Abstract
In the olfactory organ of the squid, Lolliguncula brevis there are five morphological types of olfactory receptor neurons (ORNs). Previous work to characterize odor sensitivity of squid ORNs was performed on only two of the five types in dissociated primary cell cultures. Here we sought to establish the odorant responsiveness of all five types. We exposed live squid or intact olfactory organs to excitatory odors plus the activity marker, agmatine (AGB), an arginine derivative that enters cells through non-selective cation channels. An antibody against AGB was used to identify odorant-activated neurons. We were able to determine the ORN types of AGB-labeled cells based on their location in the epithelium, morphology and immunolabeling by a set of metabolites: arginine, aspartate, glutamate, glycine and glutathione. Of 389 neurons identified from metabolite-labeled tissue, 3% were type 1, 32% type 2, 33% type 3, 15% type 4 and 17% type 5. Each ORN type had different odorant specificity with type 3 cells showing the highest percentages of odorant-stimulated AGB labeling. Type 1 cells were rare and none of the identified type 1 cells responded to the tested odorants, which included glutamate, alanine and AGB. Glutamate is a behaviorally attractive odorant and elicited AGB labeling in types 2 and 3. Glutamate-activated AGB labeling was significantly reduced in the presence of the adenylate cyclase inhibitor, SQ22536 (80 μM). These data suggest that the five ORN types differ in their relative abundance and odor responsiveness and that the adenylate cyclase pathway is involved in squid olfactory transduction.
Keywords: Squid, Olfaction, Activity labeling
Introduction
In general, olfactory receptor neurons (ORNs) are bipolar-shaped cells with apical dendrites containing cilia and basal unmyelinated axons, which synapse in a glomerular structure in the brain. In cephalopods, multiple morphological variations on this general theme have been described (Woodhams and Messenger, 1974; Emery, 1975; Wildenburg and Fioroni, 1989; Emery, 1992). In the squid, Lolliguncula brevis, the olfactory epithelium (OE) consists of five morphologically distinct ORN types (types 1-5) and one or more types of epithelial or support cells (Emery, 1975). The odorant sensitivity and signaling of squid ORNs have been tested on dissociated cells from the OE, however, the fragility of three of the ORN types has resulted in data from only the type 2 and type 4 ORNs (Lucero et al., 1992; Danaceau and Lucero, 1998). Type 2 and type 4 ORNs are sensitive to a variety of odorants such as glutamate, betaine (Danaceau and Lucero, 2000), dopamine and diluted squid ink (Lucero et al., 1992). It has been shown behaviorally that cephalopods are attracted to amino acids such as L-serine, L-alanine, Larginine, L-proline, L-glutamate and glycine (Anraku et al., 1998). In contrast, tetraethylammonium, tetrodotoxin and squid ink are behaviorally aversive and elicit escape responses (Gilly and Lucero, 1992). Measuring of ventilation cycles suggests that cuttlefish use distance chemoreception to sense features in their environment such as food, conspecifics, ink and predators (Boyle, 1983; Boal and Golden, 1998).
Based on studies showing that sensory neurons with different structures are receptive to different classes of odorants (bile acids, amino acids) (Hansen et al., 2003), 2003), and express different transduction molecules (Hansen et al., 2004; Hansen et al., 2005; Mobley et al., 2007), we hypothesized that squid ORN types have different but overlapping odor specificities. In order to study the odor responsiveness of all of the squid ORN types, we used the activity marker, agmatine (AGB), on intact squid olfactory organs. AGB is decarboxylated arginine (1-amino-4-guanidobutane), a cationic compound that has the ability to carry charge and pass through some non-selective cation channels (Yoshikami, 1981; Marc, 1999b). Odorant activation of non-selective cation channels allows externally applied AGB to accumulate intracellularly where it cross-links to amino side-chains of proteins during glutaraldehyde fixation (Steinbusch et al., 1978; Steinbusch et al., 1982; Kuzirian et al., 1986; Marc, 1999b). Several studies have used AGB as a marker of neuronal excitation (Michel et al., 1999; Marc, 1999a; Marc, 1999b;Steullet et al., 2000;Edwards and Michel, 2002;Marc and Jones, 2002;Lipschitz and Michel, 2002;Wirsig-Wiechmann et al., 2002;Sakata et al., 2003;Acosta et al., 2005;Marc et al., 2005;Schubert et al., 2006). Immunostaining with an anti-AGB antibody allows visualization of odorant-activated neurons across the entire tissue. A second technique termed “metabolite profiling” (Marc et al., 1990;Marc et al., 1995;Marc et al., 1998;Marc and Cameron, 2001;Marc and Jones, 2002;Marc and Jones, 2003) involves immunolabeling consecutive sections with antibodies against metabolite molecules such as glutamate or glutathione (GSH). Each cell type has a unique metabolite profile based on the combination of steady state levels of free metabolites. This metabolite profiling technique bypasses the need to identify unique proteins expressed in each cell type. Here we measured the total number of neurons in squid OE, used metabolite profiling and morphological information to identify the neuronal types, and determined the odorant specificity using activity-dependent AGB immunoreactivity. We tested only excitatory odorants to determine whether only one cell type or multiple cell types would respond. We found that each squid ORN type had different but overlapping odor specificity.
Immunohistochemical studies show that proteins for both the adenylate cyclase (AC) pathway and the phospholipase C (PLC) pathway are expressed heterogeneously among the ORN types and are co-localized in subsets of ORNs (Mobley et al., 2007). We hypothesized that inhibiting AC and therefore cAMP production would prevent AGB from entering ORNs. Addition of the AC inhibitor SQ22536 to odorant solutions significantly decreased odorant-activated AGB labeling, implicating the AC pathway in odorant-dependent activity labeling of squid ORNs.
Our data suggest that the morphologically distinct ORN types in squid OE have unique odor sensitivities to amino acid stimuli. We also present evidence to support our hypothesis that the AC pathway is involved in the signal transduction of odorants in squid ORNs. This is the first report of odorant responses in the squid ORN type 3.
Materials and Methods
Animals
Juvenile L. brevis (n = 26; mantle length ∼10cm) were caught in the Galveston Bay by the National Resource Center for Cephalopods (University of Texas Marine Biomedical Institute, Galveston, TX). Animals were held for 3-5 days and shipped overnight to Utah. Experiments commenced upon arrival.
In vivo AGB exposure
Animals (n = 25) were placed in a small aquarium net and transferred between two 500 ml opaque chambers. The exposure chamber contained 20 mM AGB (or artificial sea water as a control) in artificial seawater (ASW: (in mM) 340 NaCl, 10 KCl, 35 MgCl, 10 Hepes, 10 CaCl2, 17.5 glucose, pH 7.4, ∼800 mOsm) either with or without an odorant (100 μM alanine, glutamate or 50 μM glutamate). The rinse chamber was filled with ASW. Each squid was placed in the exposure chamber for 10 sec intervals every minute for 20 min or continuously for 120 sec. When tested, the adenylate cyclase inhibitor SQ 22536 (80 μM) was added to the exposure chamber; the same exposure protocol was followed as previously stated. Immediately following the last rinse, animals were decapitated and heads were placed in cold 1% paraformaldehyde/2.5% glutaraldehyde at 4°C overnight.
Immunohistochemistry
Olfactory organs were dissected from the fixed tissue, dehydrated in a methanol and acetone series, embedded in Eponate plastic and cured at 60-65°C overnight. Fifty nm sections cut on a Leica Ultracut T microtome (Leica, Vienna, Austria) were collected on Teflon-coated spot slides (Erie Scientific, Portsmouth, NH). The sections were deplasticized with 25% sodium ethoxide in anhydrous ethanol for 7 min, rinsed with 100% methanol (3×2 min), then double distilled (dd) H2O (5 min) and dried. Individual sections were stained with one of the following rabbit polyclonal primary antibodies (final dilution): anti-AGB (1:2000), anti-arginine (1:500), anti-aspartate (1:2000), anti-glutamate (1:32000), anti-glycine (1:4000) or anti-GSH (1:4000) (antibodies were a generous gift from Dr. Robert Marc, Univ. of Utah) overnight at room temperature. Primary antibodies were diluted in 0.1 M PBS containing 1% goat serum and 0.05% thimerosal (pH 7.4). Dot blot analysis showed that the aspartate, glutamate, glycine (Marc et al., 1990), arginine (Marc et al., 1995) and AGB (Marc, 1999) antibodies are at least 1,000-fold less cross-reactive to other structurally related antigens. Low cross-reactivity for GSH (<1:1000) is reported by the manufacturer (Signature Immunologics, Inc., SLC, UT). Elimination of any of the primary antibodies from the procedure resulted in no immunoreactivity. Secondary gold-conjugated goat anti-rabbit antibodies were applied for one hour followed by visualization with silver intensification (0.14% silver nitrate, 43 mM hydroquinone, 64 mM citric acid, 141 mM sodium citrate; 3 min at 32°C). Intensification was quenched by 5% acetic acid followed by a 10 min wash in ddH2O. Blocking non-specific staining has been shown to be unnecessary (Marc et al., 1995). Slides were coverslipped with Eponate plastic and cured at 65°C.
For hematoxylin and eosin (H&E), one freshly dissected olfactory organ was placed in 4% paraformaldehyde for 2 hrs. Tissue was cryoprotected in 15% and 30% sucrose in 0.1 M phosphate buffered saline (PBS) containing 20% sucrose and 0.2% picric acid overnight at 4°C. The tissue was embedded in Tissue-Tek O.C.T. Compound (Sakura, Torrance, CA), and then flash frozen with liquid nitrogen. Serial sections (10 μm thick) of the entire organ were collected on Superfrost/Plus slides (VWR, West Chester, PA).
H&E staining procedure was conducted on 10 μm thick frozen sections in order to evaluate the number of nuclei and cilia containing invaginations (cilia pockets) across an entire olfactory organ. The H&E procedure was from http://science.peru.edu/gregarina/html/harris.html (Jun. 16, 2006) as described by Mobley, et al. (2007). Briefly, sections were hydrated in 50% ethanol (EtOH), then ddH2O. Slides were stained with Harris Hematoxylin-Modified (Sigma, St. Louis, MO) rinsed with ddH2O and destained with 1% acid EtOH. Slides were exposed to ammonia water until sections turned bright blue, then rinsed with ddH2O. Slides were dehydrated in an EtOH series, cleared in a graded xylene/ EtOH series as follows: 50% xylene/EtOH, 100% xylene, Zelmer's Eosin-xylol, 100% xylene. Slides were coverslipped using Vectamount (Vector, Burlingame, CA).
Optical Imaging
A set of serial sections immunostained for the six metabolites listed above was captured with an Axiocam CCD camera at 20X or 40X on a Zeiss upright Axioplan2 microscope as 8-bit grayscale images. Images were registered digitally using PCI Geomatica 8.0 (PCI Remote Sensing, Richmond Hill, Ontario, Canada). Although similar exposure times and light levels were used during image capture, small differences in background intensities were measured from a region without tissue and used to set equivalent background levels across images. Images used for RGB have been inverted so that black = no labeling, and white = labeling.
Quantitative Analysis
Counting AGB labeled cells
We analyzed one 40X image (345.7 × 273.9 μm) of squid OE from animals used in the AGB labeling experiments (n = 15 squid). Images were chosen for their inclusion of cells with identifiable morphology. Images were analyzed for the number of AGB labeled ORNs using UTHSCSA ImageTool (developed at the University of Texas Health Science Center at San Antonio, Texas and available from the Internet by anonymous FTP from maxrad6.uthscsa.edu). ORNs were chosen for analysis based on having an identifiable morphology, with the portion of the cell that could be AGB labeled visible (type 1 and 2 somas; type 3, 4 and 5 cilia pockets). Using Image Pro Plus (MediaCybernetics, Silver Spring, MD), areas of interest (AOIs) were placed on the ORNs over the relevant areas of the cell (soma or cilia pockets). AOI placements were saved and applied to each image in a set to obtain pixel intensity measurements for each AOI. Mean pixel intensities of AGB-negative AOIs were measured and only those AOIs with an average value greater than two standard deviations above the mean were considered AGB-positive. The mean pixel intensity of AOIs counted as AGB-positive were therefore significantly higher than the mean for all AOIs (student's t-test, P<0.05). Because the results are expressed as percentages, a nonparametric term, we used chi-squared tests to identify a correlation between ORN type and odor sensitivity as measured by the number of AGB-labeled cells.
For experiments involving the adenylate cyclase inhibitor SQ22536, images of squid OE (n = 15 olfactory organs) were captured at 20X. The OE was masked to determine the total epithelial area. We analyzed three OE sections per olfactory organ and three olfactory organs per stimulus. A threshold criterion of two standard deviations above the average intensity, measured from a large unlabeled portion of the epithelium, was used to determine the percent of AGB-labeled area. The objects (somas or cilia pockets), greater than a 100-pixel area (∼7 μm2), were counted as labeled neurons. We did not include the apical cilia in our measurements, because control experiments showed some non-specific labeling. One-way ANOVA followed by Tukey's posthoc pairwise comparison was used to determine significance between groups.
Counting H&E labeled cells
We captured images of each H&E labeled section at 20X and used Adobe Photoshop Elements (1.0) to merge images of sections that were larger than a single image. All nuclei and cilia pockets in the upper (type 3), middle (type 4) and lower (type 5) thirds of the OE were marked using a separate image layer in Adobe Photoshop CS. Total marks from each image were counted using Image Pro Plus object analysis.
Results
Identification of cell types
Physiological studies of the specificity and selectivity of squid ORNs have been limited to the type 2 and type 4 neurons that retain morphology following dissociation. To survey odorant-stimulated activity across all cell types in intact squid olfactory organs, we used AGB activity labeling. We registered images from 50 nm sections labeled with antibodies against small metabolites and AGB from squid olfactory organs that had been exposed to AGB in the presence or absence of an odor. Several parameters were used to determine the cell type of the neurons: location in the epithelium, morphology, and the immunoreactivity of L-arginine, L-aspartate, L-glutamate, glycine, GSH, and AGB (Fig. 1A). Type 1 ORNs are the most rare and difficult to identify (Fig. 1B-E, L). They are found in the apical half of the epithelium and have a narrow invagination at the distal pole where several cilia originate (Emery, 1975). The type 2 cells have a small cell body in the middle or basal layer of the OE, and a long dendrite ending in a knob-like structure at the apical surface (Fig. 1D-E). ORN types 3, 4 and 5 have large invaginations filled with cilia called cilia pockets, which retain access to the external environment. The cilia within the invaginations contain signal transduction proteins (Lucero et al., 2000;Mobley et al., 2007) and have been shown to be the cellular region necessary for odor transduction (Danaceau and Lucero, 2000). The type 3 cilia pocket is located in the apical half of the OE and is connected to its soma in the mid to basal layer of the OE by a narrow isthmus (Fig. 1F-G). Type 4 ORNs have a large cilia pocket that can span the height of the OE, with the soma found at the base of the cell (Fig. 1H-I). Type 5 ORNs have a round cilia pocket found at the base of the epithelium (Fig. 1J-K), and a thin process that extends to the surface (not shown). These cell types are not homogeneously distributed throughout the olfactory organ (Lucero et al., 2000).
Figure 1.
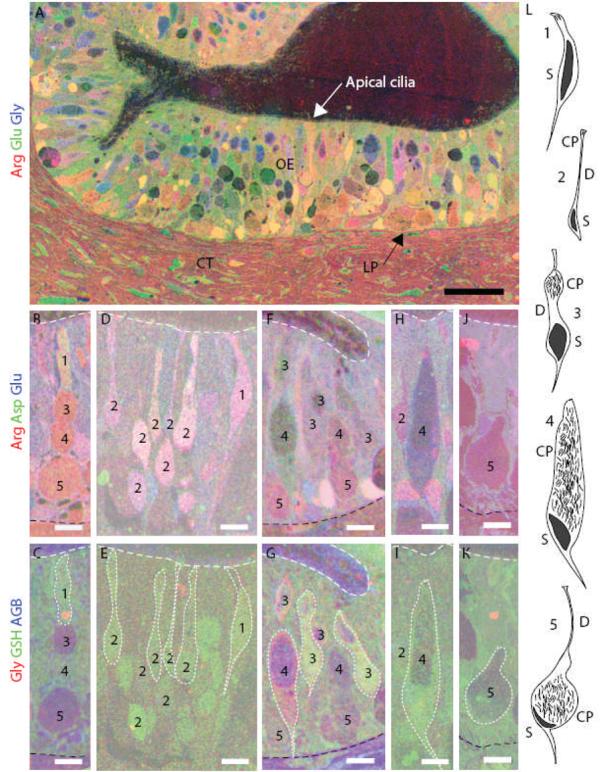
Identification of the five morphological types of squid ORNs using metabolite profiles and location. (A) Thin sections through the squid OE were immunostained and superimposed. An RGB image shows levels of arginine (red), glutamate (green) and glycine (blue) in neurons and epithelial (support) cells. A basement membrane (arrow) separates the OE from the lamina propia (LP) and connective tissue (CT) (scale bar = 50 μm). (B,D,F,H,J) arginine = red, aspartate = green and glutamate = blue. (C,E,G,I,K) the same cells as in B, D, F, H and J; glycine = red, GSH = green, AGB = blue. White dotted line outlines some of the ORNs. (B-C) Type 1 ORN. (D-E) Type 2 ORNs. (F-G) Type 3 ORNs. (H-I) Type 4 ORN. (J-K) Type 5 ORN. White dashed line indicates the apical edge of the OE. Black dashed line indicates the basal limit of the OE in C, G and K. (L) Illustrations of the morphology of the five ORN types (redrawn from Emory 1975). CP, cilia pocket; D, dendrite; S, soma. Black region in each cell is the nucleus. (B-K), scale bars = 10 μm.
Metabolite profiles for each ORN type (Sitthichai et al., 2003) were used to visualize and identify the ORNs (Fig. 1). Briefly, elevated levels of GSH identified type 1 ORNs (Fig. 1C), high arginine, aspartate and glutamate levels identified type 2 ORNs (Fig. 1D), elevated glycine and GSH distinguished type 3 and 4 ORNs (Fig. 1G) from other ORNs, while high arginine levels identified type 5 ORNs (Fig. 1J). By combining morphology with location and metabolite labeling, we were able to distinguish five ORN types in squid OE.
Quantification of cell types
The relative abundance of each ORN type was first ascertained from 389 identified cells within representative sections from 15 squid OE (Fig. 2A). The type 2 and 3 ORNs were the most abundant, 32% and 33%, respectively, 15% were type 4 and 17% were type 5 ORNs (Fig. 2B). Type 1 ORNs were relatively rare (3%). To confirm our findings and account for non-sensory support cells, an entire olfactory organ was serially sectioned at 10 μm intervals and stained with H&E. Nuclei and cilia pockets within the OE were counted in every section and the general location of each cilia pocket (upper, middle and lower epithelium) was recorded (Fig. 2C). There were 37,778 nuclei and 7,668 cilia pockets, suggesting that type 3, 4 and 5 ORNs account for 20% of all cells. Type 3 (upper) accounted for 37% of the cilia pockets, type 4 (middle) accounted for 36% of the cilia pockets, and type 5 (lower) accounted for 27% of the cilia pockets. The histology sections gave us a more accurate count of cilia pockets but did not distinguish between nuclei of type 1 and type 2 cells, which lack cilia pockets, or support cell nuclei. In metabolite-labeled tissue, the ratio of identified type 1 ORNs to the sum of types 3, 4 and 5 ORNs was 1:25 and the ratio of type 2 ORNs to the sum of types 3, 4, and 5 ORNs was 1:2. Thus, we estimated the numbers of type 1 and 2 ORNs in our H & E sections by multiplying the total number of cilia pockets (7668) by the ratios derived from the metabolite-labeled tissue (0.04 × 7668 = 307 type 1 cells; 0.5 × 7668 = 3834 type 2 cells). We calculated that type 1 ORNs represent <1% while type 2 neurons represent 10% of the cells in the squid OE. Based on these data the ORNs account for 31% (11,809) of the nuclei, with the remaining 69% (25,969) nuclei presumably attributed to non-sensory support cells (Fig. 2D). While simple counts of identifiable cells suggest type 2 and 3 ORNs are equally numerous, our calculations indicate that type 2 ORNs outnumber the type 3 ORNs.
Figure 2.
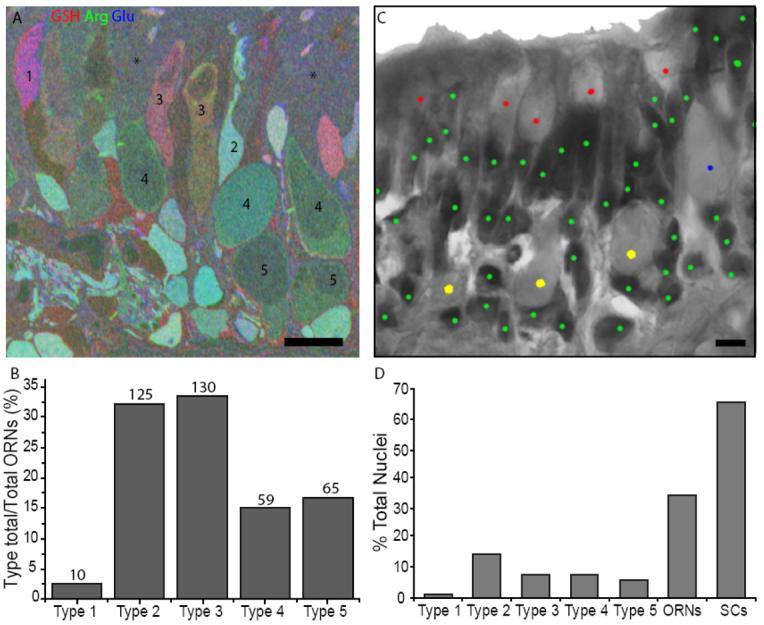
Distribution of cells in the squid olfactory organ. (A) An RGB image from a metabolite-labeled squid OE section was used to identify and count ORN types. Numbers identify the ORN types, asterisks indicate areas with indistinguishable cells (see Discussion), darker regions within cells are cilia pockets. (B) The number of identified ORNs for each type were divided by the total number of identified ORNs and expressed as a percentage. (C) A representative H&E image from a squid OE section used to count nuclei (dark stain) and cilia pockets (light stain). Cilia pockets were divided into upper, middle and lower epithelium locations. Green dots, nuclei; red dots, upper cilia pockets (type 3); blue dot, middle cilia pocket (type 4); yellow dots, lower cilia pockets (type 5). (D) The percentages of cilia pockets to nuclei for ORN types 3, 4, and 5 and estimates for ORN types 1 and 2 were based on cell counts in B (see text). The total number of cilia pockets plus type 1 and 2 nuclei divided by total nuclei yields the estimated percentage of ORNs compared to support cells (SCs).
AGB labeling of ORNs
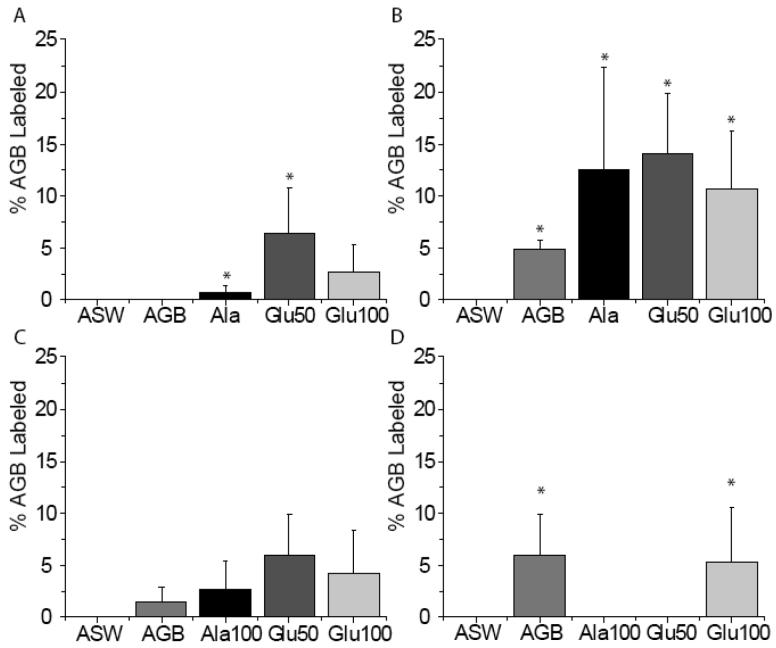
Live squid exposed to the activity marker AGB in the absence of odorants showed an increase in AGB immunoreactivity compared to artificial seawater (ASW) alone, suggesting that AGB can act as an odorant (Fig. 3A-B). Inclusion of an odorant (100 μM alanine, 50 or 100 μM glutamate) previously shown to be behaviorally relevant (Anraku et al., 1998) or physiologically excitatory (Danaceau and Lucero, 2000) further increased the number of cells labeled with AGB over AGB exposure alone (Fig. 3C). Within the 389 identified neurons from 15 animals, 78 cells were AGB labeled. A chi-squared test of the behaviorally relevant odorants showed a significant correlation between AGB labeling and olfactory organs stimulated with 100 μM alanine + AGB, 50 μM glutamate + AGB and AGB alone (p < 0.05; Fig. 4A). The significant increase in activity-dependent labeling in the presence of 100 μM alanine or 50 μM glutamate suggests the presence of odorant receptors with non-overlapping odorant specificities.
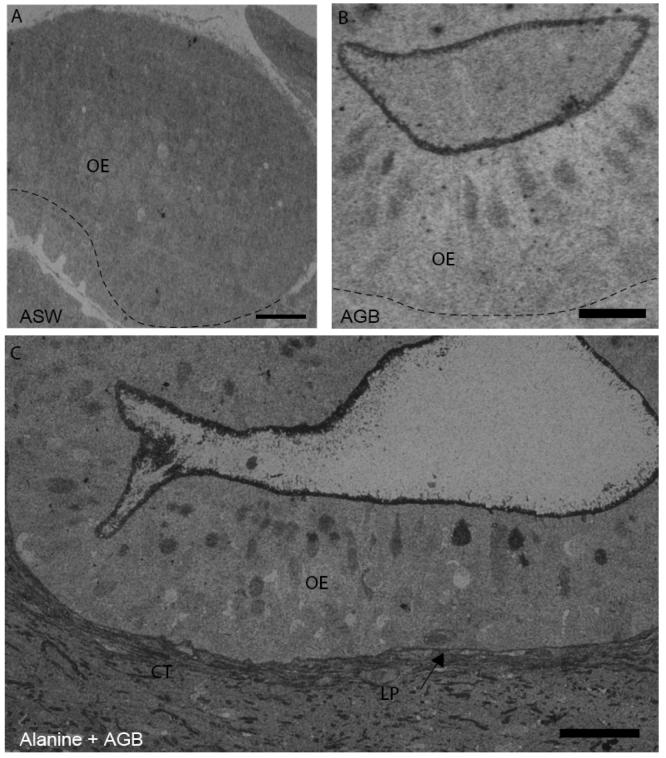
Figure 3.
Odorants increase AGB labeling in squid OE. (A) Shown is an example of non-specific immunostaining with anti-AGB of squid OE from an animal exposed to ASW. Scale bar = 100 μm. (B) Positive anti-AGB labeling was observed in an animal exposed to 20 mM AGB. Scale bar = 50 μm. (C) Squid ORNs show anti-AGB labeling in an animal exposed to 20 mM AGB + 100 μM alanine. Scale bar = 50 μm. CT = connective tissue, LP = lamina propria, arrow indicates basement membrane; dashed lines in A & B = basement membrane.
Figure 4.
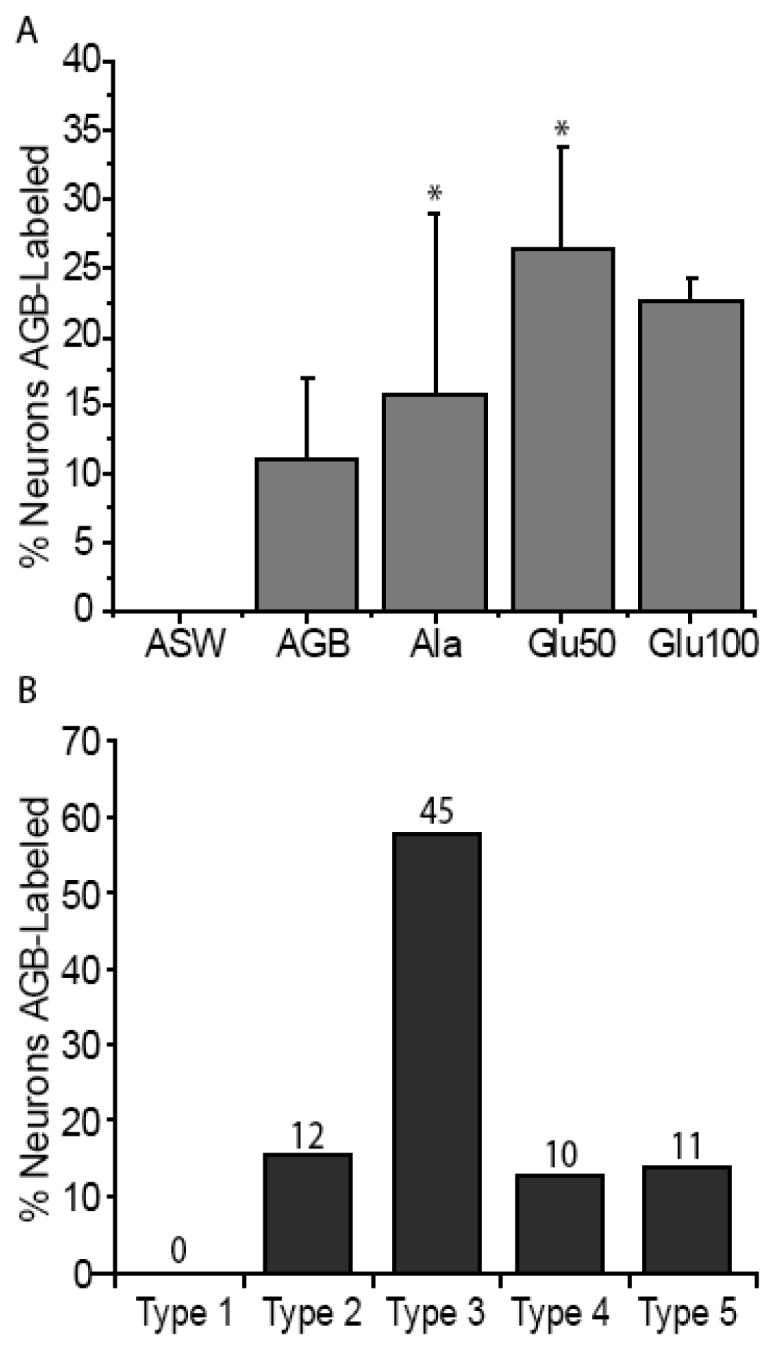
Odor responsiveness and proportions from 389 squid ORNs. (A) Odors induce different percentages of AGB labeling; the mean ± S.E. percentages of the 389 identified ORNs labeled with AGB in response to the odorant is shown. There is a significant association between odorant (Ala and Glu50) and AGB labeling (asterisks; p < 0.05 chi-squared test). For each condition n = 3 olfactory organs. (B) The numbers indicate the AGB-labeled neurons within each type. The total number of cells for each cell type in B is divided by the 78 cells labeled. Type 3 cells had the highest percentage of AGB labeling in response to odorant stimulation.
An analysis of the AGB labeling of all 389 identified neurons, regardless of odorant or application paradigm showed that type 3 cells had the largest proportion of labeled cells (58%; Fig. 4B). These data suggest that type 3 ORNs were the most broadly tuned to the panel of odorants tested. Types 2, 4, and 5 showed some responsiveness to the odorants tested however we did not find any responsive type 1 cells.
We asked whether odor is significantly associated with AGB labeling for each ORN type using the chi-squared test (Fig. 5). We were unable to find any type 1 ORNs that responded to AGB or the other odorants tested. However, due to the infrequency of type 1 ORNs, we cannot rule out their ability to respond to our odorant panel. Type 2 ORN exposure to 100 μM alanine or 50 μM glutamate was significantly associated with AGB labeling (p < 0.05) (Fig. 5A). Type 3 ORN exposure to every odorant tested was significantly associated with AGB labeling (p < 0.05) (Fig. 5B). Type 4 ORNs did not have a significant association between our tested odors and increased AGB labeling. Type 5 ORN exposure to AGB alone or 100 μM glutamate was significantly associated with AGB labeling (p < 0.05), exposure to100 μM alanine or 50 μM glutamate did not result in AGB labeling. While the type 5 cilia pocket has been described as obstructed (Emery, 1975), the presence of AGB labeling suggests that AGB gained access to the cilia pocket. Collectively, these data indicate that all four of the identified morphological ORN types differ from each other in their odor sensitivity.
Figure 5.
AGB labeling is significantly correlated with odor exposure. Shaded bars show the mean ± S.E. of identified neurons that were AGB-labeled in the presence of the stimulus. (A) Type 2 ORNs exposed to Ala and Glu50 have a significant association with AGB labeling. (B) Type 3 ORNs exposed to AGB, Ala, Glu50 and Glu100 have a significant association with AGB labeling. (C) Type 4 ORNs do not have a significant association with AGB labeling after exposure to the odorant panel. (D) Type 5 ORNs exposed to AGB and Glu100 have a significant association with AGB labeling. Asterisks indicate significant association between odorant and AGB labeling for that particular cell type at p < 0.05 (chi-squared test). For each condition n = 3 different olfactory organs (15 squid).
Role of adenylate cyclase in odor responsiveness
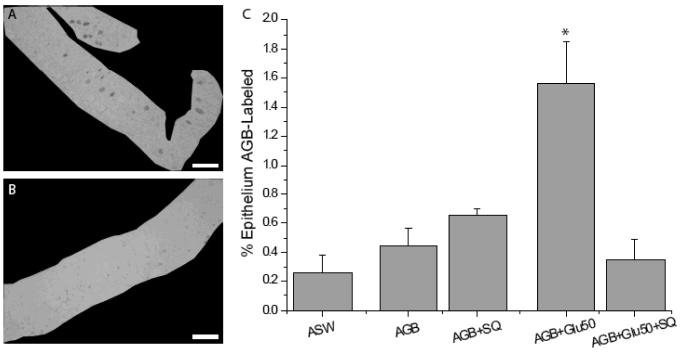
We showed previously that Gαs/olf is present in the cilia of squid ORNs (Mobley et al., 2007) suggesting a role for the AC pathway (Jones and Reed, 1989; Menco et al., 1992) in squid odorant transduction. To examine a functional role of the AC pathway in squid odor transduction, we measured the area of odorant-induced, AGB-labeled olfactory epithelium in the presence or absence of the adenylate cyclase inhibitor SQ22536 (SQ), and did not limit the analysis to identifiable cell types (Fig. 6A-B). Compared to ASW controls, AGB almost doubled the percentage of AGB labeling, however this increase was not statistically significant (0.26 ± 0.11% ASW versus 0.44 ± 0.12% AGB; ANOVA with posthoc Tukey HSD, p > 0.05; n = 3 sections from 3 squid for each condition: ASW, AGB, AGB + SQ, AGB + 50 μM glutamate, AGB + 50 μM glutamate + SQ) (Fig. 6C). Addition of 80 μM SQ to AGB treatment did not significantly alter the percentage of labeled epithelium compared to AGB alone (0.65 ± 0.04% AGB; p = 0.18). In the presence of AGB, 50 μM glutamate elicited a significant increase in the percentage of labeled epithelium (1.56 ± 0.28%; p < 0.05) (Fig. 6C) compared to AGB alone. The addition of 80 μM SQ to the 50 μM glutamate stimulation reduced the percentage of epithelial labeling (0.34 ± 0.14%; p= 0.02) to levels that were not significantly different from AGB alone (p = 0.99), AGB + SQ (p = 0.65), or ASW (p = 1.0). These data suggest that an AC pathway transduces the response to glutamate and that AGB enters glutamate-stimulated squid ORNs through channels gated either directly or indirectly by cAMP.
Figure 6.
Inhibition of the AC pathway reduces the percentage of glutamate-induced AGB labeling. (A) AGB immunostaining of squid OE exposed to 50 μM glutamate and 20 mM AGB is shown masked off to measure the epithelial area. (B) AGB labeling is minimal in squid OE exposed to 50 μM glutamate (Glu50), 20 mM AGB and 80 μm SQ22536 (SQ). (C) Summary of AC inhibitor experiments. AGB+Glu50 significantly increased AGB labeling compared to AGB alone. The presence of SQ significantly reduced AGB+Glu50-activated AGB labeling. The asterisk indicates significant difference from all other conditions (ANOVA with posthoc Tukey HSD; p < 0.05). For each condition n = 3 sections each from 3 different animals. Scale bars = 100 μm.
Discussion
We used histological and immunocytochemical techniques to investigate the distribution and odor sensitivity of ORNs in squid OE. Although our previous work showed that type 2 and type 4 neurons are odor sensitive, the current studies are the first to show odorant-induced activation of the type 3 ORNs. Interestingly, we found that although the type 2 ORNs were the most abundant, the type 3 ORNs were the most responsive to our panel of odorants using the AGB activity-dependent labeling method. We found that each ORN type has a different sensitivity to the odors we tested and that the odorant glutamate activates an AC dependent pathway.
Distribution of ORN types across the squid olfactory organ
We provide estimates of the abundance of each cell type in the squid OE using two different approaches, which together, overcame the disadvantages associated with each approach. We counted every nucleus and identifiable cilia pocket in every serial section of a squid olfactory organ. Using H&E staining it is not possible to determine whether a nucleus belongs to a type 1, type 2 or support cell. Immunohistochemistry against a set of small metabolites provided a snapshot of all cells of the tissue at the time of fixation, an advantage over using an antibody to one single protein. A combination of morphology, location of cilia pocket or soma in the epithelium and metabolite profiles identified the types of cells present. However, a portion of each metabolite-labeled image had regions where there were no distinguishable cell types (asterisks in Fig. 2A). Comparison with our H&E preparations indicated that the indistinct regions of metabolite-labeled tissue were the somas from the epithelial support cells. Thus, by combining immunohistochemistry and histology, we were able to identify and quantify the ORN types and their distributions in the squid olfactory organ.
The ORNs with cilia pockets, (types 3, 4, and 5), make up ∼20% of the cells in the squid olfactory organ, while the estimated total ORNs account for ∼34% of cells in the squid olfactory organ. These percentages likely underestimate the number of neurons because it is more accurate to identify nuclei than cilia pockets and we did not mark a cilia pocket unless clearly identified. In addition, the number of type 2 ORNs was estimated from subsampling sections of OE and any variations in type 2 ORN densities across the entire olfactory organ were not considered in the analysis. We also determined the relative percentages of ORN types containing cilia pockets compared to all cells in an entire olfactory organ and found that types 3, 4 and 5 have similar numbers (7%, 7% and 6% respectively; Fig. 2D). Thus, the higher percentage of odor responsive type 3 neurons is not due to a higher percentage of type 3 cells compared to types 4 and 5 (Fig. 2B).
Odorant sensitivity and signaling of squid ORNs
The responses of individual ORNs to odor stimulation were measured using the activity dependent marker, AGB. A few cells of each ORN type were labeled when exposed to AGB alone. This suggests that either AGB is an odorant specifically recognized by subsets of cells within each ORN type or that the AGB molecule enters these cells through some non-specific mechanism. The observation that only a small fraction of the cells within any ORN type were labeled by AGB suggests that labeling is a result of AGB activating odorant receptors (Ressler et al., 1993;Vassar et al., 1994;Lipschitz and Michel, 1999). Although AGB itself may act as an odorant, the addition of glutamate (50 μM) or alanine significantly increased the AGB labeling of ORNs, indicating that cells responded to AGB, amino acid or both. Exposure to 100 μM glutamate did not significantly increase labeling possibly due to receptor desensitization before enough AGB entered the cells. Indeed, whether we averaged the number of labeled cells for the various stimulation conditions (Fig. 5B) or measured the relative area of labeled epithelium (Fig. 6C), we observed significant increases in AGB labeling in the presence of alanine or glutamate compared to AGB alone. Thus, the AGB labeling technique allows visualization of odorant-stimulated cells among all of the identified cell types in a section of OE.
Using a limited odorant set, we have demonstrated that ORN types 2-5 have unique but overlapping odorant specificity. Detailed analysis of identified ORN types showed variability in odor-stimulated AGB labeling (Fig. 5). For example, type 5 was the only type that did not show AGB labeling after exposure to alanine, suggesting that at least the type 5 ORNs express different odorant receptors than types 2, 3, and 4. In the presence of alanine, very few type 2 ORNs were AGB-labeled. Type 3 ORNs were not the most abundant cells in the OE, however, they showed a larger number of AGB labeled cells than the other ORN types (Fig. 2D, 4B). This observation suggests a greater sensitivity of type 3 cells to the odorants tested, which may be due to either higher receptor expression or a large population of type 3 cells with receptors for the odorants tested. Type 4 ORNs did not show significant increases in AGB labeling due to AGB alone or additional odors (Fig. 5C). However, calcium imaging or patch clamp studies have shown that type 4 ORNs respond to filtered squid ink, 5-10 mM betaine, 50 μM and 10 mM dopamine, 10 mM L-Dopa, 10 mM caffeine (a component of squid ink) and 100 μM-100 mM glutamate (Lucero et al., 1992;Piper and Lucero, 1999;Danaceau and Lucero, 2000). It is possible that the slower stimulation protocol used in AGB studies precluded observation of the rapidly desensitizing glutamate responses observed in patch clamp studies of type 4 cells.
ORN morphologies are consistent with multiple transduction pathways
The observation that morphologically different types of squid ORNs respond to the same odorants is similar to the result of correlational studies on fish ORNs, in which the morphologically distinct microvillar and ciliated cells respond to amino acids, yet differ in their specificity for nucleotides and bile acids (Hansen et al., 2003;Hansen et al., 2004). Interestingly, ciliated and microvillar mammalian ORNs can also respond to the same odorants (Elsaesser et al., 2005), and mouse ORNs have been demonstrated to respond to pheromones, as well as volatile odorants (Spehr et al., 2006). These data suggest that different cell morphologies may express similar classes of odorant receptors however there is evidence for differential expression of the transduction pathways among the ORN types.
Previous work in our lab has shown that in squid, type 3, 4 and 5 ORNs express both Gαolf and Gαq, while the type 2 ORNs express Gαq and PLC140 (Mobley et al., 2007). Our data is consistent with other studies showing a correlation between the structure of the cell and transduction protein expression (Hansen et al., 2004;Hansen et al., 2005). For example, in catfish, ciliated ORNs express Gαolf, microvillar ORNs express Gαq, and the crypt cells express Gαo (Hansen et al., 2003).
The AC and PLC pathways have been established as signal transduction pathways in the olfactory systems of mammals (Boekhoff et al., 1990;Breer et al., 1990;Ronnett et al., 1993), amphibians (Nakamura and Gold, 1987;Schild and Lischka, 1994), fish (Restrepo et al., 1990;Miyamoto et al., 1992) and crustaceans (Michel and Ache, 1992;Fadool and Ache, 1992). We report here evidence for the AC pathway in the ORNs of squid. The AC inhibitor SQ significantly reduced glutamate-stimulated AGB labeling. In other olfactory systems, activation of AC increases cAMP which opens a calcium permeant cyclic nucleotide gated (CNG) channel (Nakamura and Gold, 1987;Bruch and Teeter, 1990;Kolesnikov et al., 1990;Firestein et al., 1991a;Firestein et al., 1991b;Zufall et al., 1994). However, as there is no evidence that AGB permeates CNG channels (Michel, 1999), the entering calcium may be activating an AGB-permeant non-selective cation channel. In the absence of glutamate, AGB-stimulated labeling was not significantly reduced by SQ suggesting that AGB does not activate the AC pathway. Previous work suggests that the PLC pathway is also present in squid ORNs (Lucero and Piper, 1994;Mobley et al., 2007) therefore, we hypothesize that AGB is activating the PLC pathway. Collectively, these data suggest that squid ORNs use both the AC and PLC pathways. Our findings show that squid are a useful model system for studying olfactory signal transduction and mixture interactions.
Acknowledgements
Thanks to F. Burki for help processing and analyzing the H&E tissue, and Drs. B. Jones and R. Marc for valuable technical advice. This research was funded by NRSA 1 F31 DC006793-01 to ASM and NIH NINDS P01 NS07938 to MTL and WCM.
Footnotes
Associate Editor: Kurt Albertine, University of Utah
Reference List
- 1.Acosta ML, Fletcher EL, Azizoglu S, Foster LE, Farber DB, Kalloniatis M. Early markers of retinal degeneration in rd/rd mice. Mol Vis. 2005;11:717–728. [PubMed] [Google Scholar]
- 2.Anraku K, Ohta M, Sasaki T, Marui T, Kawamura G. The effect of amino acids on feeding behavior of cuttlefish Sepia esculenta. Jpn J Taste Smell Res. 1998;5:519–522. [Google Scholar]
- 3.Boal JG, Golden DK. Distance chemoreception in the common cuttlefish, Sepia officinalis (Mollusca, Cephalopoda) J Exp Mar Biol Ecol. 1998:3226–3236. [Google Scholar]
- 4.Boekhoff I, Strotmann J, Raming K, Tareilus E, Breer H. Odorant-sensitive phospholipase C in insect antennae. Cellular Signalling. 1990;2:49–56. doi: 10.1016/0898-6568(90)90032-6. [DOI] [PubMed] [Google Scholar]
- 5.Boyle PR. Ventilation rate and arousal in the octopus. J Exp Mar Biol Ecol. 1983;69:129–136. [Google Scholar]
- 6.Breer H, Boekhoff I, Tareilus E. Rapid kinetics of second messenger formation in olfactory transduction. Nature. 1990;345:65–68. doi: 10.1038/345065a0. [DOI] [PubMed] [Google Scholar]
- 7.Bruch RC, Teeter JH. Cyclic AMP links amino acid chemoreceptors to ion channels in olfactory cilia. Chem Senses. 1990;15:419–430. [Google Scholar]
- 8.Danaceau JP, Lucero MT. Betaine activates a hyperpolarizing chloride conductance in squid olfactory receptor neurons. J Comp Physiol [A] 1998;183:225–235. doi: 10.1007/s003590050250. [DOI] [PubMed] [Google Scholar]
- 9.Danaceau JP, Lucero MT. Mixture interactions of glutamate and betaine in single squid olfactory neurons. Journal of Comparative Physiology A. 2000;186:57–67. doi: 10.1007/s003590050007. [DOI] [PubMed] [Google Scholar]
- 10.Edwards JG, Michel WC. Odor-stimulated glutamatergic neurotransmission in the zebrafish olfactory bulb. J Comp Neurol. 2002;454:294–309. doi: 10.1002/cne.10445. [DOI] [PubMed] [Google Scholar]
- 11.Elsaesser R, Montani G, Tirindelli R, Paysan J. Phosphatidyl-inositide signalling proteins in a novel class of sensory cells in the mammalian olfactory epithelium. European Journal of Neuroscience. 2005;21:2692–2700. doi: 10.1111/j.1460-9568.2005.04108.x. [DOI] [PubMed] [Google Scholar]
- 12.Emery DG. The histology and fine structure of the olfactory organ of the squid Lolliguncula brevis Blainville. Tissue & Cell. 1975;7:357–367. doi: 10.1016/0040-8166(75)90011-7. [DOI] [PubMed] [Google Scholar]
- 13.Emery DG. Fine structure of olfactory epithelia of gastropod molluscs. Microsc Res Tech. 1992;22:307–324. doi: 10.1002/jemt.1070220402. [DOI] [PubMed] [Google Scholar]
- 14.Fadool DA, Ache BW. Plasma membrane inositol 1,4,5-trisphosphate-activated channels mediate signal transduction in lobster olfactory receptor neurons. Neuron. 1992;9:907–918. doi: 10.1016/0896-6273(92)90243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firestein S, Darrow B, Shepherd GM. Activation of the sensory current in salamander olfactory receptor neurons depends on a G protein-mediated cAMP second messenger system. Neuron. 1991a;6:825–835. doi: 10.1016/0896-6273(91)90178-3. [DOI] [PubMed] [Google Scholar]
- 16.Firestein S, Zufall F, Shepherd GM. Single odor-sensitive channels in olfactory receptor neurons are also gated by cyclic nucleotides. J Neurosci. 1991b;11:3565–3572. doi: 10.1523/JNEUROSCI.11-11-03565.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilly WF, Lucero MT. Behavior responses to chemical stimulation of the olfactory organ in the squid Loligo opalescens. Journal of Experimental Biology. 1992;162:209–229. [Google Scholar]
- 18.Hansen A, Anderson KT, Finger TE. Differential distribution of olfactory receptor neurons in goldfish: Structural and molecular correlates. Journal of Comparative Neurology. 2004;477:347–359. doi: 10.1002/cne.20202. [DOI] [PubMed] [Google Scholar]
- 19.Hansen A, Rolen SH, Anderson K, Morita Y, Caprio J, Finger TE. Correlation between olfactory receptor cell type and function in the channel catfish. Journal of Neuroscience. 2003;23:9328–9339. doi: 10.1523/JNEUROSCI.23-28-09328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen A, Rolen SH, Anderson K, Morita Y, Caprio J, Finger TE. Olfactory receptor neurons in fish: structural, molecular and functional correlates. Chem Senses. 2005;30(Suppl 1):i311. doi: 10.1093/chemse/bjh239. [DOI] [PubMed] [Google Scholar]
- 21.Kolesnikov SS, Zhainazarov AB, Kosolapov AV. Cyclic nucleotide-activated channels in the frog olfactory receptor plasma membrane. FEBS Lett. 1990;266:96–98. doi: 10.1016/0014-5793(90)81515-p. [DOI] [PubMed] [Google Scholar]
- 22.Kuzirian A, Meyhofer E, Hill L, Neary JT, Alkon DL. Autoradiographic measurement of tritiated agmatine as an indicator of physiologic activity in Hermissenda visual and vestibular neurons. J Neurocytol. 1986;15:629–643. doi: 10.1007/BF01611862. [DOI] [PubMed] [Google Scholar]
- 23.Lipschitz DL, Michel WC. Physiological evidence for the discrimination of L-arginine from structural analogues by the zebrafish olfactory system. J Neurophysiol. 1999;82:3160–3167. doi: 10.1152/jn.1999.82.6.3160. [DOI] [PubMed] [Google Scholar]
- 24.Lipschitz DL, Michel WC. Amino acid odorants stimulate microvillar sensory neurons. Chem Senses. 2002;27:277–286. doi: 10.1093/chemse/27.3.277. [DOI] [PubMed] [Google Scholar]
- 25.Lucero MT, Horrigan FT, Gilly WF. Electrical responses to chemical stimulation of squid olfactory receptor cells. Journal of Experimental Biology. 1992;162:231–249. [Google Scholar]
- 26.Lucero MT, Huang W, Dang T. Immunohistochemical evidence for the Na+/Ca2+ exchanger in squid olfactory neurons. Philosophical Transactions of the Royal Society of London, B. 2000;355:1215–1218. doi: 10.1098/rstb.2000.0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucero MT, Piper DR. IP3 and cyclic nucleotides elicit opposite membrane potential changes in squid olfactory receptor neurons. Chemical Senses. 1994;19(5):509. [Google Scholar]
- 28.Marc RE. Kainate activation of horizontal, bipolar, amacrine, and ganglion cells in the rabbit retina. J Comp Neurol. 1999a;407:65–76. [PubMed] [Google Scholar]
- 29.Marc RE. Mapping glutamatergic drive in the vertebrate retina with a channel-permeant organic cation. J Comp Neurol. 1999b;407:47–64. doi: 10.1002/(sici)1096-9861(19990428)407:1<47::aid-cne4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 30.Marc RE, Cameron D. A molecular phenotype atlas of the zebrafish retina. J Neurocytol. 2001;30:593–654. doi: 10.1023/a:1016516818393. [DOI] [PubMed] [Google Scholar]
- 31.Marc RE, Jones BW. Molecular phenotyping of retinal ganglion cells. J Neurosci. 2002;22:413–427. doi: 10.1523/JNEUROSCI.22-02-00413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marc RE, Jones BW. Phenotyping neurons with pattern recognition of molecular mixtures. Symposium on Signal Processing and its Applications. 2003:S1571. [Google Scholar]
- 33.Marc RE, Kalloniatis M, Jones BW. Excitation mapping with the organic cation AGB2+ Vision Res. 2005;45:3454–3468. doi: 10.1016/j.visres.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 34.Marc RE, Liu WL, Kalloniatis M, Raiguel SF, Van Haesendonck E. Patterns of glutamate immunoreactivity in the goldfish retina. J Neurosci. 1990;10:4006–4034. doi: 10.1523/JNEUROSCI.10-12-04006.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marc RE, Murry RF, Basinger SF. Pattern recognition of amino acid signatures in retinal neurons. J Neurosci. 1995;15:5106–5129. doi: 10.1523/JNEUROSCI.15-07-05106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marc RE, Murry RF, Fisher SK, Linberg KA, Lewis GP, Kalloniatis M. Amino acid signatures in the normal cat retina. Invest Ophthalmol Vis Sci. 1998;39:1685–1693. [PubMed] [Google Scholar]
- 37.Michel WC. Cyclic nucleotide-gated channel activation is not required for activity-dependent labeling of zebrafish olfactory receptor neurons by amino acids. Biol Signals Recept. 1999;8:338–347. doi: 10.1159/000014607. [DOI] [PubMed] [Google Scholar]
- 38.Michel WC, Ache BW. Cyclic nucleotides mediate an odor-evoked potassium conductance in lobster olfactory receptor cells. J Neurosci. 1992;12:3979–3984. doi: 10.1523/JNEUROSCI.12-10-03979.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michel WC, Steullet P, Cate HS, Burns CJ, Zhainazarov AB, Derby CD. High-resolution functional labeling of vertebrate and invertebrate olfactory receptor neurons using agmatine, a channel-permeant cation. J Neurosci Methods. 1999;90:143–156. doi: 10.1016/s0165-0270(99)00077-1. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto T, Restrepo D, Cragoe EJJ, Teeter JH. IP3 and cAMP-induced responses in isolated olfactory receptor neurons from the channel catfish. J Membrane Biol. 1992;127:173–183. doi: 10.1007/BF00231505. [DOI] [PubMed] [Google Scholar]
- 41.Mobley AS, Gandham M, Lucero MT. Evidence for multiple signaling pathways in single squid olfactory receptor neurons. J Comp Neurol. 2007;501:231–242. doi: 10.1002/cne.21230. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura T, Gold GH. A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature. 1987;325:442–444. doi: 10.1038/325442a0. [DOI] [PubMed] [Google Scholar]
- 43.Piper DR, Lucero MT. Calcium signalling in squid olfactory receptor neurons. Biological Signals and Receptors. 1999;8:329–337. doi: 10.1159/000014606. [DOI] [PubMed] [Google Scholar]
- 44.Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 45.Restrepo D, Miyamoto T, Bryant BP, Teeter JH. Odor stimuli trigger influx of calcium into olfactory neurons of the channel catfish. Science. 1990;249:1166–1168. doi: 10.1126/science.2168580. [DOI] [PubMed] [Google Scholar]
- 46.Ronnett GV, Cho H, Hester LD, Wood SF, Snyder SH. Odorants differentially enhance phosphoinositide turnover and adenylyl cyclase in olfactory receptor neuronal cultures. J Neurosci. 1993;13:1751–1758. doi: 10.1523/JNEUROSCI.13-04-01751.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakata Y, Olson JK, Michel WC. Assessment of neuronal maturation and acquisition of functional competence in the developing zebrafish olfactory system. Methods Cell Sci. 2003;25:39–48. doi: 10.1023/B:MICS.0000006852.16798.34. [DOI] [PubMed] [Google Scholar]
- 48.Schild D, Lischka FW. Amiloride-insensitive cation conductance in Xenopus laevis olfactory neurons: a combined patch clamp and calcium imaging analysis. Biophys J. 1994;66:299–304. doi: 10.1016/s0006-3495(94)80804-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubert SN, Houck LD, Feldhoff PW, Feldhoff RC, Woodley SK. Effects of androgens on behavioral and vomeronasal responses to chemosensory cues in male terrestrial salamanders (Plethodon shermani) Horm Behav. 2006;50:469–476. doi: 10.1016/j.yhbeh.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 50.Sitthichai AA, Lucero MT, Michel WC. A cross species comparison of metabolic markers in olfactory epithelium. Chem Senses. 2003;28:556. [Google Scholar]
- 51.Spehr M, Kelliher KR, Li XH, Boehm T, Leinders-Zufall T, Zufall F. Essential role of the main olfactory system in social recognition of major histocompatibility complex peptide ligands. J Neurosci. 2006;26:1961–1970. doi: 10.1523/JNEUROSCI.4939-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steinbusch HW, Verhofstad AA, Joosten HW. Localization of serotonin in the central nervous system by immunohistochemistry: description of a specific and sensitive technique and some applications. Neuroscience. 1978;3:811–819. doi: 10.1016/0306-4522(78)90033-7. [DOI] [PubMed] [Google Scholar]
- 53.Steinbusch HW, Verhofstad AA, Joosten HW. Antibodies to serotonin for neuroimmunocytochemical studies. J Histochem Cytochem. 1982;30:756–759. doi: 10.1177/30.8.6749970. [DOI] [PubMed] [Google Scholar]
- 54.Steullet P, Cate HS, Derby CD. A spatiotemporal wave of turnover and functional maturation of olfactory receptor neurons in the spiny lobster Panulirus argus. J Neurosci. 2000;20:3282–3294. doi: 10.1523/JNEUROSCI.20-09-03282.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 56.Wildenburg G, Fioroni P. Ultrastructure of the olfactory organ during embryonic development and at the hatchling stage of Loligo vulgaris Lam. J Ceph Biol. 1989;1:56–70. [Google Scholar]
- 57.Wirsig-Wiechmann CR, Houck LD, Feldhoff PW, Feldhoff RC. Pheromonal activation of vomeronasal neurons in plethodontid salamanders. Brain Res. 2002;952:335–344. doi: 10.1016/s0006-8993(02)03369-3. [DOI] [PubMed] [Google Scholar]
- 58.Woodhams PL, Messenger JB. A note on the ultrastructure of the Octopus olfactory organ. Cell Tiss Res. 1974;152:255–260. doi: 10.1007/BF00224699. [DOI] [PubMed] [Google Scholar]
- 59.Yoshikami D. Transmitter sensitivity of neurons assayed by autoradiography. Science. 1981;212:929–930. doi: 10.1126/science.6262911. [DOI] [PubMed] [Google Scholar]
- 60.Zufall F, Firestein S, Shepherd GM. Cyclic nucleotide-gated ion channels and sensory transduction in olfactory receptor neurons. Annu Rev Biophys Biomol Struct. 1994;23:577–607. doi: 10.1146/annurev.bb.23.060194.003045. [DOI] [PubMed] [Google Scholar]