Abstract
A common request of proteomics core facilities is protein identification. However, in some instances primary sequence information for the protein in question is not present in public databases. In other cases, the amino acid sequence of a protein may differ in some way from the sequence predicted from the gene sequence in a database as a result of gene mutation, gene splicing, and/or multiple posttranslational modifications. Thus, it may be necessary to determine the sequence of one or more peptides de novo in order to identify and/or adequately characterize the protein of interest. The primary goal of this study was to give participating laboratories an opportunity to evaluate their proficiency in sequencing unknown peptides that are not included in any published database. Samples containing 3–6 pmol each of five synthetic peptides with amino acid sequences that were not present in public databases were sent to 106 laboratories. One nonstandard amino acid was present in one of the peptides. From a comparison of the results obtained by different strategies, participating laboratories will be able to gauge their own capabilities and establish realistic expectations for the approaches that can be used for this determination.
Keywords: de novo peptide sequencing, post-translational modification, Edman sequencing, mass spectrometry
INTRODUCTION
Proteomics core laboratories are often presented with unknown proteins to be identified. Sometimes, these are not identifiable by commonly used strategies that involve proteolytic digestion, tandem mass spectrometry (MS) analysis, and database searching. There are several reasons why this approach might not be successful. The peptides derived from the protein might be modified in some way that is not being considered by the database search program being used, it might not have a required sequence characteristic (e.g., a C-terminal Lys or Arg from a tryptic digest), or it might come from an organism for which the primary sequence is not known. Sometimes a homologous protein can be identified, but this requires that the sequences have a sufficiently high degree of similarity. For example, if an unknown protein is 95% identical to a known one, there is approximately a 60% probability that a 20-residue peptide from the unknown protein will have at least one substitution compared to the corresponding known peptide—i.e., 1–(0.95)20. Alternative approaches may be required to obtain the needed sequence(s). The primary goal of the 2005 Association of Biomolecular Resource Facilities (ABRF) Proteomics Research Group (PRG) study was to give participating laboratories a chance to evaluate their capabilities in the following areas: (a) determination of peptide sequence; (b) identification of unusual amino acids; and (c) use of software to assist in the interpretation of de novo sequence data.
The sequences of the peptides synthesized for this study are shown in Table 1. No specific approaches for determining the sequences were recommended, although it was anticipated that tandem mass spectrometry and possibly Edman sequencing would be employed. Each of the laboratories that requested a sample was provided with a mixture consisting of 3–6 pmol each of the five synthetic peptides shown in Table 1; the sequences of these peptides were not present in any public database. The sample was supplied as a dried pellet that could be dissolved in most common aqueous solutions; one peptide (A1) proved somewhat difficult to dissolve. As with any “real-life” sample, there were minor contaminants present. There was either a Lys or an Arg at the C-terminus of each peptide, analogous to tryptic peptides; one peptide had a double “missed cleavage” and another contained two hydroxyproline (Hyp) residues. Participants were asked to return experimental evidence for each sequence they determined in addition to completing a Web-based questionnaire.
TABLE 1.
Amino Acid Sequences of the Five Peptides in the PRG05 Sample
| Peptide | No. | Mr (Da) | MH+ | Sequence |
|---|---|---|---|---|
| T50 | 1 | 1192.8276 | 1193.8349 | LGAILkkLIPk |
| A2 | 2 | 1395.6610 | 1396.6683 | AYTFNMGqHSLk |
| J1 | 3 | 1463.7665 | 1464.7738 | vYkPHypASHypSPvYk |
| A3 | 4 | 1504.7316 | 1505.7389 | GvPGADIFYEANPR |
| A1 | 5 | 2327.1340 | 2328.1413 | FPHvANSGEWPDLvYvvNER |
Monoisotopic mass values are listed.
Hyp, hydroxyproline; Mr, relative molecular mass.
METHODS
Synthesis
The peptides were synthesized and purified at the following locations: A1, A2, and A3 at the HHMI Mass Spectrometry Laboratory at University of California, Berkeley; T50 at the NYU Protein Chemistry Laboratory; and J1 at the Macromolecular Structure Facility, Michigan State University. The synthetic peptides were analyzed by reversed-phase high performance liquid chromatography (HPLC) and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) to verify purity.
Composition analysis
Amino acid analysis was conducted on small portions of A2, A3, and T50, individually dissolved in the appropriate volume of water to yield 1 mg/mL stock solutions. For each of these three peptides, 3 μL of the stock solution was added to an amino acid analysis tube. The blank contained 3 μL of 1% acetic acid. The samples were dried in a vacuum centrifuge, sealed, and analyzed in duplicate for amino acid content using a Waters AccQtag AAA column in conjunction with a Waters 2690 HPLC equipped with a Waters 2475 fluorometer.
Sample distribution
For distribution to requesting laboratories, the appropriate volume corresponding to 3–6 pmol of each peptide was added to a 0.5-mL polypropylene tube and the peptide mixture was dried in a vacuum centrifuge. Dried samples were sent to 76 laboratories in North America, 20 in Europe, and 10 in other countries.
RESULTS
Sequence data were submitted by 40 laboratories, corresponding to a return rate of 38%, which was similar to that of other recent PRG studies.1,2 A summary of the study results, organized according to instrument configuration and ionization method, is shown in Table 2. A compilation of all results received is shown in Table 3. The following approaches were used: MS alone (35); Edman degradation (1); Edman degradation plus MS (4).
TABLE 2.
Summary of Instrument Configuration and Ionization Mode Utilization
| Number of Laboratories | Average Score1 | |
|---|---|---|
| Mass analyzer2 | ||
| Single instrument used | ||
| q/TOF | 12 | 40.2 |
| Ion trap | 8 | 19.5 |
| TTOF | 7 | 42.5 |
| One or more instruments used3 | ||
| q/TOF + | 19 | 41.3 |
| Ion trap + | 14 | 26.7 |
| TTOF + | 9 | 45.4 |
| Ionization mode | ||
| ES | 21 | 33.2 |
| MALDI | 8 | 45.4 |
| ES and MALDI | 4 | 27.4 |
Average total score for all peptides analyzed in which the indicated instrument or ionization mode was used. The peptide score represents the sum of consecutive correct residues as follows: score = xc + yN + zM, where the number of consecutive correct residues starting at the C-terminus is xc, at the N-terminus is yN and in the middle is zM. Lack of differentiation between isobaric or nearly isobaric residues was scored as follows: Ile/Leu, 0.5; Gln/Lys, 0.5 Gln/Lys/Hyp, 0.3. Detailed results are shown in Table 3.
q/TOF, quadrupole time-of-flight; ion trap; 3D or linear trap; TTOF, tandem time-of-flight; ES, electrospray; MALDI, matrix-assisted laser desorption ionization time-of-flight mass spectrometry.
This category represents each instance of the use of the indicated instrument. A number of laboratories reported use of more than one mass spectrometer to generate sequence information; however, details were not provided about which specific instruments were used for each sequence analysis. For this table, if a specific instrument was listed, it was included in the appropriate category.
TABLE 3.
Summary of Results
| Identifier | Total Score | Peptide Sequence (first choice) and Score | |||
|---|---|---|---|---|---|
| Score | T50 | Score | A2 | ||
| 70.0 | 11.0 | LGAILkkLIPk | 12.0 | AYTFN MGqHSLk | |
| 13579A | 66.0 | 7.0 | kLILqkLIPk | 12.0 | AYTFNMGqHSLk |
| 72079 | 64.0 | 8.5 | (L/I)GA(L/I)(L/I)kk(L/I)(L/I)Pk | 11.5 | AYTFN MoxGqHS(L/I)k |
| 715 | 64.0 | 9.0 | LGAILkIqIPk | 12.0 | AYTFNMGqHSLk |
| 26019 | 62.3 | 7.5 | XGA(I/L)(I/L)(q/k)k(I/L)(I/L)Pk | 11.5 | AYTFNMGqHS(I/L)k |
| 65214 | 61.5 | 6.0 | kHyp(L/I)(L/I)kk(I/L)(I/L)Pk | 11.5 | AYTFNMoxGqHS(L/I)k |
| 46011 | 58.0 | 7.0 | LGALLkkLLPk | 12.0 | AYTFNMGqHSLk |
| 12800 | 52.5 | 4.5 | vvR(I/L)kkHypHypPk | 8.0 | (AY)TFEGMLHSLk |
| 78364 | 52.0 | 7.5 | (I/L)GA(I/L)(I/L)(k/q)(k/q)(I/L)(I/L)Pk | 9.5 | (AY)TFNMox(k/q G)HS(I/L) k |
| 51565 | 51.0 | 4.0 | kLLkHypkLLPk | 9.0 | HPTFNMGqHSHypk |
| 30109 | 48.8 | 5.0 | q(I/L/Hyp)(I/L/Hyp)Lkk(I/L/Hyp)(I/L/Hyp)Pk | 9.5 | PHTFNMGqHS(i/l)k |
| 11010 | 48.3 | 7.0 | LXAILkkLIDL | 10.0 | AYTFNMGqH(L/I)Sk |
| 47223 | 44.5 | 6.0 | ga(l/i)(l/i)psgag(l/i)(l/i)pk | 1.0 | ag(l/i)spgvsm(l/i)hpck |
| 55000 | 42.0 | 6.0 | kHyp(I/L)(I/L)kk(I/L)(I/L)Pk | 1.0 | YATFNMGqHS(I/L) k |
| 51952 | 41.0 | 2.0 | (k/q)(I/L)(I/L)(I/L)(k/q)(k/q)(I/L)(I/L)Pk | 11.0 | AYTFNMoxG(k/q)HS(I/L)k |
| 99999 | 41.0 | 5.0 | (I/L)(qk)(I/L)(I/L)(qk)(qk)(I/L)(I/L)Pk | 11.0 | AYTFNMoxG(q/k)HS(I/L)k |
| 73108 | 40.0 | 2.0 | kLLkLGALLPk | 11.0 | AYTFNMGkHSLk |
| 17999 | 40.0 | 6.0 | (I/L)GA(I/L)(I/L)(q/k)(I/L)AG(I/L/Hyp)Pk | 9.5 | YATFNMG(q/k)HSLk |
| 98166 | 38.0 | 7.5 | (I/L)GA(I/L)(I/L)(k/q)(k/q)(I/L)(I/L)Pk | 10.5 | AYTFNFG(k/q)HSHypk |
| 27406 | 38.0 | 7.0 | (I/L)GA(I/L)(q/k)(q/k)(I/L)(I/L)Pk | 11.0 | AYTFNMG(k/q)HS(L/I)k |
| 91741 | 34.5 | 3.5 | (q/k)(I/L)Rvvk(I/L)(I/L)P(q/k) | 8.5 | HPTFNMG(q/k)HS(I/L)(q/k) |
| 91573 | 34.0 | 1.0 | TFNMoxGqHS(I/L)k | 11.5 | AYTFNMoxGqHS(I/L)k |
| 70091 | 31.0 | 6.0 | LGAIk[467.2]Pk | 1.0 | [221.0]PA[427.0]NFTSk |
| 19351 | 30.0 | 0.0 | 6.0 | AYTFNM | |
| 27974 | 29.5 | 0.0 | [242.6](I/L)(q/k)(I/L)(q/k)(I/L)[356.5] | 12.0 | AYTFNMGqHSLk |
| 17017 | 26.0 | 4.5 | (q/k)Hyp(I/L)(q/k)(q/k)(I/L)(I/L)Pk-NH2 | ||
| 12144 | 25.0 | 0.0 | aygplvpvslppr | 2.0 | agplascppvyk |
| 32466 | 22.0 | 1.0 | kD(i/l)(i/l)qkasyk | 11.5 | AYTFNMGqHS(I/L)k |
| 78544 | 21.3 | 0.0 | vY(q/k)APS(L/I)SAPYR | 0.0 | [235]T(Mox/F)(114)(Mox/F)[185] |
| 1467 | 19.5 | 5.0 | lgalIkGA(L/I)(L/I)Pk | 1.0 | (L/I)(q/k)SHPSMNFTSk |
| 52104 | 19.5 | 1.0 | (I/L)PASHypSPvYk | ||
| 80053 | 19.5 | ||||
| 54321 | 18.5 | 0.0 | 0.0 | ||
| 87458 | 10.5 | 0.0 | (RD)(P q/k)L(F/Mox)YEAN[315.01] | 4.5 | (YA/FS/HP/Mc)T(Mox/F)NMG(q/k)H(EA/Tv/cP)k |
| 1605 | 8.8 | 0.0 | (k/q)Hyp(I/L)(I/L)G(k/q)AHyp(I/L)Pk | 6.5 | YATFNMNAS(I/L)k |
| 12345 | 5.0 | 0.0 | vXkPLAkHypIPvN | 5.0 | AYTFHypMIFHXLykr |
| 11747 | 1.0 | 1.0 | FSTFNMSYASMk | ||
| 7974 | 0.0 | 0.0 | kN(I/L/Hyp) | ||
| 49495 | 0.0 | ||||
| 47551 | 0.0 | 0.0 | vYkPHypASHypSPvYk(k) | ||
| 11089 | 0.0 | ||||
| Identifier | Total Score | Peptide Sequence (first choice) and Score | |||
| Score | J1 | Score | A3 | ||
| 70.0 | 13.0 | vYkPHypASHypSPvYk | 14.0 | GvPGADIFYEANPR | |
| 13579A | 66.0 | 13.0 | vYkPHypASHypSPvYk | 14.0 | GvPGADIFYEANPR |
| 72079 | 64.0 | 13.0 | vYkPHypASHypSPvYk | 13.5 | GvPGAD(L/I)FYEANPR |
| 715 | 64.0 | 13.0 | vYkPHypASHypSPvYk | 14.0 | GvPGADIFYEANPR |
| 26019 | 62.3 | 12.3 | vYkP(I/L/Hyp)ASHypSPvYk | 12.0 | GvPGADXFYEAGGPR |
| 65214 | 61.5 | 13.0 | vYkPHypASHypSPvYk | 11.5 | RPGAD(L/I)FYEANPR |
| 46011 | 58.0 | 5.0 | vYkP(ps)S(tv/ea/cp/sl)(qc)qk | 14.0 | GvPGADIFYEANPR |
| 12800 | 52.5 | 11.0 | vykplaslspvyk | 13.0 | GvPGADLFYEANPR |
| 78364 | 52.0 | 7.0 | FD(k/q)P(I/L)AS(I/L)SPvYk | 10.5 | RPGAD(I/L)(F/Mox)YEANPR |
| 51565 | 51.0 | 9.0 | vYqPLASLSPvYk | 11.0 | RPGADLFYEANPR |
| 30109 | 48.8 | 12.0 | vYkPIAHypSP vYk | 12.0 | RPGADIFYEANPR |
| 11010 | 48.3 | 13.0 | vYkPHypASHypSPvYk | 12.0 | (vG)PGADIFYEANPR |
| 47223 | 44.5 | 7.0 | vy(k/q)p(l/i)apcpsvyk | 14.0 | GvPGADIFYEANPR |
| 55000 | 42.0 | 5.0 | k[134]kP(I/L)AS(I/L)SHS(cysP)k | 11.5 | vGPGAD(I/L)FYEANPR |
| 51952 | 41.0 | 12.0 | vY(q/k)PHypAS(l/Hyp)SPvYk | 7.0 | rP(k/q)DLFyEAnpR |
| 99999 | 41.0 | 11.0 | vYkP(I/L)AS(I/L)SPvYk | 11.0 | GvPGAD(I/L)FYcgPGPR |
| 73108 | 40.0 | 11.0 | vYkPASLSPvYk | 7.0 | GvPSLRFYEPGkR |
| 17999 | 40.0 | 11.0 | vYk P(I/L)AS(I/L)SvPYk | 13.5 | GvPGAD(I/L)FYEANPR |
| 98166 | 38.0 | 9.5 | vY(k/q)P(I/L)ASs(i/l)PvYk | 10.5 | RPGAD(I/L)FYEAggPR |
| 27406 | 38.0 | 8.5 | (Yv)(k /q)P(I/L)AS[279.12]vYk | 11.5 | vGPGAD(I/L)FYEANPR |
| 91741 | 34.5 | 3.5 | PHGvPIASPcPvY(q/k) | 14.0 | GvPGADIFYEANPR |
| 91573 | 34.0 | 10.0 | vYqP(I/L)AS(I/L)SpvYk | 11.5 | [156]PGAD(I/L)FYEANPR |
| 70091 | 31.0 | 10.0 | vYkPHypASHypGPk Yk | 14.0 | GvPGADIFYEANPR |
| 19351 | 30.0 | 12.0 | vYkPLASHypSPvYk | 12.0 | (vG)PGADIFYEANPR |
| 27974 | 29.5 | 6.0 | vYkP(I/L)AS(I/L)SHDPR | 9.5 | RP(q/k)D(I/L)FYEANPR |
| 17017 | 26.0 | 13.0 | vvYkPHypASHypSPvYk | 8.5 | RPqD(I/L)FYEANPR |
| 12144 | 25.0 | 11.0 | vYkPLASLSPvYk | 12.0 | gvpgadlfyeaggpr |
| 32466 | 22.0 | 1.0 | cmTFNkgfhsLk | 8.5 | RPqD(I/L)FYEANPR |
| 78544 | 21.3 | 9.8 | vY(q/k)P(I/L/Hyp)AS[200]PvYk | 11.5 | RPGAD(I/L)FYEANPR |
| 1467 | 19.5 | 1.0 | [649]vasapqdk | 4.0 | RPqD(I/L)FYEANPR |
| 52104 | 19.5 | 12.5 | vY(k/q)PHypASHypSPvYk | 6.0 | PqDLFYEAGGPR + neutral loss of 157 |
| 80053 | 19.5 | ||||
| 54321 | 18.5 | 8.0 | vYkP(L/I)ASHcRYk, +cys(ox) [+16,32,48] | 10.5 | RPGAD(L/I)FYEAGGPR |
| 87458 | 10.5 | 0.0 | [262.32](q/k)P(LA/PS)S(Tv/cD/LS/EA)[487.19] | 6.0 | (q/k P)DL(F/Mox)YEANPR |
| 1605 | 8.8 | 1.0 | vqNNMoxYEANPR | 1.3 | ED(I/L/Hyp)(k/q)TNHPk |
| 12345 | 5.0 | 0.0 | LXAIAkSLSEA | 0.0 | SPLvNDGqEXk |
| 11747 | 1.0 | ||||
| 7974 | 0.0 | ||||
| 49495 | 0.0 | 0.0 | (vY)(q/k)PSP(I/L)S(Pv)Y(k/q) | ||
| 47551 | 0.0 | ||||
| 11089 | 0.0 | ||||
| Identifier | Total Score | Peptide Sequence (first choice) and Score | Ionization Method | Ionization Type | |
| Score | A1 | ||||
| 70.0 | 20.0 | FPHvANSGEWPDLvYvvNER | |||
| 13579A | 66.0 | 20.0 | FPHvANSGEWPDLvYvvNER | MALDI | TTOF |
| 72079 | 64.0 | 17.5 | FPHvANSWWPD(L/I)vYvvNER | ES | q/TOF |
| 715 | 64.0 | 16.0 | FPHvANSWWPD(I/L)vYvv(k/q)DR | ES, E+ | q/TOF |
| 26019 | 62.3 | 19.0 | FPHvANSGEWPDXvYvvNER | ES | q/TOF |
| 65214 | 61.5 | 19.5 | FPHvANSGEWPD(L/I)vYvvNER | MALDI | q/TOF, TTOF, RTOF |
| 46011 | 58.0 | 20.0 | FPHvANSGEWPDLvYvvNER | MALDI | RTOF, TTOF (PSD) |
| 12800 | 52.5 | 16.0 | FPHvANSTTPDLvYvvG(GE)R | MALDI | TTOF |
| 78364 | 52.0 | 17.5 | (I/L)(M)HvANSGEWPD(I/L)vYvvNER | ES | LIT |
| 51565 | 51.0 | 18.0 | FPHvANSWWPDLvYvvNER | MALDI | TTOF, PSD |
| 30109 | 48.8 | 10.3 | [568.3]PSWWPD(I/L/Hyp)vYvvNER | MALDI | TTOF, q/TOF |
| 11010 | 48.3 | 5.3 | [235.19]fv[214.05]pfw[212.15](I/L/Hyp)vYvv[243.15]R | E+ | 3DIT, q/TOF |
| 47223 | 44.5 | 16.5 | FPHvanswadpd(l/i)vyvvnER | MALDI | TTOF (PSD) |
| 55000 | 42.0 | 18.5 | FPHvANSGEADPD(I/L)vYvvNER | ES, MALDI | LIT, q/TOF |
| 51952 | 41.0 | 9.0 | [938.57]WpdLvYvv[243.14]R | ES | q/TOF |
| 99999 | 41.0 | 3.0 | (cS)(I/L)NvvYv(I/L)DP[1110.5] | ES | q/TOF |
| 73108 | 40.0 | 9.0 | PDLvYGFFWPDLvYvvGWR | ES | q/TOF |
| 17999 | 40.0 | ES | q/TOF | ||
| 98166 | 38.0 | 0.0 | TFNFg(k/q)HSHypk | ES | 3DIT, q/TOF |
| 27406 | 38.0 | 0.0 | AYTFNMoxG(q/k)HS(L/I)k | ES | q/TOF |
| 91741 | 34.5 | 5.0 | FNFASEGWWLvLvYvvRDk | MALDI | TTOF |
| 91573 | 34.0 | 0.0 | T(I/L)(I/L)vNGvMYF[400] | ES | q/TOF, 3DIT |
| 70091 | 31.0 | 0.0 | [678.2]t[424.1]wegs(i/l/Hyp)av[381.3] | ES, E+ | LIT |
| 19351 | 30.0 | ES, E+ | q/TOF, LIT-FT | ||
| 27974 | 29.5 | 2.0 | (?)vvYv (I/L)DPW(?) | ES | q/TOF |
| 17017 | 26.0 | ES, MALDI, E+ | 3DIT, PSD | ||
| 12144 | 25.0 | ES | q/TOF | ||
| 32466 | 22.0 | 0.0 | ES, MALDI | 3DIT | |
| 78544 | 21.3 | ES | LIT | ||
| 1467 | 19.5 | 8.5 | RPqD(I/L)FYEANPR | ES | 3DIT |
| 52104 | 19.5 | ES, MALDI | q/TOF | ||
| 80053 | 19.5 | 19.5 | FPHvANSGEWPD(I/L)vYvvNER | ||
| 54321 | 18.5 | ES | q/TOF | ||
| 87458 | 10.5 | 0.0 | (YA/FS/HP/Mc)(F/Mox)AYYvLDPW[920.54] | MALDI | TTOF |
| 1605 | 8.8 | 0.0 | MoxDqPHypASAEDEDk | ES | LIT |
| 12345 | 5.0 | E+ | |||
| 11747 | 1.0 | ES | LIT | ||
| 7974 | 0.0 | ES | 3DIT | ||
| 49495 | 0.0 | ||||
| 47551 | 0.0 | ||||
| 11089 | 0.0 | ||||
The peptide score represents the sum of consecutive correct residues as follows: score = xc + yN + zM, where the number of consecutive correct residues starting at the C-terminus is xc; at the N-terminus, yN; and in the middle, zM. Lack of differentiation between isobaric or nearly isobaric residues was scored as follows: Ile/Leu, 0.5; Gln/ Lys, 0.5; Gln/Lys/Hyp, 0.3. The correct sequence is shown on the first results line. All methods and instruments used by a laboratory are listed together; in a few cases, different methods/instruments were used for different peptides. Groups that used Edman in addition to mass spectrometry are indicated by E+. The collision energy used depended on the instrument type and is not specified in the table. Some groups also used PSD and one used EcD, as noted. Additional details can be found at http://www.abrf.org/index.cfm/group.show/Proteomics.34.htm.
Abbreviations: 3DIT, 3-dimensional ion trap; E+, Edman used in addition to MS; EcD, electron capture dissociation; ES, electrospray; Hyp, hydroxyproline; LIT, IT-TOF, linear ion trap, ion trap/time-of-flight; LIT-FT, linear ion trap/Fourier transform hybrid; MALDI, matrix-assisted laser desorption ionization; Mr, relative molecular mass; Mox, oxidized Met; PSD, post-source decay; q/TOF, quadrupole/time-of-flight; RTOF, reflectron time-of-flight; TTOF, tandem time-of-flight.
The majority of laboratories reported the correct nominal peptide masses; peptide A2 was often found to contain an oxidized Met. Differences in sample preparation and use of derivatization prior to analysis did not seem to influence the success rate for sequencing, although one group used a variety of derivatization strategies and obtained the correct sequence for four of the five peptides.
Static nanoelectrospray worked as well as on-line fractionation by capillary HPLC. Laboratories using a tandem time-of-flight (TOF/TOF) mass spectrometer generally had a slightly higher success rate in obtaining the correct sequences for these peptides. These instruments typically use MALDI ionization; for this study it was not possible to assess the relative importance of ionization mode versus instrument type as related to the TOF/TOF results. In addition, the scores for laboratories reporting use of both an ion trap and another type of instrument were notably higher than those using a trap alone. Some level of manual interpretation was used by all laboratories; software alone did not appear to be sufficient to provide complete sequences. It is clear that there is a wide range of capabilities and levels of expertise among the participating laboratories. Moreover, it is important to note that the total number of responses was not very large. Therefore, it is not possible to formulate statistically rigorous conclusions about the capabilities of any specific approach or instrument used based on the results of this study.
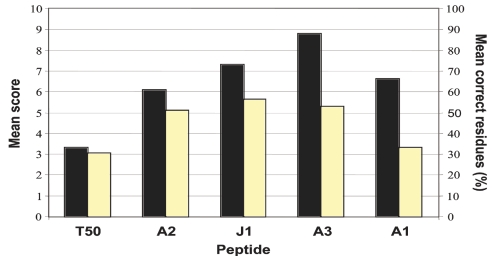
The success rates for sequencing the individual peptides varied (Table 2 and Figure 1). This is most likely due to differences in the sequences. The internal Lys residues combined with the multiple Leu and Ile (scored as 0.5 if not distinguished) undoubtedly contributed to the low scores for peptide T50. Peptide A1 was the longest and, therefore, expected to be more difficult.
FIGURE 1.
Success rate for individual peptides. Solid bars denote mean score obtained by all labs for a given peptide. Empty bars denote mean correct number of amino acid residues obtained by all labs for a given peptide.
DISCUSSION
The purpose of this study was to evaluate the capabilities of core laboratories to determine the sequences of peptides not found in any published database. Overall, the results show that this is an area that is difficult for many core laboratories. A sufficient amount of each of the peptides was supplied such that sample quantity should not have been a limitation (although solubility issues might have caused problems for sequencing of peptide A1). Peptides T50 and A1 were the most challenging, probably due to specific sequence features of those peptides.
In general, laboratories that reported using more than one type of instrument did slightly better than those that used only a single instrument. It is possible that facilities with multiple instruments might have a larger staff with more overall expertise. Too few cases in which Edman sequencing was used were reported to draw any conclusions. However, quantity limitations and time constraints made it generally less feasible to separate the peptides sufficiently for Edman analysis.
Although there are a variety of computer programs that are designed to perform de novo sequencing, the versions that were available at the time of this study did not appear to be capable of determining the sequences of the study peptides. The peptides used in this study were, by design, not naturally occurring sequences. In many “real” cases, a partial sequence obtained by mass spectrometry followed by database searching, even with errors in the partial sequence obtained by mass spectrometry, can be linked to a protein by a BLAST search. But that would require that a protein of sufficient homology be present in a published database. While that strategy would not be successful for the synthetic peptides provided in this study, it should be routinely considered.
It is clear that manual interpretation was necessary in order to determine the sequences of the peptides in this study. Commercially available instruments can usually provide sufficient tandem MS information to determine the sequences of most unknown peptides. However, it is critically important not only to acquire the spectra with the requisite mass accuracy and resolution, but also to be skilled in data interpretation. For example, there are two Hyp residues in peptide J1. The residue mass of Hyp (113.04768) is 36.4 mmu less than that of Leu/Ile (113.08406). Using some commercial instruments, it is possible to measure collision-induced dissociation fragment masses with sufficient accuracy to distinguish between these residues.
Finally, expertise in de novo sequencing is clearly essential, regardless of whether the data are acquired by mass spectrometry or Edman analysis or both. Whereas proteins that are present in a published database can be identified on a routine basis by scientists who are not experts in interpretation of mass spectra, the same cannot be said for proteins for which sequences are not included in any database. The results of this study provide excellent justification for core laboratories to have not only state-of-the-art instrumentation but also personnel with expertise in instrument operation and data analysis.
CONCLUSIONS
The average success rate in this study was relatively low, indicating that in 2005, most core laboratories did not have the capability to perform de novo sequencing. (Note that this study addressed issues that are very different from identifying a protein that is in a database.)
MALDI ionization and TOF/TOF mass analyzers appeared to be more successful than the alternatives, but too few laboratories participated in this study to reach any firm conclusions.
No individual sample preparation or derivatization strategy was notably more successful than others.
Laboratories that used more than one type of instrument were slightly more successful than those that only used a single type of instrument.
Software available in 2005 for de novo sequencing was not sufficient on its own for successful sequence analysis of the test peptides.
Expertise in MS and MS/MS data acquisition and manual interpretation was essential for success.
ACKNOWLEDGMENTS
We thank David S. King of the HHMI Mass Spectrometry Laboratory at the University of California, Berkeley, for synthesis and purification of peptides A1, A2, and A3; Joe Leykam at the Macro-molecular Structure Facility at Michigan State University for synthesizing peptide J1 and for the amino acid analyses; Ron Beavis and Janet Brostowin at the NYU Protein Chemistry Laboratory for the synthesis of peptide T50; Vivek Shetty, Chongfeng Xu, and Yun Lu of the NYU Protein Analysis Facility for mass spectrometry analysis of the samples; Dawn Maynard of the NIMH at the National Institutes of Health for mailing and receiving correspondence and for ensuring that the participants remained anonymous; and Debra Diana of the NYU Skirball Institute of Biomolecular Medicine for receiving confirmatory data.
REFERENCES
- 1.Arnott D, Gawinowicz MA, Grant RA, Neubert TA, Packman LC, Speicher KD. ABRF-PRG03: Phosphorylation site determination. J Biomol Tech. 2003;14:205–215. [PMC free article] [PubMed] [Google Scholar]
- 2.Arnott D, Gawinowicz MA, Kowalak JA, Lane WS, Speicher KS, Turck CW, et al. ABRF-PRG04: Differentiation of protein isoforms. J Biomol Tech. 2007;18:124–134. [PMC free article] [PubMed] [Google Scholar]