore at the same stage (Fig. 3). These expression profiles are consistent with the previous reports. In promoter analyses of Osc6 (Tsuchiya et al. 1994) and RA8 (Jeon et al. 1999), weak or spotted GUS signals, respectively, were observed in the microspores, in addition to a predominant GUS signal in the tapetum. Wang et al. (2003) reported the possible expression of OsRAFTIN1 in both microspores and tapetum. In our study, hybridization patterns of two probes corresponding to UDT1 (CI515529, Fig. 3) showed a similar expression profile to the GUS assay of the T-DNA-tagged udt1 mutant reported by Jung et al. (2005), who suggested that UDT1 might function in early meiosis and tapetum development. The TDR gene showed higher expression in the tapetum and relatively lower expression in the microspore, mainly at the MEI and TET stages (Fig. 3), consistent with RNA in situ analysis performed by Li et al. (2006), which showed that TDR was expressed predominantly in the tapetum at the meiosis and tetrad stage.
YY1 and Osc4 were expressed almost identically at each stage of microspore and tapetum development, while YY2 expression showed opposite trends at the TET and UN stages of each cell type (Fig. 3). In a previous report (Hihara et al. 1996), the existence of the YY1 transcript in microspores before the uninuclear microspore stage was not described, although RNA in situ analysis hinted at YY1 expression in uninuclear microspores. In the case of Osc4 and YY2, Tsuchiya et al. (1994) and Hihara et al. (1996) showed tapetum-specific expression of these genes by RNA in situ hybridization. These previous data for Osc4, YY1 and YY2 differ from our results, indicating the difficulty in detecting small amounts of transcripts in the microspores (discussed below). Through LM-microarray technology, we are now able to determine the expression profiles of these genes as follows: Osc4 is expressed in both microspores and tapetum cells at the MEI to UN stages, YY1 is expressed at the MEI and TET stages of both cell types, and expression of YY2 is higher at the TET stage in the microspores but higher at the UN stage in the tapetum (Fig. 3).
The expression patterns of OsGEN-L and OsCP1 were distinct from those of all the other genes discussed here, namely higher expression in the microspores and relatively lower expression in the tapetum (Fig. 3). OsGEN-L was expressed in the MEI and TET microspores as well as in the tapetum at the same stages. Expression in microspores was observed from the MEI stage and peaked at the TET stage; the same profile was observed in the tapetum although its overall expression level was relatively lower than in microspores (Fig. 3). In previous reports, the promoter activity of OsGEN-L was found to be up-regulated during the post-meiotic stage in most of the diploid and haploid cells of the anthers, while knock-down OsGEN-L-RNAi plants displayed defects in early microspore development (Moritoh et al. 2005), all consistent with our 44K LM-microarray data. OsCP1 was expressed predominantly in the UN microspore and weakly in UN tapetum (Fig. 3), which might correspond to the temporal pattern of promoter activity in anther locules revealed by histochemical GUS assay (Lee et al. 2004). Although this previous report did not characterize a precise expression pattern of OsCP1 in terms of the developmental stages of rice anther, an oscp1 mutant displayed pollen degradation after the microspore stage, indicating that the functional role of OsCP1 is to support development and/or maturation of pollen grains. Our data from the 44K LM-microarray are consistent with this conclusion and, moreover, determine the OsCP1expression profile more precisely, demonstrating the potential of LM-microarray technology for fine-scale gene expression analysis.
Cell type-specific gene expression profiles of the anther-specific genes reported in the 4K microarray
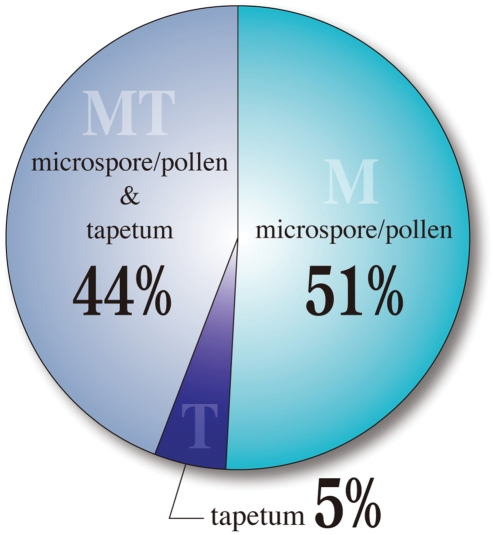
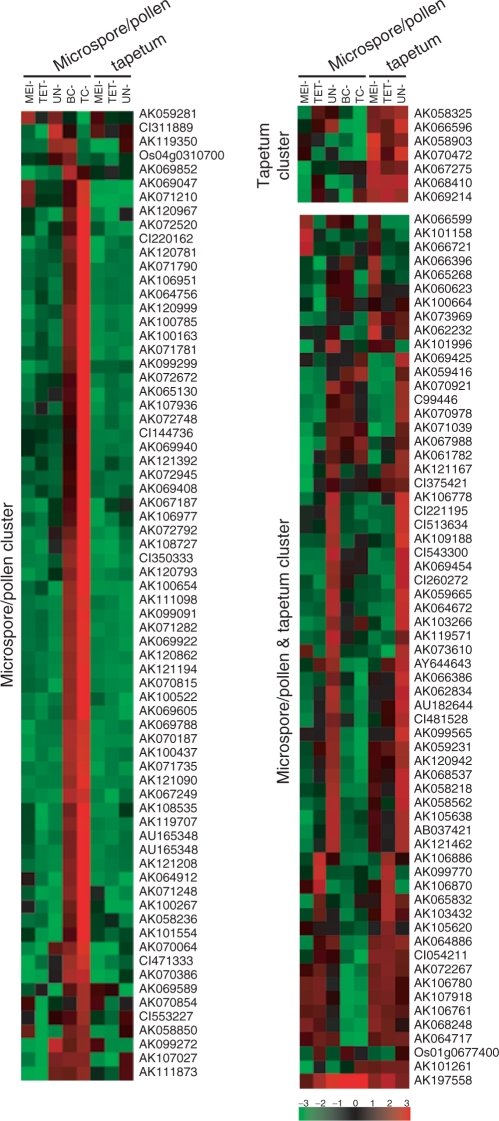
Overall, the 140 probes of the 44K LM-microarray, corresponding to the anther-specific genes identified in the 4K microarray analysis of Endo et al. (2004), were classified into 71 genes expressed specifically in male gametophytes (51%), seven genes expressed specifically in the tapetum (5%) and 62 expressed in both tissues (44%) (Fig. 4, Supplementary Tables S1, S2). The relatively small number of tapetum-specific genes identified is a reflection of the stages of the original 4K microarray analysis, which focused on the late developmental stages of anthers (from uninuclear microspore to tricellular pollen).
Fig. 4.
Classification of 140 anther-specific genes from the previous 4K microarray in the 44K LM-microarray. Expression profiles of the 44K LM-microarray for the genes reported by Endo et al. (2004) were classified into three spatial expression patterns; microspore/pollen-specific, tapetum-specific and expressed in both tissues.
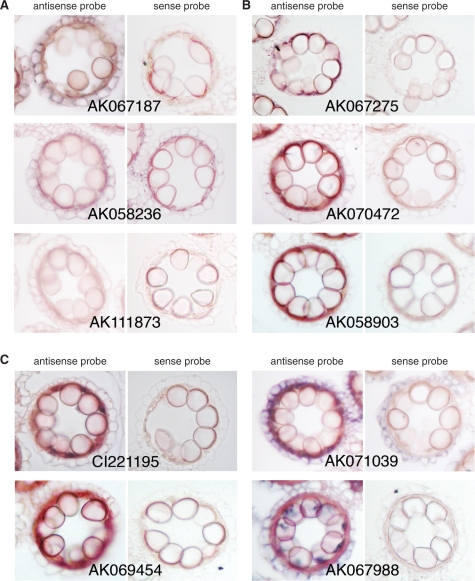
We further investigated the spatial expression patterns of 21 of the 140 probes by RNA in situ analysis: representative results are shown in Fig. 5. RNA in situ hybridization of anther cross-sections with non-RI probes tend to give faint signals in microspores, while high background signals are often obtained in the tapetum. This makes it difficult to identify small amounts of transcripts in microspores by RNA in situ analysis, presumably explaining the absence of detectable microspore expression of Osc4, YY1 and YY2 in previous reports (Tsuchiya et al. 1994, Hihara et al. 1996). This technical challenge may be common to many genes expressed in the microspore/pollen. In addition, it is a laborious task to conduct RNA in situ analyses for many genes (probes). Therefore, the LM-microarray is a useful tool to determine transcriptomes in the microspores both precisely and efficiently.
Fig. 5.
Localization of the anther-specific transcripts in rice anthers. Representative results of RNA in situ hybridization of microspore/pollen-specific genes (A), tapetum-specific genes (B) and genes expressed in both microspore/pollen and tapetum (C). DIG-labeled antisense and sense (control) RNA probes were hybridized to the cross-sections of rice anthers containing immature microspores.
We performed a cluster analysis of the 140 probes in the 44K LM-microarray, and classified the data according to spatial and temporal expression patterns (Fig. 6). The anther transcriptome can be precisely separated into male gametophyte and tapetum transcriptomes. Since cell type-specific RNAs were isolated from the cross-sections, the developmental stage of samples was strictly determined, as was precise discrimination of the cells. Fig. 6 shows that a large number of genes in the pollen/microspore cluster were expressed in the late developmental stages (BC and TC), suggesting that maturation of pollen requires various special transcripts that accumulate in the male gametophyte. This might be related to the large-scale gene repression that occurs during the transition from bicellular to tricellular pollen in Arabidopsis (Honys and Twell 2004). In the process of male gamete development, from pollen mother cell to mature pollen, the tapetum cells provide factors necessary for differentiation, development and maturation of the male gametophyte, and then begin their degradation at the bicellular pollen stage. After this, the pollen must independently produce the substances and nutrients essential for its survival, development and maturation. It has been suggested that mature pollen already contains all the transcripts necessary for the events of pollination and fertilization, e.g. pollen germination, penetration into the stigma, pollen tube elongation and double fertilization (Becker et al. 2003). An up-regulation of a large number of genes in the late developmental stage would reflect these biological features of pollen. In the pollen/microspore and tapetum cluster, synchronous expression in both cell types (e.g. high expression at the UN microspores and UN tapetum) was also a notable feature (Fig. 6). Although pollen/microspore and tapetum have independent roles, they may function by sharing some aspects of these biological events. As shown in Fig. 6, a wide variety of genes are involved in male gamete development, so it is likely that the spatial and temporal balance of their expression is critical for accurate pollen development.
Fig. 6.
Heat map view of the previously reported anther-specific genes differentially expressed in five stages of pollen/microspores and three stages of tapetum cells in the 44K LM-microarray. Cluster analysis of expression profiles of the 140 probes corresponding to the anther-specific genes characterized in the 4K array (Endo et al. 2004). Red indicates higher expression while green represents lower expression.
Our comparative study of the 44K LM-microarray and the previous 4K-microarray suggested that the 44K gene expression profiles of male gametes and tapetum in O. sativa are valid, and can be used as reliable data supporting further analyses in these specialized tissues (Hobo et al. 2008, Hirano et al. 2008). Indeed, comprehensive dissection of the gene networks involved in male gametophyte and anther development, including understanding the interaction between gametophytic and sporophytic tissues, should now be possible using the 44K LM-microarray data.
Materials and Methods
Plant materials
Rice (O. sativa L. ssp. japonica cv. Nipponbare) plants were grown in a greenhouse under normal conditions. Developing anthers of various stages were collected by confirming their developmental stage by microscopy, using one of six anthers from each flower. The stage classification of anthers is described in detail in the Results and Discussion section.
Sample preparation for LM
The anthers were fixed in Farmer's fixative (ethanol : acetate = 3 : 1) overnight at 4°C. Dehydration and paraffin embedding were performed as described by Inada and Wildermuth (2005) by using a microwave processor. Paraffin-embedded sections were cut to a thickness of 16 μm and mounted on PEN membrane glass slides (Molecular Devices, Ontario, Canada) for LM. To remove the paraffin, slides were immersed in 100% xylene (twice), 50% xylene/50% ethanol and 100% ethanol (v/v) for 5 min each, and then air-dried completely at room temperature. Three or four individual flowers were used for each LM experiment. LM was performed using the Veritas Laser Microdissection System LCC1704 (Molecular Devices). Selected areas were captured by an infrared laser (IR laser) onto CapSure Macro LCM Caps (Molecular Devices), and were subsequently cut by a UV laser (Fig. 2). The target cells that fused to the LCM cap were collected by removing the cap from the tissue section.
Microarray analysis
Total RNAs were extracted from LM cells with a PicoPure™ RNA isolation kit (Molecular Devices). Total RNA was quantified with a Quant-iT™ RiboGreen RNA reagent and kit (Invitrogen, San Diego, CA, USA). A rice 44K oligo microarray (Agilent Technologies, Palo Alto, CA, USA) which contains ∼42,000 oligonucleotides synthesized based on the nucleotide sequence and full-length cDNA data of the Rice Annotation Project (RAP) was used. Fluorescent probe labeling using the oligo-dT-T7 strand-specific amplification method, hybridization and scanning were performed according to the manufacturer's instructions with slight modification (Agilent Technologies). Microarray data were statistically analyzed for variance stabilization (VSN) and mean – SD (Z) scaling, using R software (http://www.r-project.org/). Cluster analysis for gene categorization was performed with Cluster and TreeView software (Eisen et al. 1998).
RNA in situ hybridization
Both antisense and sense probes were synthesized using a T3/T7 digoxigenin (DIG) RNA labeling kit (Roche, Basel, Switzerland) according to the manufacturer's instructions. Subsequent in situ hybridization was performed according to Endo et al. (2004) and Masuko et al. (2006), and tissue sections were observed under a light microscope (Eclipse E800 microscope system; Nikon, Tokyo, Japan).
Supplementary Material
Supplementary Material are available at PCP Online.
Funding
The Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), Grants-in-Aid for Special Research on Priority Areas (Nos. 18075003 and 18075012 to M.W., 18075005 to N.T., 18075013 to N.K., 19043015 to K.Y.); the Japan Society for Promotion of Science (JSPS) Grants-in-Aid for the 21st Century Center of Excellence Program (to M.W.); the Ministry of Agriculture, Forestry and Fisheries of Japan (MAFF) grant for the Integrated Research Project for Plant, Insect, and Animal using Genome Technology (Nos. IPG-0012 to M.N., IPG-0018 to M.K.K., IPG-0019 to M.W.); Research Fellowship for Postdoctoral Fellowships for Young Researchers of JSPS (K.S.).
Supplementary Material
Acknowledgments
The authors thank Professor Makoto Matsuoka (Nagoya University) for his valuable comments. The authors are also grateful to Hisae Kamakura (University of Tokyo), Ayako Chiba (Iwate University) and Kosuke Matsumoto, Masumi Miyano, Hiromi Shoji, Yuta Tsunaga and Ayumi Yamakawa (Tohoku University) for technical assistance.
Glossary
Abbreviations:
- DIG
digoxigenin
- GUS
β-glucuronidase
- IR
infrared
- LM
laser microdissection
References
- Amagai M, Ariizumi T, Endo M, Hatakeyama K, Kuwata C, Shibata D, et al. Identification of anther-specific genes in a cruciferous model plant, Arabidopsis thaliana, by using a combination of Arabidopsis macroarray and mRNA derived from Brassica oleracea. Sex. Plant Reprod. 2003;15:213–222. [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, et al. Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 2004;39:170–1814. doi: 10.1111/j.1365-313X.2004.02118.x. [DOI] [PubMed] [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, et al. Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J. 2002;32:401–408. doi: 10.1046/j.1365-313x.2002.01423.x. [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 2003;133:713–725. doi: 10.1104/pp.103.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lashbrook CC. Laser capture microdissection of plant cells from tape-transferred paraffin sections promotes recovery of structurally intact RNA for global gene profiling. Plant J. 2006;48:628–637. doi: 10.1111/j.1365-313X.2006.02886.x. [DOI] [PubMed] [Google Scholar]
- Cai S, Lashbrook CC. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 2008;146:1305–1321. doi: 10.1104/pp.107.110908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K. Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J. 2005;42:111–123. doi: 10.1111/j.1365-313X.2005.02355.x. [DOI] [PubMed] [Google Scholar]
- Day RC, Grossniklaus U, Macknight RC. Be more specific! Laser-assisted microdissection of plant cells. Trends Plant Sci. 2005;10:397–406. doi: 10.1016/j.tplants.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Dembinsky D, Woll K, Saleem M, Liu Y, Fu Y, et al. Transcriptomic and proteomic analysis of pericycle cells of the maize primary root. Plant Physiol. 2007;145:575–588. doi: 10.1104/pp.107.106203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat. Genet. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Bostein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Matsubara H, Kokubun T, Masuko H, Takahata Y, Tsuchiya T, et al. The advantages of cDNA microarray as an effective tool for identification of reproductive organ-specific genes in a model legume, Lotus japonicus. FEBS Lett. 2002;514:229–237. doi: 10.1016/s0014-5793(02)02371-2. [DOI] [PubMed] [Google Scholar]
- Endo M, Tsuchiya T, Saito H, Matsubara H, Hakozaki H, et al. Identification and molecular characterization of novel anther-specific genes in japonica rice, Oryza sativa L. by using cDNA microarray. Genes Genet. Syst. 2004;79:213–226. doi: 10.1266/ggs.79.213. [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, Benning C, Ohlrogge J. Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 2000;124:1570–1581. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM. Anther development: basic principles and practical applications. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Hara C, Uchimiya H. Isolation and characterization of two cDNA clones for mRNAs that are abundantly expressed in immature anthers of rice (Oryza sativa L.). Plant Mol. Biol. 1996;30:1181–1193. doi: 10.1007/BF00019551. [DOI] [PubMed] [Google Scholar]
- Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, Shim R, et al. Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 2008;49:1429–1450. doi: 10.1093/pcp/pcn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo T, Suwabe K, Aya K, Suzuki G, Yano K, et al. Various spatiotemporal expression profiles of anther-expressed genes in rice. Plant Cell Physiol. 2008;49:1417–1428. doi: 10.1093/pcp/pcn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 2003;132:640–652. doi: 10.1104/pp.103.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada N, Wildermuth MC. Novel tissue preparation method and cell-specific marker for laser microdissection of Arabidopsis mature leaf. Planta. 2005;221:9–16. doi: 10.1007/s00425-004-1427-y. [DOI] [PubMed] [Google Scholar]
- Itoh J-I, Nonomura K-I, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, et al. Rice plant development. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- Jeon J-S, Chung Y-Y, Lee S, Yi G-H, Oh B-G, An G. Isolation and characterization of an anther-specific gene, RA8, from rice (Oryza sativa L.). Plant Mol. Biol. 1999;39:35–44. doi: 10.1023/a:1006157603096. [DOI] [PubMed] [Google Scholar]
- Jung K-H, Han M-J, Lee Y-S, Kim Y-W, Hwang I, Kim M-J, et al. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 2005;17:2705–2722. doi: 10.1105/tpc.105.034090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, et al. Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell. 2004;16:33–44. doi: 10.1105/tpc.017327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003;132:27–35. doi: 10.1104/pp.102.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell. 1990;2:1201–1224. doi: 10.1105/tpc.2.12.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L-F, Chen W, Lai Y, Suo J, Kong Z, et al. Monitoring of gene expression profiles and isolation of candidate genes involved in pollination and fertilization in rice (Oryza sativa L.) with a 10 K cDNA microarray. Plant Mol. Biol. 2004;54:471–487. doi: 10.1023/B:PLAN.0000038254.58491.c7. [DOI] [PubMed] [Google Scholar]
- Lee S, Jung K-H, An G, Chung Y-Y. Isolation and characterization of a rice cysteine protease gene, OsCP1 using T-DNA gene-trap system. Plant Mol. Biol. 2004;54:755–765. doi: 10.1023/B:PLAN.0000040904.15329.29. [DOI] [PubMed] [Google Scholar]
- Li N, Zhang D-S, Liu H-S, Yin C-S, Li X-x, et al. The rice Tapetum Degeneration Retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18:2999–2014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Morrw DJ, Fernandes J, Walbot V. Comparative profiling of the sense and antisense transcriptome of maize lines. Genome Res. 2006;7:R22. doi: 10.1186/gb-2006-7-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A, Kumar VD, Amway M, Browse J. Microarray and differential display identify genes involved in jasmonate-dependent anther development. Plant Mol. Biol. 2003;52:775–786. doi: 10.1023/a:1025045217859. [DOI] [PubMed] [Google Scholar]
- Masuko H, Endo M, Saito H, Hakozaki H, Park J-I, et al. Anther-specific genes, which expressed through microsporogenesis, are temporally and spatially regulated in model legume, Lotus japonicus. Genes Genet. Syst. 2006;81:57–62. doi: 10.1266/ggs.81.57. [DOI] [PubMed] [Google Scholar]
- McCormick S. Male gametophyte development. Plant Cell. 1993;5:1265–1275. doi: 10.1105/tpc.5.10.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. Control of male gametophyte development. Plant Cell. 2004;16:S142–S153. doi: 10.1105/tpc.016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritoh S, Miki D, Akiyama M, Kawahara M, Izawa T, Maki H, et al. RNAi-mediated silencing of OsGEN-L (OsGEN-like), a new member of the RAD2/XPG nuclease family, cause male sterility by defect of microspore development in rice. Plant Cell Physiol. 2005;46:699–715. doi: 10.1093/pcp/pci090. [DOI] [PubMed] [Google Scholar]
- Nakazono M, Qui F, Brsuk LA, Schnable PS. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell type: identification of genes expressed differentially in epidermal cell or vascular tissues of maize. Plant Cell. 2003;15:583–596. doi: 10.1105/tpc.008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Tausta SL, Gandotra N, Liu T. Laser microdissection of plant tissue: what you see is what you get. Annu. Rev. Plant Biol. 2006;57:181–201. doi: 10.1146/annurev.arplant.56.032604.144138. [DOI] [PubMed] [Google Scholar]
- Ohtsu K, Smith MB, Emrich SJ, Borsuk LA, Zhou R, et al. Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.). Plant J. 2007a;52:391–404. doi: 10.1111/j.1365-313X.2007.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsu K, Takahashi H, Schnable PS, Nakazono M. Cell type-specific gene expression profiling in plants by using a combination of laser microdissection and high-throughput technologies. Plant Cell Physiol. 2007b;48:3–7. doi: 10.1093/pcp/pcl049. [DOI] [PubMed] [Google Scholar]
- Richmond T, Somerville S. Chasing the dream: plant EST microarray. Curr. Opin. Plant Biol. 2000;3:108–116. doi: 10.1016/s1369-5266(99)00049-7. [DOI] [PubMed] [Google Scholar]
- Rubinelli P, Hu Y, Ma H. Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol. Biol. 1998;37:607–619. doi: 10.1023/a:1005964431302. [DOI] [PubMed] [Google Scholar]
- Scott R, Dagless E, Hodge R, Paul W, Soufleri I, Draper J. Patterns of gene expression in developing anthers of Brassica napus. Plant Mol. Biol. 1991;17:195–207. doi: 10.1007/BF00039494. [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG. Stamen structure and function. Plant Cell. 2004;16:S46–S60. doi: 10.1105/tpc.017012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen A-M, Krober S, Unte US, Huijser P, Dekker K, Saedler H. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 2003;33:413–423. doi: 10.1046/j.1365-313x.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- Spencer MWB, Casson SA, Lindsey K. Transcriptional profiling of the Arabidopsis embryo. 2007;42:560–565. doi: 10.1104/pp.106.087668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Shiba H, Iwano M, Shimosato H, Che F-S, Kai N, et al. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl Acad. Sci. USA. 2000;97:1920–1925. doi: 10.1073/pnas.040556397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Toriyama K, Ejiri S, Hinata K. Molecular characterization of rice genes specifically expressed in the anther tapetum. Plant Mol. Biol. 1994;26:1737–1746. doi: 10.1007/BF00019488. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Toriyama K, Nasrallah ME, Ejiri S. Isolation of genes abundantly expressed in rice anthers at the microspore stage. Plant Mol. Biol. 1992;20:1189–1193. doi: 10.1007/BF00028907. [DOI] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchi-Tanaka M, Shimada Y, Nakazono M, et al. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 2006;47:427–444. doi: 10.1111/j.1365-313X.2006.02795.x. [DOI] [PubMed] [Google Scholar]
- Wang A, Xia Q, Xie W, Datla R, Selvaraj G. The classic Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc. Natl Acad. Sci. USA. 2003;100:14487–14492. doi: 10.1073/pnas.2231254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liang Y, Li C, Xu Y, Lan L, Zhao D, et al. Microarray analysis of gene expression involved in anther development in rice (Oryza sativa L.). Plant Mol. Biol. 2005;58:721–737. doi: 10.1007/s11103-005-8267-4. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Hatakeyama K, Takada Y, Hinata K. Molecular aspects of self-incompatibility in Brassica species. Plant Cell Physiol. 2001;42:560–565. doi: 10.1093/pcp/pce075. [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILE1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001;28:27–39. doi: 10.1046/j.1365-313x.2001.01125.x. [DOI] [PubMed] [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F. Isolation, characterization and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol. 2005;139:1255–1267. doi: 10.1104/pp.105.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nakayama K, Hayashi T, Yazaki J, Kishimoto N, Kikuchi S, et al. cDNA microarray analysis of rice anther genes under chilling stress at the microsporogenesis stage revealed two genes with DNA transposons castaway in the 5′-flanking region. Biosci. Biotechnol. Biochem. 2004;68:1315–1323. doi: 10.1271/bbb.68.1315. [DOI] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Wilson ZA. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell. 2007;19:3530–3548. doi: 10.1105/tpc.107.054981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui R, Iketani S, Mikami T, Kubo T. Antisense inhibition of mitochondrial pyruvate dehydrogenase E1α subunit in anther tapetum causes male sterility. Plant J. 2003;34:57–66. doi: 10.1046/j.1365-313x.2003.01704.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Madi S, Borsuk L, Nettleton D, Elschire RJ, Buckner B, et al. Laser microdissection of narrow sheath mutant uncovers novel gene expression in the shoot apical meristem. PLoS Genet. 2007;3:1040–1052. doi: 10.1371/journal.pgen.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.