Abstract
Currently, there are no effective biomarkers for ovarian cancer prognosis or prediction of therapeutic response. The objective of this study was to examine a panel of 10 serum biochemical parameters for their ability to predict response to chemotherapy, progression and survival of ovarian cancer patients. Sera from ovarian cancer patients were collected prior and during chemotherapy and were analysed by enzyme-linked immunosorbent assay for CA125, kallikreins 5, 6, 7, 8, 10 and 11, B7-H4, regenerating protein IV and Spondin-2. The odds ratio and hazard ratio and their 95% confidence interval (95% CI) were calculated. Time-dependent receiver-operating characteristic (ROC) curves were utilised to evaluate the prognostic performance of the biomarkers. The levels of several markers at baseline (c0), or after the first chemotherapy cycle (rc1), predicted chemotherapy response and overall or progression-free survival in univariate analysis. A multiparametric model (c0 of CA125, KLK5, KLK7 and rc1 of CA125) provided predictive accuracy with area under the ROC curve (AUC) of 0.82 (0.62 after correction for overfitting). Another marker combination (c0 of KLK7, KLK10, B7-H4, Spondin-2) was useful in predicting short-term (1-year) survival with an AUC of 0.89 (0.74 after correction for overfitting). All markers examined, except KLK7 and regenerating protein IV, were powerful predictors of time to progression (TTP) among chemotherapy responders. Individual and panels of biomarkers from the kallikrein family (and other families) can predict response to chemotherapy, overall survival, short-term (1-year) survival, progression-free survival and TTP of ovarian cancer patients treated with chemotherapy.
Keywords: human tissue kallikreins, serine proteases, ovarian cancer, biomarkers, multiparametric analysis, prediction of therapy
Ovarian cancer is the most lethal gynaecologic malignancy, ranking fifth in mortality among all carcinomas in women (American Cancer Society, 2007; Jemal et al, 2007). The high mortality rate has not improved substantially over the past years, despite our better understanding of the molecular events underlying malignancy and the availability of improved surgical techniques and novel chemotherapeutic agents (Marsden et al, 2000; American Cancer Society, 2007). The major reason for the poor prognosis of ovarian cancer is that over 75% of patients have disseminated disease (International Federation of Gynaecology and Obstetrics (FIGO) stage III or IV) at diagnosis (Marsden et al, 2000). These patients have a 5-year survival rate less than 30%, whereas patients with localised disease at diagnosis have an over 80% survival rate within 5 years (Nguyen et al, 1993; Bast, 2003).
One screening method for ovarian cancer is based on serum levels of CA125, a tumour-associated glycoprotein of unknown function. Nowadays, measurement of serum CA125 is considered essential for monitoring response to treatment (Van der Berg et al, 1988; Tuxen et al, 1995; Rustin et al, 1996) but has a limited value for general population screening (Tuxen et al, 1995; Zanotti and Kennedy, 1999). Furthermore, the clinical value of routinely measuring CA125 during follow-up after initial treatment is uncertain due to its limitations. One important limitation of serum CA125 is that its levels are not elevated in 50% of women with stage I disease and up to 30% of patients with more advanced disease (Tuxen et al, 1995). Its value as a screening test is even more uncertain, due to its poor specificity. Serum CA125 is elevated in a proportion of patients with diverse malignancies, such as colon, breast, endometrial, lung, liver and pancreatic tumours (Tuxen et al, 1995). Its levels can also be elevated during menstruation or pregnancy, as well as in other benign gynaecological conditions, such as endometriosis, peritonitis or cirrhosis, especially with ascites production (Tuxen et al, 1995, Meyer and Rustin, 2000).
Novel methods and markers with improved sensitivity and specificity are needed for early detection of ovarian cancer (Mills et al, 2001; Bast, 2003). High-throughput technologies, such as gene microarrays and proteomic approaches (Rai et al, 2002; Adib et al, 2004; Hibbs et al, 2004; Lu et al, 2004; Santin et al, 2004; Zhang et al, 2004), despite their limitations thus far (Diamandis, 2004; van der Merwe et al, 2007) have identified several molecular markers of early disease. Some promising novel ovarian cancer biomarkers include mesothelin (Huang et al, 2006) and human epididymis protein 4 (HE4) (Drapkin et al, 2005). In addition, among the novel markers are the human tissue kallikreins (kallikrein proteins (KLKs); reviewed in Borgono et al (2004) and Borgono and Diamandis (2004)).
Kallikreins are a family of 15 secreted serine proteases encoded by conserved genes, which are tandemly localised on chromosome 19q13.4 (Borgono et al, 2004). The kallikrein proteins are widely expressed in different tissues, during physiological or pathological conditions (Borgono et al, 2004; Shaw and Diamandis, 2007). Furthermore, several kallikreins are coexpressed in tissues or fluids (Borgono and Diamandis, 2004), suggesting their involvement in proteolytic cascades (Borgono and Diamandis, 2004; Pampalakis and Sotiropoulou, 2007; Yoon et al, 2007). Clinical studies have associated kallikreins with several types of cancers and especially hormone-associated carcinomas, such as breast, prostate and ovarian (Borgono et al, 2004; Borgono and Diamandis, 2004). The most recognised kallikrein is KLK3, also known as prostate-specific antigen, which is an established biomarker for detecting or monitoring prostate cancer progression and response to treatment (Magklara et al, 1999; Stephan et al, 2002).
Kallikreins 4, 5, 6, 7, 8, 9, 10, 11, 13, 14 and 15 are highly expressed in ovarian cancer patients or in ovarian cancer cell lines, at the mRNA and/or protein level (Tanimoto et al, 1999; Shvartsman et al, 2003; Borgono et al, 2004; Lu et al, 2004; Ni et al, 2004; Santin et al, 2004). Four recent studies verified the overexpression of several kallikreins in ovarian cancer tissues (Yousef et al, 2003), in disseminated ovarian cancer cells isolated from ascites fluid of ovarian cancer patients (Oikonomopoulou et al, 2006b) and in ovarian cancer effusions (Shaw and Diamandis, 2007; Shih et al, 2007). Some kallikreins, such as KLK6 (Hoffman et al, 2002) and KLK10 (Luo et al, 2001), broadcast unfavourable prognosis, whereas others, such as KLK8 (Borgono et al, 2006), have been associated with favourable prognosis.
In this study, we investigated the serum levels of kallikreins 5, 6, 7, 8, 10 and 11 in relation to the clinical response of ovarian cancer patients to chemotherapy, the survival outcome (overall and progression-free survival), as well as the time to progression (TTP) of the disease. We also included the novel cancer markers B7-H4 (Simon et al, 2007a, 2007b), regenerating protein IV (Reg-IV) and Spondin-2 (Simon et al, 2007b), which have been reported to be differentially expressed in ovarian cancer. We performed multiparametric analysis, including or excluding CA125, in an effort to improve the predictive and/or prognostic value of this panel for ovarian cancer.
Materials and methods
Ovarian cancer patients and specimens
Ninety-eight patients with primary epithelial ovarian cancer were included in this study, ranging in age from 22 to 77 years, with a median of 50 years. Patients with stage I–IV ovarian carcinomas (FIGO stage I: n=14; stage II: n=5; stage III: n=73; stage IV: n=6) were included in this study. Patients received the adjuvant chemotherapy regimen CBDCA/CFA or Taxol/CBDCA.
Among the 95 patients with known clinical response, according to WHO criteria, 58 (61%) patients had good response, categorised as complete remission (CR, n=29), partial remission (PR, n=21) or stable disease (SD, n=8) and 37 patients had poor clinical response with progressive disease (PD). Among the 50 CR and PR patients, 27 experienced disease progression, with a median TTP of 6 months (ranging from 3 to 24 months). Patients were monitored for survival and disease progression with a median follow-up of 24 months (ranging from 2 to 36 months). For 45 patients, the final end point within the time limits of this study was death.
Blood was drawn before the first chemotherapy and centrifuged within an hour for serum collection. Serum samples were stored at −80 °C until analysis. CA125 values were measured for all patients, utilising a Roche Elecsys method (Roche, Mississauga, ON, Canada).
Quantification of biomarkers in serum
We quantified protein levels of KLKs 5, 6, 7, 8, 10 and 11, as well as B7-H4, Reg-IV and Spondin-2, in the serum of ovarian cancer patients by sandwich-type enzyme-linked immunosorbent assays, as previously described (Supplementary Table 1, Simon et al, 2007a, 2007b and our unpublished data). KLKs were quantified in Toronto (Dr Eleftherios P, Diamantis, Mount Sinai Hospital, Toronto, ON, Canada) and the other three biomarkers at diaDexus Inc. (San Francisco, CA, USA). All assays are highly sensitive and specific for each of the measured biomarkers.
Statistical and clinical evaluation of the results
End points
For our analysis, we considered four end points: (a) clinical response, which was subcategorised as good (CR, PR, SD) vs poor response (PD); (b) overall survival, defined as time from first chemotherapy to death from any cause; (c) progression-free survival, defined as time from first chemotherapy to any progression or death from any cause; and (d) time from the first chemotherapy to progression (TTP) among CR and PR patients. For TTP, death was a competing risk and was treated as censored data.
Biomarkers
Serum samples were obtained before therapy (baseline specimen; c0) and before each chemotherapy cycle (c1–cn). The markers B7-H4, Spondin-2 and Reg-IV were measured only at c0, due to limited sample availability. As 59 patients had at least two measurements for each marker and 32 patients had at least three measures available, we considered marker concentration before and after the first treatment cycle (c0, c1) in logarithmic scale, relative changes in tumour marker after the first and the second cycle of chemotherapy (rc1=log (c0/c1); rc2=log (c1/c2)) and sometimes the ratios of these changes, (rc1/rc2), when sample sizes were deemed sufficient.
The relationships between biomarkers with patient and tumour characteristics were examined with the Kruskal–Wallis test, a non-parametric method for examining differences among multiple groups. Spearman's rank correlation coefficient was utilised to assess the correlations among biomarkers. The primary outcome for survival analyses was the progression-free survival, defined as the time from diagnosis to ovarian cancer recurrence or death from any cause. Patients, who were alive and did not meet any events, as defined by these end points, were censored at the time the last vital status was ascertained. Kaplan–Meier curves were utilised to present the survival probabilities as a function of time among groups of patients, defined by the tertile of the marker values, and log-rank tests were utilised to examine the overall difference among the curves. Cox regression model was applied to evaluate the hazard ratios (HRs) of biomarkers on overall and progression-free survival. Clinical parameters, including age and stage of disease, were adjusted in multivariate Cox proportional hazard models. Logistic regression was performed to calculate the odds ratio (OR) that defines the relation between biomarkers and response to therapy, where the outcome is response (partial response or complete response) vs no response (no change or progression). Both HR and OR were calculated on log-transformed biomarkers and were represented with their 95% confidence interval (95% CI) and two-sided P-values.
To further evaluate the diagnostic or prognostic usefulness of the markers for dichotomous classification, we considered receiver-operating characteristic (ROC) curve analysis. If by convention larger values of a biomarker were associated with adverse outcome, a cutoff point was utilised to define a positive marker-based test result, that is positive if the marker value exceeded a certain cutoff point. For a marker measured on continuous scales, the ROC curve was evaluated for all possible cutoff points. For binary outcome, that is response to chemotherapy, the ROC curve quantified the discriminatory ability of a marker for distinguishing between responders and non-responders. For TTP analysis, where the disease outcome is not concurrent with the test and the accuracy is a function of time, time-dependent ROC techniques for censored survival times were considered (Heagerty et al, 2000). We compared the true positive fraction, P (marker>cutoff point; death within t year) and false positive fraction, P (marker>cutoff point; survived beyond t year), across all possible cutoff points, and for t equal to 1 year and 5 years, respectively. For each ROC curve, we calculated the area under the curve (AUC), which ranges from 0.5 (for a non-informative marker) to 1 (for a perfect marker) and corresponds to the probability that a randomly selected patient who dies within t years has a higher marker value than a randomly selected patient who survived. Bootstrap method was utilised to calculate the CIs for AUC.
The ROC analysis was first conducted on individual markers and then in combination to explore the potential that a panel of markers can provide improved performance. We considered an algorithm that renders a single composite score using the linear predictor fitted from a binary regression model. This algorithm has been justified to be optimal under the linearity assumption (Eguchi and Copas, 2002; McIntosh and Pepe, 2002) in the sense that ROC curve is maximised (i.e., best sensitivity) at every threshold value. In particular, a weighted logistic regression which is appropriate for censored failure time data was utilised (Zheng et al, 2006) for deriving the prognostic index. A stepwise regression procedure was utilised to select markers within the panel, sometimes along with clinical variables.
As an independent validation series was not available for this study, the predictive accuracy of the composite scores was evaluated based on re-sampling of the original data. Specifically, we randomly split the data into a learning set and a test set. The learning set included two of three of the observations and the test set one of three of the observations. Using the learning set, we first performed model selection from which the final selected model gave rise to the linear combination rule. We then calculated two ROC curves for the linear score, one using data from the learning set and the other from the training set. The vertical differences between the two ROC curves gave the overestimation of the sensitivities at given specificities. The whole procedure was repeated 200 times and these differences were averaged to yield an estimate of the expected overestimation. For each type of the ROC analyses mentioned above, we present both the original ROC curves and the ROC curves that are corrected for overestimation.
All statistical analyses were performed using software SAS 9.1 (SAS Institute Inc., Cary, NC, USA) and S-PLUS 7.0 (Insightful Corp., Seattle, WA, USA).
Results
Associations of biomarkers with clinicopathological factors
Significant correlations (P<0.05) were observed between most of the biomarkers, in particular among CA125 and all markers except KLK7 (Supplementary Table 2). Supplementary Table 3 presents distributions of individual markers, stratified by age and stage. Lower levels of CA125, B7-H4 and Spondin-2 were significantly associated with older age (all P<0.05), whereas higher values of markers tended to be associated with higher clinical stages for CA125, KLK5, KLK6, KLK10, KLK11, B7-H4 and Spondin-2 (all P<0.05).
Associations of biomarkers with response to chemotherapy
We considered the association of serially measured biomarkers with clinical response to chemotherapy (good: CR, PR or SD vs poor: PD) using logistic regression models (Table 1). Univariately, statistically significant predictors of response included baseline measures of CA125, KLK6, KLK8, KLK10, B7-H4 and Reg-IV (all P<0.05), with a lower baseline level of the marker associated with favourable response. In addition, changes in CA125 from first chemotherapy cycle compared to baseline also appeared to predict response (OR=1.84, 95% CI (1.12, 3.03), P=0.017). Similarly, rapid decreases in KLK5 level after the first chemotherapy cycle also appeared to associate with favourable response (OR=1.86, 95% CI (1.09, 3.18), P=0.022). As age and stage were not significantly related to chemotherapy response, we did not consider further multivariate regression models adjusting for age or stage of disease.
Table 1. Univariate logistic regression for chemotherapy response (good vs poor).
| Clinical variable | N (%) | OR | CI | P | |
|---|---|---|---|---|---|
| Age (years) | |||||
| ⩽50 | 52 (54%) | 1 | |||
| >50 | 45 (46%) | 1.09 | (0.47, 2.51) | 0.842 | |
| Stage | |||||
| I or II | 19 (19%) | 1 | |||
| III or IV | 79 (81%) | 0.89 | (0.32, 2.53) | 0.833 | |
|
Poor
|
Good
|
||||
| Markers | N | Mean (s.d.)a | N | Mean (s.d.) | P ** |
| CA125 | |||||
| Baseline (c0) | 37 | 5.488 (2.227) | 57 | 4.401 (1.854) | 0.023 |
| rc1=log(c0/c1) | 34 | 0.121 (1.069) | 53 | 0.753 (1.105) | 0.003 |
| rc2=log(c1/c2) | 20 | 0.339 (0.473) | 34 | 0.587 (1.191) | 0.986 |
| rc1/rc2 | 20 | 0.181 (1.215) | 33 | 0.223 (1.338) | 0.279 |
| KLK5 | |||||
| Baseline (c0) | 37 | −1.776 (1.629) | 58 | −1.657 (1.03) | 0.466 |
| rc1=log(c0/c1) | 35 | −0.141 (1.016) | 54 | 0.372 (0.903) | 0.016 |
| rc2=log(c1/c2) | 22 | 0.111 (0.482) | 34 | 0.217 (0.584) | 0.860 |
| rc1/rc2 | 22 | −0.141 (1.216) | 34 | 0.134 (1.149) | 0.140 |
| KLK6 | |||||
| Baseline (c0) | 37 | 3.077 (0.808) | 58 | 2.562 (0.743) | 0.002 |
| rc1=log(c0/c1) | 35 | 0.175 (0.668) | 53 | 0.084 (0.645) | 0.932 |
| rc2=log(c1/c2) | 22 | 0.036 (0.568) | 34 | 0.082 (0.466) | 0.827 |
| rc1/rc2 | 22 | 0.286 (1.191) | 34 | 0.003 (0.927) | 0.627 |
| KLK7 | |||||
| Baseline (c0) | 37 | 1.283 (0.501) | 58 | 1.071 (0.579) | 0.0747 |
| rc1=log(c0/c1) | 35 | 0.013 (0.374) | 53 | 0.012 (0.537) | 0.956 |
| rc2=log(c1/c2) | 22 | 0.016 (0.291) | 34 | 0.078 (0.319) | 0.568 |
| rc1/rc2 | 22 | 0.01 (0.581) | 34 | −0.16 (0.557) | 0.450 |
| KLK8 | |||||
| Baseline (c0) | 37 | 2.54 (0.653) | 58 | 2.246 (0.569) | 0.017 |
| rc1=log(c0/c1) | 35 | 0.062 (0.466) | 53 | 0.019 (0.511) | 0.673 |
| rc2=log(c1/c2) | 22 | −0.013 (0.405) | 34 | 0.114 (0.364) | 0.302 |
| rc1/rc2 | 22 | 0.07 (0.737) | 34 | −0.15 (0.717) | 0.227 |
| KLK10 | |||||
| Baseline (c0) | 37 | 1.463 (0.864) | 58 | 1.022 (0.947) | 0.013 |
| rc1=log(c0/c1) | 35 | 0.126 (0.511) | 53 | 0.198 (0.74) | 0.805 |
| rc2=log(c1/c2) | 22 | 0.073 (0.218) | 34 | 0.076 (0.351) | 0.481 |
| rc1/rc2 | 22 | 0.073 (0.709) | 34 | 0.108 (0.7) | 0.663 |
| KLK11 | |||||
| Baseline (c0) | 36 | 0.052 (0.774) | 57 | −0.199 (0.737) | 0.196 |
| rc1=log(c0/c1) | 34 | 0.001 (0.632) | 53 | 0.192 (0.677) | 0.058 |
| rc2=log(c1/c2) | 22 | 0.129 (0.358) | 34 | 0.066 (0.306) | 0.574 |
| rc1/rc2 | 21 | 0.03 (0.901) | 34 | 0.176 (0.527) | 0.161 |
| B7-H4 | |||||
| Baseline (c0) | 35 | 1.216 (1.006) | 52 | 0.719 (0.976) | 0.009 |
| Reg-IV | |||||
| Baseline (c0) | 34 | −0.246 (0.517) | 49 | −0.518 (0.521) | 0.027 |
| Spondin-2 | |||||
| Baseline (c0) | 34 | 4.398 (0.551) | 49 | 4.209 (0.542) | 0.101 |
CI=confidence interval; OR=odds ratio.
Logarithmically transformed values.
Wilcoxon Rank Sum test.
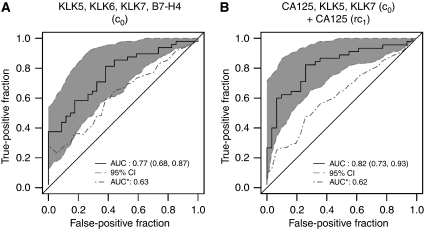
Receiver-operating characteristic curve analysis was utilised to evaluate the clinical usefulness of these markers for predicting response to chemotherapy. When considered separately, these markers only had moderate discriminatory capacity for separating good from poor responders. For example, the baseline level of KLK6 had an AUC of 0.69 (95% CI (0.58, 0.80)) (Table 1), whereas changes of CA125 after the first cycle had an AUC of 0.69 (95% CI (0.58, 0.81)). We further explored if the combination of a panel of markers could improve this diagnostic accuracy. We employed a stepwise model selection procedure that took all the baseline markers into consideration. Using the linear predictors from the final model involving KLK5, KLK6, KLK7 and B7-H4, we obtained an ROC curve with an AUC of 0.77 (95% CI (0.68, 0.87)). However, due to sample-size limitation, when correcting for the uncertainty in both model selection and ROC-estimation cross-validation procedure, the analysis resulted in a corrected AUC of only 0.63 (Figure 1A). We also considered building a logistic regression model using information on both baseline concentration and relative changes after each chemotherapy cycle. The final selected multivariate model, which included baseline measures of CA125, KLK5 KLK7 and the relative change at the first chemotherapy for CA125, resulted in an ROC curve that had an AUC of 0.82 (95% CI (0.73, 0.93)). However, due to limited sample size and even greater uncertainty involved in the model-fitting procedure, the corrected AUC was 0.62 (Figure 1B). Therefore, with the current sample size, we cannot conclude if including longitudinal information can improve the predictive accuracy.
Figure 1.
ROC curves for chemotherapy response using marker panels. ROC curves for the ‘combined’ marker, without (‘original’, solid line) and with correction for overfitting (‘corrected’, dashed line). The correction for overfitting was done by the cross-validation procedure described under Materials and methods. (A) Combination using only baseline values (c0) of KLK5, KLK6, KLK7 and B7-H4. (B) Combination using baseline values (c0) of CA125, KLK5 and KLK7 and relative changes (rc1=log (c0/c1)) of CA125 from baseline to the first chemotherapy. Data for good vs poor response are further described in Table 1 and the combined model is described in Supplementary Table 4.
Associations of biomarkers with overall survival
The Kaplan–Meier estimates of time to overall survival for patients stratified in three risk groups, as defined by tertiles of the level of an individual marker, indicated that many of the markers have good prognostic values (Supplementary Figure 1). In addition, we also saw good separations of Kaplan–Meier curves when patients were classified by both stage (I/II vs III/IV) and marker concentrations (upper, defined as >median value; vs lower, defined as <median value) (Supplementary Figure 1), suggesting that these markers may provide useful prognostic information in addition to clinical staging criteria.
The unadjusted and adjusted HRs estimated from Cox proportional hazard models are presented in Table 2. Higher values of baseline measures of CA125, KLK5, KLK6, KLK10, B7-H4 and Spondin-2 were all significantly associated with worse overall survival. For example, the HR for Spondin-2 was 2.67 (95% CI (1.43, 4.96), P=0.002). The prognostic values of many of the kallikreins, B7-H4 and Spondin-2 were stronger than those of CA125 (based on HR values), which had a HR of 1.34 (95% CI (1.17, 1.53), P<0.001). Adjustment for clinical variables including age, stage and response to chemotherapy was also performed. When considering this multivariate model, some markers became insignificant but some of them retained their significance. For example, an HR of 1.28 (95% CI (1.02, 1.60), P=0.034) was observed for KLK5 and 1.69 (95% CI (1.14, 2.49), P=0.008) for B7-H4, indicating that these two markers may provide additional prognostic value beyond age, clinical stage or response to chemotherapy. Changes in some marker concentrations after each chemotherapy treatment cycle also appeared to predict survival. In particular, rc1 of KLK6 significantly predicted time to survival in the univariate Cox regression models (HR=1.77, 95% CI (1.07, 2.95), P=0.028); however, none of the subsequent measurements remained a significant independent predictor in the multivariate model (with the exception of KLK6 rc1/rc2 ratio).
Table 2. Cox regression for overall survival outcome.
|
Univariate
|
Multivariate
|
||||||
|---|---|---|---|---|---|---|---|
| Clinical variable | N (%) | HR | CI | P | HR | CI | P |
| Age (years) | |||||||
| ⩽50 | 52 (54%) | 1 | 1 | ||||
| >50 | 45 (46%) | 0.56 | (0.3, 1.06) | 0.074 | 0.63 | (0.33, 1.21) | 0.166 |
| Stage | |||||||
| I or II | 19 (19%) | 1 | 1 | ||||
| III or IV | 79 (81%) | 2.49 | (0.98, 6.34) | 0.056 | 2.04 | (0.78, 5.36) | 0.146 |
| Chemotherapy response | |||||||
| Poor | 37 (37%) | 1 | 1 | ||||
| Good | 58 (61%) | 0.18 | (0.09, 0.35) | <0.001 | 0.19 | (0.1, 0.37) | <0.001 |
| Markers | |||||||
| CA125 | |||||||
| Baseline (c0) | 97 | 1.34 | (1.17, 1.53) | <0.001 | 1.16 | (0.98, 1.38) | 0.086 |
| rc1=log (c0/c1) | 90 | 0.96 | (0.71, 1.31) | 0.792 | 1.06 | (0.75, 1.51) | 0.732 |
| rc2=log (c1/c2) | 57 | 1.36 | (0.94, 1.96) | 0.102 | 1.49 | (0.94, 2.36) | 0.087 |
| rc1/rc2 | 56 | 1.01 | (0.69, 1.47) | 0.967 | 1.08 | (0.73, 1.6) | 0.685 |
| KLK5 | |||||||
| Baseline (c0) | 98 | 1.33 | (1.03, 1.71) | 0.028 | 1.28 | (1.02, 1.6) | 0.034 |
| rc1=log (c0/c1) | 92 | 0.91 | (0.64, 1.27) | 0.571 | 1.04 | (0.73, 1.48) | 0.827 |
| rc2=log (c1/c2) | 59 | 1.66 | (0.87, 3.16) | 0.125 | 1.63 | (0.76, 3.51) | 0.208 |
| rc1/rc2 | 59 | 0.95 | (0.68, 1.34) | 0.780 | 1.06 | (0.74, 1.53) | 0.748 |
| KLK6 | |||||||
| Baseline (c0) | 98 | 2.09 | (1.4, 3.11) | <0.001 | 1.36 | (0.88, 2.12) | 0.168 |
| rc1=log (c0/c1) | 91 | 1.77 | (1.07, 2.95) | 0.028 | 1.49 | (0.85, 2.6) | 0.160 |
| rc2=log (c1/c2) | 59 | 0.89 | (0.38, 2.08) | 0.786 | 0.53 | (0.24, 1.19) | 0.125 |
| rc1/rc2 | 59 | 1.57 | (1.01, 2.45) | 0.044 | 1.61 | (1.08, 2.4) | 0.020 |
| KLK7 | |||||||
| Baseline (c0) | 98 | 0.86 | (0.5, 1.48) | 0.595 | 0.59 | (0.33, 1.08) | 0.088 |
| rc1=log (c0/c1) | 91 | 0.9 | (0.46, 1.75) | 0.753 | 0.86 | (0.39, 1.91) | 0.713 |
| rc2=log (c1/c2) | 59 | 0.54 | (0.15, 2.01) | 0.359 | 0.71 | (0.19, 2.7) | 0.614 |
| rc1/rc2 | 59 | 1.51 | (0.7, 3.26) | 0.289 | 1.25 | (0.58, 2.68) | 0.570 |
| KLK8 | |||||||
| Baseline (c0) | 98 | 1.58 | (0.95, 2.64) | 0.080 | 1.16 | (0.69, 1.94) | 0.586 |
| rc1=log (c0/c1) | 91 | 1.13 | (0.57, 2.23) | 0.727 | 0.97 | (0.48, 2) | 0.942 |
| rc2=log (c1/c2) | 59 | 0.66 | (0.23, 1.9) | 0.443 | 1.48 | (0.47, 4.62) | 0.503 |
| rc1/rc2 | 59 | 1.33 | (0.78, 2.28) | 0.289 | 0.99 | (0.55, 1.78) | 0.975 |
| KLK10 | |||||||
| Baseline (c0) | 98 | 1.47 | (1.11, 1.96) | 0.008 | 1.3 | (0.89, 1.9) | 0.176 |
| rc1=log (c0/c1) | 91 | 1.04 | (0.65, 1.66) | 0.880 | 1.13 | (0.64, 2.01) | 0.669 |
| rc2=log (c1/c2) | 59 | 1.08 | (0.3, 3.89) | 0.901 | 0.57 | (0.11, 2.95) | 0.505 |
| rc1/rc2 | 59 | 1.31 | (0.67, 2.54) | 0.432 | 1.56 | (0.83, 2.93) | 0.169 |
| KLK11 | |||||||
| Baseline (c0) | 95 | 1.46 | (0.93, 2.28) | 0.097 | 1.18 | (0.72, 1.91) | 0.513 |
| rc1=log (c0/c1) | 89 | 0.98 | (0.61, 1.57) | 0.917 | 1.07 | (0.63, 1.79) | 0.811 |
| rc2=log (c1/c2) | 59 | 1.38 | (0.45, 4.18) | 0.572 | 1.09 | (0.36, 3.33) | 0.881 |
| rc1/rc2 | 57 | 1.14 | (0.64, 2.04) | 0.659 | 1.22 | (0.73, 2.03) | 0.453 |
| B7-H4 | |||||||
| Baseline (c0) | 89 | 2.04 | (1.48, 2.81) | <0.001 | 1.69 | (1.14, 2.49) | 0.008 |
| Reg-IV | |||||||
| Baseline (c0) | 85 | 1.56 | (0.89, 2.74) | 0.119 | 1 | (0.52, 1.92) | 0.996 |
| Spondin-2 | |||||||
| Baseline (c0) | 85 | 2.67 | (1.43, 4.96) | 0.002 | 1.65 | (0.81, 3.35) | 0.167 |
CI=confidence interval; HR=hazard ratio.
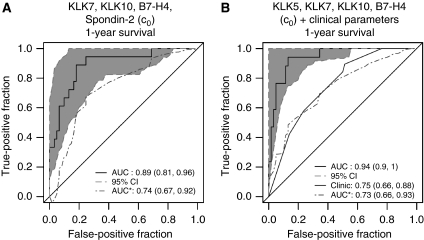
To assess whether the markers have good capacity in discriminating between subjects who progress before a given time t and those who survive beyond t, the time-dependent ROC method was utilised to evaluate the prognostic accuracy of the biomarkers. We plotted ROC curves at 1 year to investigate whether they have the accuracy to separate short-term survivors (failed by 1 year vs alive beyond 1 year). Although univariately, the predictive accuracy of each individual marker was quite low (data not shown), it was interesting to see whether we could identify a marker panel that would offer improved prognostic accuracy. We utilised a weighted logistic regression for survival data that aimed to directly find a linear combination of markers with which the area under the time-dependent ROC curve is maximised at each t. We found that a combination of markers may have the potential for improved prognostic accuracy. A linear combination rule considering KLK7, KLK10, B7-H4 and Spondin-2 yielded an ROC curve for survival at 1 year after chemotherapy with an AUC of 0.89 (95% CI (0.81, 0.96)) (Figure 2A). After correcting for overfitting, the prognostic performance was retained with an AUC of 0.74. Addition of clinical data (age, stage and response to chemotherapy) to the selected marker panel further improved the unadjusted AUC to 0.94 (Figure 2B). No improvement was observed for 2-year mortality even with a combination of a panel of markers (data not shown). These results suggest that there may be a good potential for the panel of markers to achieve good short-term (within 1-year) prognostic performance.
Figure 2.
ROC curves for overall survival at 1-year post-chemotherapy, using a panel of biomarkers, without (‘original’, solid line) and with correction for overfitting (‘corrected’, dashed line). The correction for overfitting was done by the cross-validation procedure described under Materials and methods. Data for overall survival are further described in Table 2 and the combined model is described in Supplementary Table 4. (A) Combination using baseline values (c0) of KLK7, KLK10, B7-H4 and Spondin-2. (B) Combination using baseline values (c0) of KLK5, KLK7, KLK10 and B7-H4 and the clinical parameters age, stage and response to chemotherapy.
Associations of biomarkers with progression-free survival
We performed Kaplan–Meier analysis of TTP for our patient groups, concluding that many of the markers have prognostic values (Supplementary Figure 2). The curves also showed a good separation when patients were divided into groups, according to marker concentrations (upper, defined as >median value; vs lower, defined as <median value).
The unadjusted HRs for progression-free survival are listed in Table 3. Higher values of baseline measures of CA125, KLK5, KLK6, KLK10, KLK11, B7-H4 and Spondin-2 were all significantly associated with worse progression-free survival. For example, the HR for Spondin-2 was 2.21 (95% CI (1.3, 3.76), P=0.003). The prognostic values of many of the kallikreins, as well as of the markers B7-H4 and Spondin-2, were stronger than those of CA125 (based on HR values). Adjustment for clinical variables, including age, stage and response to chemotherapy, resulted in HRs of 1.23 (95% CI (1.07, 1.42), P=0.004) for CA125, 1.33 (95% CI (1.08, 1.64), P=0.008) for KLK5, 1.44 (95% CI (1.06, 1.97), P=0.022) for KLK10, 1.67 (95% CI (1.10, 2.55), P=0.017) for KLK11 and 1.71 (95% CI (1.25, 2.35), P=0.001) for B7-H4. Changes in marker concentrations after the chemotherapy cycles also appeared to predict progression-free survival. In particular, rc1 of CA125, KLK10 and KLK11 significantly predicted TTP in the multivariate Cox regression models (all P-values <0.05) that adjust for age, stage and chemotherapy response, indicating that decreases in these markers after the first chemotherapy cycle may be associated with an adverse event in the future.
Table 3. Cox regression for progression-free survival outcome.
|
Univariate
|
Multivariate
|
||||||
|---|---|---|---|---|---|---|---|
| Clinical variable | N (%) | HR | CI | P | HR | CI | P |
| Age (years) | |||||||
| ⩽50 | 52 (54%) | 1 | 1 | ||||
| >50 | 45 (46%) | 0.59 | (0.35, 1) | 0.049 | 0.68 | (0.4, 1.17) | 0.166 |
| Stage | |||||||
| I or II | 19 (19%) | 1 | 1 | ||||
| III or IV | 79 (81%) | 3.43 | (1.47, 8.01) | 0.004 | 2.98 | (1.26, 7.03) | 0.013 |
| Chemotherapy response | |||||||
| Poor | 37 (37%) | 1 | 1 | ||||
| Good | 58 (61%) | 0.56 | (0.33, 0.94) | 0.030 | 0.62 | (0.37, 1.06) | 0.079 |
| Markers | |||||||
| CA125 | |||||||
| Baseline (c0) | 97 | 1.31 | (1.17, 1.46) | <0.001 | 1.23 | (1.07, 1.42) | 0.004 |
| rc1=log (c0/c1) | 90 | 1.4 | (1.09, 1.78) | 0.007 | 1.31 | (1.01, 1.69) | 0.042 |
| rc2=log (c1/c2) | 57 | 1.56 | (1.18, 2.05) | 0.002 | 1.47 | (1.09, 1.98) | 0.013 |
| rc1/rc2 | 56 | 0.92 | (0.67, 1.27) | 0.605 | 0.94 | (0.7, 1.28) | 0.715 |
| KLK5 | |||||||
| Baseline (c0) | 98 | 1.41 | (1.14, 1.74) | 0.001 | 1.33 | (1.08, 1.64) | 0.008 |
| rc1=log (c0/c1) | 92 | 1.19 | (0.9, 1.59) | 0.224 | 1.16 | (0.88, 1.54) | 0.301 |
| rc2=log (c1/c2) | 59 | 2.16 | (1.28, 3.63) | 0.004 | 1.74 | (0.97, 3.1) | 0.062 |
| rc1/rc2 | 59 | 0.95 | (0.71, 1.27) | 0.728 | 1.06 | (0.78, 1.45) | 0.701 |
| KLK6 | |||||||
| Baseline (c0) | 98 | 1.54 | (1.11, 2.15) | 0.011 | 1.23 | (0.85, 1.77) | 0.265 |
| rc1=log (c0/c1) | 91 | 1.56 | (1.01, 2.39) | 0.044 | 1.18 | (0.74, 1.9) | 0.491 |
| rc2=log (c1/c2) | 59 | 1.2 | (0.54, 2.65) | 0.659 | 0.8 | (0.37, 1.72) | 0.560 |
| rc1/rc2 | 59 | 1.25 | (0.83, 1.87) | 0.292 | 1.32 | (0.91, 1.89) | 0.140 |
| KLK7 | |||||||
| Baseline (c0) | 98 | 1.19 | (0.74, 1.91) | 0.478 | 1.03 | (0.64, 1.66) | 0.897 |
| rc1=log (c0/c1) | 91 | 1.33 | (0.73, 2.45) | 0.354 | 1.28 | (0.68, 2.4) | 0.441 |
| rc2=log (c1/c2) | 59 | 0.69 | (0.23, 2.09) | 0.509 | 0.68 | (0.22, 2.08) | 0.497 |
| rc1/rc2 | 59 | 1.32 | (0.69, 2.53) | 0.408 | 1.34 | (0.7, 2.56) | 0.376 |
| KLK8 | |||||||
| Baseline (c0) | 98 | 1.53 | (0.98, 2.39) | 0.061 | 1.33 | (0.84, 2.11) | 0.231 |
| rc1=log (c0/c1) | 91 | 1.3 | (0.72, 2.35) | 0.380 | 1.21 | (0.65, 2.24) | 0.549 |
| rc2=log (c1/c2) | 59 | 0.91 | (0.37, 2.25) | 0.834 | 1.28 | (0.47, 3.5) | 0.627 |
| rc1/rc2 | 59 | 1.11 | (0.69, 1.79) | 0.664 | 1.05 | (0.62, 1.78) | 0.847 |
| KLK10 | |||||||
| Baseline (c0) | 98 | 1.6 | (1.21, 2.12) | 0.001 | 1.44 | (1.06, 1.97) | 0.022 |
| rc1=log (c0/c1) | 91 | 1.6 | (1.12, 2.27) | 0.010 | 1.56 | (1.04, 2.33) | 0.030 |
| rc2=log (c1/c2) | 59 | 3.06 | (0.86, 10.87) | 0.084 | 1.84 | (0.46, 7.39) | 0.391 |
| rc1/rc2 | 59 | 1.07 | (0.59, 1.94) | 0.817 | 1.2 | (0.67, 2.17) | 0.536 |
| KLK11 | |||||||
| Baseline (c0) | 95 | 1.94 | (1.33, 2.83) | 0.001 | 1.67 | (1.1, 2.55) | 0.017 |
| rc1=log (c0/c1) | 89 | 1.7 | (1.17, 2.46) | 0.005 | 1.56 | (1.04, 2.34) | 0.031 |
| rc2=log (c1/c2) | 59 | 2.22 | (0.91, 5.44) | 0.081 | 1.5 | (0.59, 3.84) | 0.396 |
| rc1/rc2 | 57 | 1.34 | (0.81, 2.22) | 0.248 | 1.43 | (0.87, 2.35) | 0.156 |
| B7-H4 | |||||||
| Baseline (c0) | 89 | 1.96 | (1.49, 2.59) | <0.001 | 1.71 | (1.25, 2.35) | 0.001 |
| Reg-IV | |||||||
| Baseline (c0) | 85 | 1.07 | (0.65, 1.76) | 0.804 | 0.89 | (0.52, 1.53) | 0.671 |
| Spondin-2 | |||||||
| Baseline (c0) | 85 | 2.21 | (1.3, 3.76) | 0.003 | 1.52 | (0.83, 2.76) | 0.173 |
CI=confidence interval; HR=hazard ratio.
We further evaluated the prognostic accuracy for different risk classifications of a panel of biomarkers. A linear combination of some markers (CA125, KLK7, KLK8 and KLK10 at the baseline) resulted in an ROC curve at 1 year, after initiation of treatment, with AUC 0.77 (95% CI (0.70, 0.90)). However, when corrected for overfitting, the AUC for corrected ROC curve was reduced to 0.62 (data not shown).
Associations of biomarkers with TTP among CR and PR patients
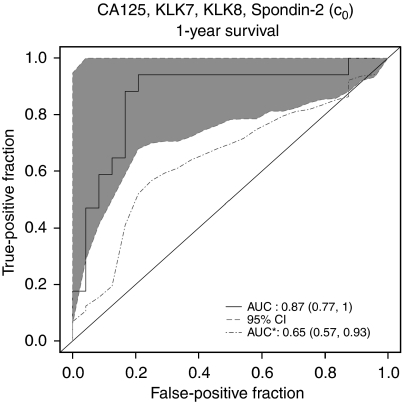
For the 50 patients who were in CR (n=29) or PR (n=21), we examined whether the markers can predict their times to progression. We investigated this question by analysing the predictive value for time to cancer progression for each individual biomarker. The results of the Cox analyses are summarised in Table 4. Younger (<50 years old) or later stage (stages III and IV) patients were associated with higher risk of progression, although the effect of age was only marginally significant. Except for KLK7 and Reg-IV, higher concentrations of all other markers at baseline were associated with higher risk of progression. For example, CA125 had an HR of 1.71 (95% CI (1.37, 2.13), P<0.001), whereas KLK5 had an HR of 2.44 (95% CI (1.59, 3.75), P<0.001) and Spondin-2 had an HR of 3.35 (95% CI (1.54, 7.26), P=0.002). In addition, changes after first chemotherapy from baseline of CA125, KLK5, KLK10 and KLK11 were significant predictors for TTP. Combining CA125, KLK7, KLK8 and Spondin-2 at baseline yielded good prognostic accuracy for predicting patients with progression by 1 year: the area under the ROC curve at 1 year after treatment was 0.87 (95% CI (0.77, 1.00)), but was reduced to 0.65 when potential overfitting was adjusted (Figure 3). Although there is potential that the prognostic accuracy could be further improved using longitudinal measures, potential of overfitting is also significant, given our sample size. We therefore did not explore this further.
Table 4. Cox regression for time to progression among CR and PR patients.
| Clinical |
Univariate
|
Multivariate
|
|||||
|---|---|---|---|---|---|---|---|
| variable | N (%) | HR | CI | P | HR | CI | P |
| Age (years) | |||||||
| ⩽50 | 27 (54%) | 1 | |||||
| >50 | 23 (46%) | 0.47 | (0.21, 1.05) | 0.067 | 0.47 | (0.21, 1.05) | 0.065 |
| Stage | |||||||
| I or II | 12 (24%) | 1 | |||||
| III or IV | 38 (76%) | 12.48 | (1.69, 92.21) | 0.013 | 12.52 | (1.69, 92.57) | 0.013 |
| Markers | |||||||
| CA125 | |||||||
| Baseline (c0) | 49 | 1.71 | (1.37, 2.13) | <0.001 | 1.52 | (1.19, 1.93) | 0.001 |
| rc1=log(c0/c1) | 47 | 1.87 | (1.37, 2.56) | <0.001 | 1.54 | (1.1, 2.14) | 0.011 |
| KLK5 | |||||||
| Baseline (c0) | 50 | 2.44 | (1.59, 3.75) | <0.001 | 2.07 | (1.29, 3.32) | 0.002 |
| rc1=log(c0/c1) | 48 | 1.5 | (1.03, 2.19) | 0.035 | 1.24 | (0.87, 1.77) | 0.239 |
| KLK6 | |||||||
| Baseline (c0) | 50 | 2.42 | (1.36, 4.32) | 0.003 | 1.95 | (1.12, 3.41) | 0.018 |
| rc1=log(c0/c1) | 47 | 1.6 | (0.84, 3.05) | 0.151 | 1.05 | (0.53, 2.08) | 0.882 |
| KLK7 | |||||||
| Baseline (c0) | 50 | 1.52 | (0.76, 3.01) | 0.233 | 1.37 | (0.71, 2.65) | 0.345 |
| rc1=log(c0/c1) | 47 | 1.37 | (0.65, 2.88) | 0.403 | 1.16 | (0.52, 2.59) | 0.721 |
| KLK8 | |||||||
| Baseline (c0) | 50 | 2.23 | (1.09, 4.58) | 0.028 | 1.69 | (0.86, 3.31) | 0.127 |
| rc1=log(c0/c1) | 47 | 1.31 | (0.59, 2.92) | 0.509 | 1.18 | (0.48, 2.88) | 0.719 |
| KLK10 | |||||||
| Baseline (c0) | 50 | 1.47 | (1.02, 2.13) | 0.040 | 1.47 | (1.02, 2.13) | 0.041 |
| rc1=log(c0/c1) | 47 | 1.58 | (1.07, 2.32) | 0.020 | 1.52 | (0.96, 2.41) | 0.071 |
| KLK11 | |||||||
| Baseline (c0) | 49 | 3.3 | (1.85, 5.89) | <0.001 | 3.02 | (1.54, 5.9) | 0.001 |
| rc1=log(c0/c1) | 47 | 1.95 | (1.26, 3.02) | 0.003 | 1.7 | (0.99, 2.93) | 0.055 |
| OVR110 | |||||||
| Baseline (c0) | 45 | 1.89 | (1.29, 2.78) | 0.001 | 1.63 | (1.11, 2.41) | 0.014 |
| CLN101 | |||||||
| Baseline (c0) | 42 | 1.38 | (0.65, 2.94) | 0.398 | 1.4 | (0.69, 2.82) | 0.353 |
| PRO108 | |||||||
| Baseline (c0) | 42 | 3.35 | (1.54, 7.26) | 0.002 | 1.99 | (0.87, 4.57) | 0.103 |
CI=confidence interval; CR=complete remission; HR=hazard ratio; PR=partial remission.
Figure 3.
ROC curves for time to progression among CR and PR patients using a panel of biomarkers. Presented is the original ROC curve (solid line) with its 95% confidence interval (shaded area) and the ROC curve calculated using cross-validation to correct for overfitting (AUC with an asterisk, dashed line). Combined markers for included baseline measures (c0) of CA125, KLK7, KLK8 and Spondin-2 are further described in Supplementary Table 4.
In summary, the regression models utilised for ROC curve analysis for a panel of markers with different outcomes are presented in Supplementary Table 4.
Discussion
Ovarian cancer detection, prognosis and response to treatment are currently based on quantification of serum levels of CA125 and imaging modalities (Van der Berg et al, 1988; Marsden et al, 2000; Meyer and Rustin, 2000). However, the clinical value of CA125 as a marker of ovarian cancer is low, due to its expression in non-malignant or non-ovarian malignant conditions (Tuxen et al, 1995; Meyer and Rustin, 2000). Serum CA125 is presently considered as a marker of response to treatment rather than a screening/diagnostic marker (Van der Berg et al, 1988; Tuxen et al, 1995; Rustin et al, 1996; Zanotti and Kennedy, 1999). As part of the quest to identify useful biomarkers for detection and management of ovarian carcinoma, the idea of multimarker analysis has been adopted by many groups (Bose and Mukherjea, 1994; Zhang et al, 2004). Thus far, increases in the diagnostic sensitivity of CA125 by using marker panels have been moderate (5–10%), often at the expense of decreases in specificity, and vice versa (Tuxen et al, 1995; Meyer and Rustin, 2000; Bast, 2003; Zhang et al, 2004).
Members of one family of cancer-related markers, the kallikrein serine protease family, have been associated with either favourable or unfavourable prognostic value in ovarian cancer or they are candidate diagnostic markers (Borgono et al, 2004, 2006; Borgono and Diamandis, 2004; Shan et al, 2006; McIntosh et al, 2007). The clinical applicability of the other three candidate ovarian cancer markers included in this study, B7-H4 (Simon et al, 2007a, 2007b), Reg-IV and Spondin-2 (Simon et al, 2007b), is still unknown.
We found significant correlations between the serological markers used in this study. For example, CA125 correlated significantly with all the markers studied, except KLK7 (Supplementary Table 2). These data are in agreement with previous observations (Borgono et al, 2004; Borgono and Diamandis, 2004). Utilisation of such biomarkers in multiparametric analyses may prove beneficial for ovarian cancer diagnosis, prognosis and prediction of therapeutic response. As a proof of this principle, in a recent study, a panel of kallikreins (KLKs 4, 5, 6, 7, 8, 10, 11, 13 and 14) was able to confidently discriminate cancerous from benign effusions, with an area under the ROC curve of 0.99, and ovarian cancer from other cancer groups with an AUC of 0.96 (Shih et al, 2007).
In our study, the predictive power of many markers for overall (Table 2) and progression-free survival (Table 3) at univariate analysis was encouraging. For example, CA125, KLKs 5, 6, 10, B7-H4 and Spondin-2 baseline values (c0) predicted a worse overall survival (Table 2) and CA125, KLKs 5, 6, 10, 11, B7-H4 and Spondin-2 baseline values (c0) a worse progression-free survival of ovarian cancer patients (Table 3). Many of these markers were independent of clinical parameters, such as age, FIGO stage or response to treatment, when multivariate analysis was performed. Similarly to the previous study for ovarian cancer detection (Shih et al, 2007), we considered panels of these markers to predict patient survival. Such a panel, which included KLK7, KLK10, B7-H4 and Spondin-2 baseline values (c0), was significantly predictive of 1-year survival post-therapy, with an AUC of 0.89 (Figure 2A).
Moreover, our findings suggested that high baseline values (c0) of markers (CA125, KLKs 6, 8, 10, B7-H4 and Reg-IV), as well as changes of CA125 and KLK5 after the first chemotherapy cycle (rc1), individually predicted an unfavourable response to chemotherapy and a higher risk for disease progression (Table 1). A panel of markers comprising KLK5, KLK6, KLK7 and B7-H4 baseline levels (c0) resulted in an improved AUC of 0.77 for predicting response to chemotherapy (Figure 1A).
Baseline values (c0) of all markers, except of KLK7 and Reg-IV, were also significant predictors of TTP among the group of responders to chemotherapy (Table 4). Combination of CA125, KLK7, KLK8 and Spondin-2 baseline levels predicted patients who would progress within 1 year, with an ROC curve of 0.87 (Figure 3).
Notably, thirty-one of the patients included in this study had a baseline serum CA125 concentration that was close to or below the cutoff point of 30 U ml−1 and remained non-informative (no significant change) over the course of chemotherapy. For these 31 patients, a few of the examined markers, for example KLK6 (OR=0.42; P=0.004), KLK8 (OR=0.44; P=0.026), KLK10 (OR=0.59; P=0.03), B7-H4 (OR=0.60; P=0.03) and Reg-IV (OR=0.36; P=0.027) appeared to be associated with response to chemotherapy. These data suggest that some markers may be superior or complement CA125 for predicting response to chemotherapy treatment.
It has been reported that the chemopreventive agent α-difluoromethylornithine is not able to efficiently block expression of many tumorigenesis-related genes, including kallikrein 6, in activated Ki-ras-expressing Caco-2 cells (Ignatenko et al, 2004). In this regard, it is possible that kallikreins may have a function in chemotherapy-resistant tumours, as it has already been proposed for kallikreins 3 (Foekens et al, 1999) and 10 (Luo et al, 2002) in breast cancer and kallikrein 4 in ovarian cancer (Xi et al, 2004). A similar role has also been proposed for Reg-IV in gastric tumours (Mitani et al, 2007). The markers included in this study should be, therefore, validated with a larger sample size to establish their applicability for ovarian cancer prognosis and response to chemotherapy.
Amongst the markers evaluated in this study, the members of the kallikrein family have been reported to cleave extracellular matrix proteins associated with tissue remodelling (Borgono and Diamandis, 2004; Ghosh et al, 2004; Michael et al, 2005), as well as activate or disarm protease-activated receptors (Oikonomopoulou et al, 2006a, Ramachandran and Hollenberg, 2007). Furthermore, they have been examined as putative modulators of neovascularisation (Aimes et al, 2003) and potential activators of molecules associated with tumour growth, survival, invasion or metastasis (Yoi et al, 1979; Frenette et al, 1997; Takayama et al, 2001; Petraki et al, 2002; Borgono and Diamandis, 2004). In this regard, kallikreins may be valuable targets for developing novel cancer therapeutic interventions.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Adib TR, Henderson S, Perrett C, Hewitt D, Bourmpoulia D, Ledermann J, Boshoff C (2004) Predicting biomarkers for ovarian cancer using gene-expression microarrays. Br J Cancer 90: 686–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimes RT, Zijlstra A, Hooper JD, Ogbourne SM, Sit ML, Fuchs S, Gotley DC, Quigley JP, Antalis TM (2003) Endothelial cell serine proteases expressed during vascular morphogenesis and angiogenesis. Thromb Haemost 89: 561–572 [PubMed] [Google Scholar]
- American Cancer Society 2007. Facts and figures 2007: American Cancer Society
- Bast Jr RC (2003) Status of tumor markers in ovarian cancer screening. J Clin Oncol 21: 200s–205s [DOI] [PubMed] [Google Scholar]
- Borgono CA, Diamandis EP (2004) The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer 4: 876–890 [DOI] [PubMed] [Google Scholar]
- Borgono CA, Kishi T, Scorilas A, Harbeck N, Dorn J, Schmalfeldt B, Schmitt M, Diamandis EP (2006) Human kallikrein 8 protein is a favorable prognostic marker in ovarian cancer. Clin Cancer Res 12: 1487–1493 [DOI] [PubMed] [Google Scholar]
- Borgono CA, Michael IP, Diamandis EP (2004) Human tissue kallikreins: physiologic roles and applications in cancer. Mol Cancer Res 2: 257–280 [PubMed] [Google Scholar]
- Bose CK, Mukherjea M (1994) Enzymatic tumour markers in ovarian cancer: a multiparametric study. Cancer Lett 77: 39–43 [DOI] [PubMed] [Google Scholar]
- Diamandis EP (2004) Mass spectrometry as a diagnostic and a cancer biomarker discovery tool: opportunities and potential limitations. Mol Cell Proteomics 3: 367–378 [DOI] [PubMed] [Google Scholar]
- Drapkin R, von Horsten HH, Lin Y, Mok SC, Crum CP, Welch WR, Hecht JL (2005) Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometrioid ovarian carcinomas. Cancer Res 65: 2162–2169 [DOI] [PubMed] [Google Scholar]
- Eguchi S, Copas J (2002) A class of logistic-type discriminant functions. Biometrika 89: 1–22 [Google Scholar]
- Foekens JA, Diamandis EP, Yu H, Look MP, Meijer-van Gelder ME, van Putten WL, Klijn JG (1999) Expression of prostate-specific antigen (PSA) correlates with poor response to tamoxifen therapy in recurrent breast cancer. Br J Cancer 79: 888–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenette G, Tremblay RR, Lazure C, Dube JY (1997) Prostatic kallikrein hK2, but not prostate-specific antigen (hK3), activates single-chain urokinase-type plasminogen activator. Int J Cancer 71: 897–899 [DOI] [PubMed] [Google Scholar]
- Ghosh MC, Grass L, Soosaipillai A, Sotiropoulou G, Diamandis EP (2004) Human kallikrein 6 degrades extracellular matrix proteins and may enhance the metastatic potential of tumour cells. Tumour Biol 25: 193–199 [DOI] [PubMed] [Google Scholar]
- Heagerty PJ, Lumley T, Pepe MS (2000) Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 56: 337–344 [DOI] [PubMed] [Google Scholar]
- Hibbs K, Skubitz KM, Pambuccian SE, Casey RC, Burleson KM, Oegema Jr TR, Thiele JJ, Grindle SM, Bliss RL, Skubitz AP (2004) Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol 165: 397–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BR, Katsaros D, Scorilas A, Diamandis P, Fracchioli S, Rigault de la Longrais IA, Colgan T, Puopolo M, Giardina G, Massobrio M, Diamandis EP (2002) Immunofluorometric quantitation and histochemical localisation of kallikrein 6 protein in ovarian cancer tissue: a new independent unfavourable prognostic biomarker. Br J Cancer 87: 763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Cheng WF, Lee CN, Su YN, Chien SC, Tzeng YL, Hsieh CY, Chen CA (2006) Serum mesothelin in epithelial ovarian carcinoma: a new screening marker and prognostic factor. Anticancer Res 26: 4721–4728 [PubMed] [Google Scholar]
- Ignatenko NA, Zhang H, Watts GS, Skovan BA, Stringer DE, Gerner EW (2004) The chemopreventive agent alpha-difluoromethylornithine blocks Ki-ras-dependent tumor formation and specific gene expression in Caco-2 cells. Mol Carcinog 39: 221–233 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ (2007) Cancer statistics, 2007. CA Cancer J Clin 57: 43–66 [DOI] [PubMed] [Google Scholar]
- Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I, Smith D, Hartmann L, Fishman D, Berchuck A, Schmandt R, Whitaker R, Gershenson DM, Mills GB, Bast Jr RC (2004) Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res 10: 3291–3300 [DOI] [PubMed] [Google Scholar]
- Luo LY, Diamandis EP, Look MP, Soosaipillai AP, Foekens JA (2002) Higher expression of human kallikrein 10 in breast cancer tissue predicts tamoxifen resistance. Br J Cancer 86: 1790–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LY, Katsaros D, Scorilas A, Fracchioli S, Piccinno R, Rigault de la Longrais IA, Howarth DJ, Diamandis EP (2001) Prognostic value of human kallikrein 10 expression in epithelial ovarian carcinoma. Clin Cancer Res 7: 2372–2379 [PubMed] [Google Scholar]
- Magklara A, Scorilas A, Catalona WJ, Diamandis EP (1999) The combination of human glandular kallikrein and free prostate-specific antigen (PSA) enhances discrimination between prostate cancer and benign prostatic hyperplasia in patients with moderately increased total PSA. Clin Chem 45: 1960–1966 [PubMed] [Google Scholar]
- Marsden DE, Friedlander M, Hacker NF (2000) Current management of epithelial ovarian carcinoma: a review. Semin Surg Oncol 19: 11–19 [DOI] [PubMed] [Google Scholar]
- McIntosh MW, Liu Y, Drescher C, Urban N, Diamandis EP (2007) Validation and characterization of human kallikrein 11 as a serum marker for diagnosis of ovarian carcinoma. Clin Cancer Res 13: 4422–4428 [DOI] [PubMed] [Google Scholar]
- McIntosh MW, Pepe MS (2002) Combining several screening tests: optimality of the risk score. Biometrics 58: 657–664 [DOI] [PubMed] [Google Scholar]
- Meyer T, Rustin GJ (2000) Role of tumour markers in monitoring epithelial ovarian cancer. Br J Cancer 82: 1535–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael IP, Sotiropoulou G, Pampalakis G, Magklara A, Ghosh M, Wasney G, Diamandis EP (2005) Biochemical and enzymatic characterization of human kallikrein 5 (hK5), a novel serine protease potentially involved in cancer progression. J Biol Chem 280: 14628–14635 [DOI] [PubMed] [Google Scholar]
- Mills GB, Bast Jr RC, Srivastava S (2001) Future for ovarian cancer screening: novel markers from emerging technologies of transcriptional profiling and proteomics. J Natl Cancer Inst 93: 1437–1439 [DOI] [PubMed] [Google Scholar]
- Mitani Y, Oue N, Matsumura S, Yoshida K, Noguchi T, Ito M, Tanaka S, Kuniyasu H, Kamata N, Yasui W (2007) Reg IV is a serum biomarker for gastric cancer patients and predicts response to 5-fluorouracil-based chemotherapy. Oncogene 26: 4383–4393 [DOI] [PubMed] [Google Scholar]
- Nguyen HN, Averette HE, Hoskins W, Sevin BU, Penalver M, Steren A (1993) National survey of ovarian carcinoma. VI. Critical assessment of current International Federation of Gynecology and Obstetrics staging system. Cancer 72: 3007–3011 [DOI] [PubMed] [Google Scholar]
- Ni X, Zhang W, Huang KC, Wang Y, Ng SK, Mok SC, Berkowitz RS, Ng SW (2004) Characterisation of human kallikrein 6/protease M expression in ovarian cancer. Br J Cancer 91: 725–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomopoulou K, Hansen KK, Saifeddine M, Tea I, Blaber M, Blaber SI, Scarisbrick I, Andrade-Gordon P, Cottrell GS, Bunnett NW, Diamandis EP, Hollenberg MD (2006a) Proteinase-activated receptors, targets for kallikrein signaling. J Biol Chem 281: 32095–32112 [DOI] [PubMed] [Google Scholar]
- Oikonomopoulou K, Michael IP, Scorilas A, Michael IP, Grass L, Soosaipillai A, Rosen B, Murphy J, Diamandis EP (2006b) Kallikreins as markers of disseminated tumour cells in ovarian cancer – a pilot study. Tumour Biol 27: 104–114 [DOI] [PubMed] [Google Scholar]
- Pampalakis G, Sotiropoulou G (2007) Tissue kallikrein proteolytic cascade pathways in normal physiology and cancer. Biochim Biophys Acta 1776: 22–31 [DOI] [PubMed] [Google Scholar]
- Petraki CD, Karavana VN, Revelos KI, Luo LY, Diamandis EP (2002) Immunohistochemical localization of human kallikreins 6 and 10 in pancreatic islets. Histochem J 34: 313–322 [DOI] [PubMed] [Google Scholar]
- Rai AJ, Zhang Z, Rosenzweig J, Shih IeM, Pham T, Fung ET, Sokoll LJ, Chan DW (2002) Proteomic approaches to tumor marker discovery. Arch Pathol Lab Med 126: 1518–1526 [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Hollenberg MD (2007) Proteinases and signalling: pathophysiological and therapeutic implications via PARs and more. Br J Pharmacol 153(Suppl 1): S263–S282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin GJ, Nelstrop AE, Tuxen MK, Lambert HE (1996) Defining progression of ovarian carcinoma during follow-up according to CA125: a North Thames Ovary Group Study. Ann Oncol 7: 361–364 [DOI] [PubMed] [Google Scholar]
- Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, O'Brien TJ, Tian E, Cannon MJ, Shaughnessy Jr J, Pecorelli S (2004) Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. In J Cancer 112: 14–25 [DOI] [PubMed] [Google Scholar]
- Shan SJ, Scorilas A, Katsaros D, Rigault de la Longrais I, Massobrio M, Diamandis EP (2006) Unfavorable prognostic value of human kallikrein 7 quantified by ELISA in ovarian cancer cytosols. Clin Chem 52: 1879–1886 [DOI] [PubMed] [Google Scholar]
- Shaw JL, Diamandis EP (2007) Distribution of 15 human kallikreins in tissues and biological fluids. Clin Chem 53: 1423–1432 [DOI] [PubMed] [Google Scholar]
- Shih IeM, Salani R, Fiegl M, Wang TL, Soosaipillai A, Marth C, Müller-Holzner E, Gastl G, Zhang Z, Diamandis EP (2007) Ovarian cancer specific kallikrein profile in effusions. Gynecol Oncol 105: 501–507 [DOI] [PubMed] [Google Scholar]
- Shvartsman HS, Lu KH, Lee J, Lillie J, Deavers MT, Clifford S, Wolf JK, Mills GB, Bast Jr RC, Gershenson DM, Schmandt R (2003) Overexpression of kallikrein 10 in epithelial ovarian carcinomas. Gynecol Oncol 90: 44–50 [DOI] [PubMed] [Google Scholar]
- Simon I, Katsaros D, Rigault de la Longrais I, Massobrio M, Scorilas A, Kim NW, Sarno MJ, Wolfert RL, Diamandis EP (2007a) B7-H4 is over-expressed in early-stage ovarian cancer and is independent of CA125 expression. Gynecol Oncol 106: 334–341 [DOI] [PubMed] [Google Scholar]
- Simon I, Liu Y, Krall KL, Urban N, Wolfert RL, Kim NW, McIntosh MW (2007b) Evaluation of the novel serum markers B7-H4, Spondin 2, and DcR3 for diagnosis and early detection of ovarian cancer. Gynecol Oncol 106: 112–118 [DOI] [PubMed] [Google Scholar]
- Stephan C, Jung K, Diamandis EP, Rittenhouse HG, Lein M, Loening SA (2002) Prostate-specific antigen, its molecular forms, and other kallikrein markers for detection of prostate cancer. Urology 59: 2–8 [DOI] [PubMed] [Google Scholar]
- Takayama TK, McMullen BA, Nelson PS, Matsumura M, Fujikawa K (2001) Characterization of hK4 (Prostase), a prostate-specific serine protease: activation of the precursor of prostate specific antigen (pro-PSA) and single-chain urokinase-type plasminogen activator and degradation of prostatic acid phosphatase. Biochemistry 40: 15341–15348 [DOI] [PubMed] [Google Scholar]
- Tanimoto H, Underwood LJ, Shigemasa K, Yan Yan MS, Clarke J, Parmley TH, O'Brien TJ (1999) The stratum corneum chymotryptic enzyme that mediates shedding and desquamation of skin cells is highly overexpressed in ovarian tumor cells. Cancer 86: 2074–2082 [PubMed] [Google Scholar]
- Tuxen MK, Soletormos G, Dombernowsky P (1995) Tumor markers in the management of patients with ovarian cancer. Cancer Treat Rev 21: 215–245 [DOI] [PubMed] [Google Scholar]
- Van der Berg ME, Lammes FB, van Putten WL, Stoter G (1988) Ovarian cancer: the prognostic value of the serum half-life of CA125 during induction chemotherapy. Gynecol Oncol 30: 307–312 [DOI] [PubMed] [Google Scholar]
- van der Merwe DE, Oikonomopoulou K, Marshall J, Diamandis EP (2007) Mass spectrometry: uncovering the cancer proteome for diagnostics. Adv Cancer Res 96: 23–50 [DOI] [PubMed] [Google Scholar]
- Xi Z, Kaern J, Davidson B, Klokk TI, Risberg B, Tropé C, Saatcioglu F (2004) Kallikrein 4 is associated with paclitaxel resistance in ovarian cancer. Gynecol Oncol 94: 80–85 [DOI] [PubMed] [Google Scholar]
- Yoi OO, Seldin DC, Spragg J, Pinkus GS, Austen KF (1979) Sequential cleavage of proinsulin by human pancreatic kallikrein and a human pancreatic kininase. Proc Natl Acad Sci USA 76: 3612–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Laxmikanthan G, Lee J, Blaber SI, Rodriguez A, Kogot JM, Scarisbrick IA, Blaber M (2007) Activation profiles and regulatory cascades of the human kallikrein-related peptidases. J Biol Chem 282: 31852–31864 [DOI] [PubMed] [Google Scholar]
- Yousef GM, Polymeris ME, Yacoub GM, Scorilas A, Soosaipillai A, Popalis C, Fracchioli S, Katsaros D, Diamandis EP (2003) Parallel overexpression of seven kallikrein genes in ovarian cancer. Cancer Res 63: 2223–2227 [PubMed] [Google Scholar]
- Zanotti KM, Kennedy AW (1999) Screening for gynecologic cancer. Med Clin North Am 83: 1467–1487 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Bast Jr RC, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, Berchuck A, Van Haaften-Day C, Hacker NF, de Bruijn HW, van der Zee AG, Jacobs IJ, Fung ET, Chan DW (2004) Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res 64: 5882–5890 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Cai T, Feng Z (2006) Application of the time-dependent ROC curves for prognostic accuracy with multiple biomarkers. Biometrics 62: 279–287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.