Abstract
In an effort to simplify and streamline compound identification from metabolomics data generated by liquid chromatography time-of-flight mass spectrometry, we have created software for constructing Personalized Metabolite Databases with content from over 15,000 compounds pulled from the public METLIN database (http://metlin.scripps.edu/). Moreover, we have added extra functionalities to the database that (a) permit the addition of user-defined retention times as an orthogonal searchable parameter to complement accurate mass data; and (b) allow interfacing to separate software, a Molecular Formula Generator (MFG), that facilitates reliable interpretation of any database matches from the accurate mass spectral data. To test the utility of this identification strategy, we added retention times to a subset of masses in this database, representing a mixture of 78 synthetic urine standards. The synthetic mixture was analyzed and screened against this METLIN urine database, resulting in 46 accurate mass and retention time matches. Human urine samples were subsequently analyzed under the same analytical conditions and screened against this database. A total of 1387 ions were detected in human urine; 16 of these ions matched both accurate mass and retention time parameters for the 78 urine standards in the database. Another 374 had only an accurate mass match to the database, with 163 of those masses also having the highest MFG score. Furthermore, MFG calculated a formula for a further 849 ions that had no match to the database. Taken together, these results suggest that the METLIN Personal Metabolite database and MFG software offer a robust strategy for confirming the formula of database matches. In the event of no database match, it also suggests possible formulas that may be helpful in interpreting the experimental results.
Keywords: LC/TOF-MS, compound, database, urine, identification
INTRODUCTION
Historically, researchers have used custom databases of known metabolites containing mass-only information to propose identities for ions observed from liquid chromatography mass spectrometry (LC-MS) experiments. The advent of accurate mass instrumentation has made these databases even more specific than when they had been used with nominal mass instruments.1–6 However, due to the presence of compound isomers, isobaric molecular formulas, and diastereomers, mass alone cannot be used as the sole parameter in the identification process. What is required is an orthogonal physical parameter to improve the specificity of the identification—either via chromatography and/or MS/MS. Since most metabolomics studies already use chromatography, the incremental cost of incorporating retention time (RT) into the database becomes negligible.
A prerequisite for identifying unknown compounds (such as metabolites) by MS is the availability of a correct elemental composition or molecular formula. Because accurate mass measurements alone are often not enough to conclusively determine the formula of unknown compounds,7 a limited number of data-processing algorithms have been written to help predict molecular formulas from mass spectra information. Most rely on isotope patterns, calculate the total number of possible formulas for a particular ion, and exclude formulas that violate particular chemical rules.8 An example of a highly effective approach is the filtering of formulas based on a set of “Seven Golden Rules”9 that the authors claim identifies the correct formula for compounds with a match in a database, as long as the mass measurements satisfy particular criteria: 3 ppm mass accuracy and 5% absolute isotope ratio deviation.
Because database searching typically uses only the value of the monoisotopic mass and ignores additional information contained in the spectra, such as naturally occurring isotope masses, the Agilent MassHunter Workstation software was developed to include a proprietary molecular formula generator (MFG) algorithm that takes advantage of both the mass accuracy and mass-spectral information to apply additional constraints on the list of candidate molecular formulas detected by mass spectrometry. This is achieved by incorporating monoisotopic mass, isotope abundances, and spacing between isotope peak information into its calculations. The software enables the user to define the type and number of allowed elements, and to set a mass error window. For each compound, a probability score is calculated that is based on how well the isotope abundance ratios for the candidate molecular formulas match those from the experimental data. This results in a shorter list of ranked candidate molecular formulas, with the top score (highest score = 100) being more likely to be correct, and therefore increases the value of the accurate-mass analysis.
Since the number of possible molecular formulas generated by MFG grows dramatically with increasing mass, selecting the correct formula becomes a progressively more difficult task. It is therefore particularly useful for lower-mass compounds (<200 Da), enabling the investigator to select from a relatively small number of possible formulas. If no database match occurs, the MFG proposed molecular formula and RT become starting points for further research. Hence, MFG reduces ambiguity and delivers a list of candidate molecular formulas with scores based on the relative probability that each formula is the correct one. This significantly reduces data interpretation time for large data sets and increases the value of accurate mass analysis. Together with RT information, it enables more confident association with results from the database matches.
METLIN is a Web-based database that has previously been developed by the Scripps Research Institute to facilitate the identification of metabolites using accurate mass data. It includes an annotated list of structural information for known metabolites. We have collaborated with the Scripps Research Institute to develop a METLIN Personal Metabolite Database that is based on content from METLIN. We have populated a subset of this database with RTs for 78 urine standards, where RT acts as an orthogonal and complementary physical parameter for querying the database, here referred to as the METLIN urine database. The goal of this proof-of-concept experiment was to improve tentative identification of compounds that had a METLIN urine database match, by (1) incorporating RT information for querying matches to 78 urine standards, and (2) relying on mass and MFG scores to determine the quality of the remaining hits. By also including MFG scores for each analyzed compound, this approach offers a more robust workflow for matching detected compounds to those residing in a personalized database.
MATERIALS AND METHODS
Standards
A mixture of 78 metabolite standards found in urine was kindly provided by Dr. Michael Reily at Eli Lilly & Co. (Indianapolis, IN) and was analyzed by LC/ MS and used for the construction of a small database of urine standards.
Samples
Human urine was collected from adult males. A 1-mL aliquot of urine was filtered through a Microcon (Millipore, MA) 10,000 nominal molecular weight limit membrane at 5000 × g; 100 μL of the filtered urine was dried in a SpeedVac and reconstituted in a solution of 0.1% formic acid/2% acetonitrile in MilliQ water.
Instrumentation
Chromatographic separation was achieved on a 2.1 × 150 mm, 3.5-μm particle size Zorbax SB-Aq column (Agilent Technologies, Santa Clara, CA). LC parameters: solvent A was 0.1% formic acid in water and solvent B was 0.1% formic acid in acetonitrile. The flow rate was 0.4 mL/min and the solvent gradient program was 2% B at time 0, 2% B at time 5 min, 60% B at 30 min, and 95% B at 30.1 min. Stop time was 35 min and the re-equilibration time was 10 min. The autosampler temperature was maintained at 4°C; the injection volume was 2 μL and column temperature was set at 20°C.
All samples were analyzed on a 1100 Series HPLC system with binary pump, degasser, thermostatted well plate autosampler, thermostatted column compartment, coupled with a 6210 MSD TOF mass spectrometer system with dual ESI source (Agilent Technologies), operated in the positive-ion mode. ESI capillary voltage was set at 4000 V and fragmentor at 170 V. The liquid nebulizer was set to 40 psig and the nitrogen drying gas was set to a flow rate of 10 L/min. The drying gas temperature was maintained at 250°C. The acquisition rate was 1.5 spectra/ sec and a stored mass range of m/z 50–1000.
Software
MassHunter Workstation Data acquisition software (Agilent Technologies) was used to operate the instrumentation. Data was processed using MassHunter Qualitative Analysis software (Agilent Technologies). Compounds were extracted from the raw data using the Molecular Feature Extraction (MFE) algorithm in Mass Hunter Qualitative analysis software. The samples were processed using MassProfiler software (Agilent Technologies) and compound identification was performed using the METLIN Personal Metabolite Database and Molecular Formula Generation software (Agilent Technologies).
Molecular feature extraction
The MFE algorithm is a compound finding technique that locates individual sample components (molecular features), even when chromatograms are complex and compounds are not well resolved. MFE locates ions that are covariant (rise and fall together in abundance) but the analysis is not exclusively based on chromatographic peak information. The algorithm uses the accuracy of the mass measurements to group related ions—related by charge-state envelope, isotopic distribution, and/or the presence of adducts and dimers. It assigns multiple species (ions) that are related to the same neutral molecule (for example, ions representing multiple charge states or adducts of the same neutral molecule) to a single compound that is referred to as a feature. Using this approach, the MFE algorithm can locate multiple compounds within a single chromatographic peak.
When using mass spectrometry to analyze samples containing unknowns, it is often necessary to derive elemental compositions (molecular formulas) for the unknowns based on the mass spectral data. The MassHunter MFG software uses a wide range of MS information, not just accurate mass measurements, to produce a list of candidate molecular formulas that are ranked according to their relative probabilities. The MFG software saves analysts considerable time because it eliminates unlikely candidates and delivers relative ranking for the remaining candidates, which makes it easier to find the correct formulas.
The MFG software uses a slightly different scoring system when it is used in conjunction with the MFE algorithm than when it is used on raw spectral data. MFE can locate multiple covariant species from the same feature, which creates additional information to be used in the determination of the molecular formula. This information is contained within adducts and in dimers (species) that are often produced by atmospheric-pressure ion sources. When MFE-reconstructed spectra are available, MFG software calculates an abundance-weighted, combined cross-species score for each molecular formula.
RESULTS AND DISCUSSION
Data analysis workflow
Once the samples were analyzed by LC/MS, MFE extracted the data into features and the calculated neutral mass was queried against the METLIN urine database of known compounds. Figure 1 shows the workflow for finding all features in LC/MS data, and how MFG was incorporated as an additional tool to help rank the database matches. The first step in the workflow used MFE to locate the ions in the raw data that were time covariant and that had logical mass relationships. They were assembled into distinct features, each feature containing data for the related ions, a single RT, and a total abundance value. An MHD file was created for each sample that contained a list of all the features. The second step in the workflow compared two sets of MHD files (i.e., two distinct samples from one or more conditions) in MassProfiler, where a list of differential features was produced. The calculated neutral mass of each feature in the list was subsequently queried against the METLIN urine database for matching to compounds falling within the user-adjusted mass tolerance window. The METLIN urine database matched the calculated neutral mass to the monoisotpic mass value calculated from the empirical formula of compounds in the database. Additional database specificity was then generated by entering the RTs for the set of 78 urinary metabolite standards. Feature lists of urinary metabolites were generated from a single synthetic urine mixture and separately, from two human urine samples, which were queried within specific RT and mass tolerance windows, against the METLIN urine database. A concurrent MFG calculation was performed for each mass within MassProfiler, using the full isotopic information from the mass spectral data to calculate possible empirical formulas within a maximum mass window of 750 Da. This helped with identifying a best molecular formula fit to the data. Finally, the database results and the MFG results were combined and aligned to produce a list of possible compounds that fit the observed data.
Figure 1.
Data processing workflow for compound finding by MFE, generation of MHD files for comparison of compounds between samples in MassProfiler, and a comparison of matched database results and their MFG scores. DB, database; MFE, Molecular Feature Extraction; MFG, Molecular Formula Generator.
Construction of a custom METLIN Personal Metabolite Database of urine standards with RT added
A mixture of 78 urine standards of varying concentrations was analyzed by LC/MS. The RT data corresponding to each monoisotopic mass were entered into the METLIN urine database (Figure 2). Once this process was completed, both the synthetic urine standards and the human urine samples were screened against it to find masses that had both mass and RT matches. We first screened the synthetic urine mixture to determine the number of individual synthetic standards that could be detected. Table 1 shows the MassProfiler results from LC/MS analysis of the synthetic urine standard mixture. We found that when we queried this database, 46 of the 78 synthetic standards were found in at least 50% of the 15 replicate (technical replicates) samples. We performed an extracted ion chromatogram on each of the standards to confirm the presence or absence of the peak at the specified RT, and then performed MFG analysis to confirm the presence of the isotopes, their abundances, and their empirical formulas. The reason for not detecting some of the standards was partly that their very low concentrations in the mixture were beyond the dynamic range (five orders of magnitude) of the TOF analyzer. Many of the hydrophilic standards (tyrosine, threonine, nicotinic acid, glycolic acid, hydroxyproline, salicylic acid, ethanolamine phosphate, phosphoenolpyruvate, mannitol, chenodeoxychloic acid, ATP, choline bilineurine, betaine) had little retention by the C-18-based SB-aq column. Consequently, failure to sufficiently retain compounds or to separate isomers reduced the identification discrimination power of this technique. Metabolite standards falling into this category require alternative separation strategies such as aqueous normal phase chromatography (research in progress).
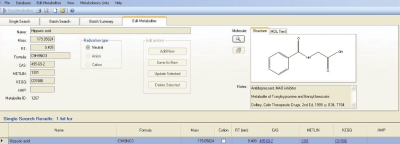
Figure 2.
The retention time for hippuric acid is added to the METLIN database by using the “edit metabolites” tab for this compound. The process was repeated for each of the 78 synthetic urine standards.
TABLE 1.
The List of 46 Synthetic Urine Standards That Were Detected in the Sample by LC/MS Analysis
| Mass | RT | Abundance | Name | Formula | CAS ID | METLIN ID | KEGG ID |
|---|---|---|---|---|---|---|---|
| 59.0378 | 1.320 | 9783 | Acetamide | C2H5NO | 60-35-5 | 3711 | |
| 75.0330 | 1.347 | 14150 | Glycine | C2H5NO2 | 56-40-6 | 20 | C00037 |
| 75.0690 | 1.062 | 3680487 | Trimethylamine N-oxide | C3H9NO | 1184-78-7 | 3773 | |
| 88.0170 | 1.282 | 63107 | Pyruvic acid | C3H4O3 | 127-17-3 | 117 | C00022 |
| 88.0536 | 3.757 | 4846123 | Isobutyric acid | C4H8O2 | 79-31-2 | 106 | C02632 |
| 89.0474 | 1.005 | 85230 | Sarcosine | C3H7NO2 | 107-97-1 | 51 | C00213 |
| 89.0480 | 1.148 | 538595 | Alanine | C3H7NO2 | 56-41-7 | 11 | C00041 |
| 90.0330 | 3.090 | 281672 | Lactic acid | C3H6O3 | 50-21-5 | 116 | C00186 |
| 92.0476 | 1.757 | 3639426 | Glycerol | C3H8O3 | 56-81-5 | 105 | C00116 |
| 103.0639 | 3.866 | 1002448 | Gamma-aminobutryic acid | C4H9NO2 | 56-12-2 | 279 | |
| 105.0429 | 1.203 | 10840 | Serine | C3H7NO3 | 56-45-1 | 30 | C00065 |
| 112.0277 | 2.314 | 4379190 | Uracil | C4H4N2O2 | 66-22-8 | 258 | |
| 113.0593 | 1.092 | 7502970 | Creatinine | C4H7N3O | 60-27-5 | 8 | C00791 |
| 115.0635 | 0.874 | 284075 | Proline | C5H9NO2 | 147-85-3 | 29 | C00148 |
| 116.0111 | 1.282 | 20984 | Fumaric acid | C4H4O4 | 110-17-8 | 3242 | C00122 |
| 118.0283 | 3.606 | 42884 | Methylmalonic acid | C4H6O4 | 516-05-2 | 3712 | |
| 126.0437 | 3.270 | 4199683 | Thymine | C5H6N2O2 | 65-71-4 | 290 | |
| 130.0635 | 4.047 | 63433 | 2-Oxoisocaproic acid | C6H10O3 | 816-66-0 | 121 | |
| 131.0697 | 1.145 | 6491475 | Creatine | C4H9N3O2 | 6020-87-7 | 7 | C00300 |
| 132.0535 | 1.001 | 138888 | Asparagine | C4H8N2O3 | 70-47-3 | 14 | C00152 |
| 132.0902 | 0.876 | 117379 | D-Ornithine | C5H12N2O2 | 6910 | ||
| 133.0375 | 1.027 | 216954 | Aspartic acid | C4H7NO4 | 56-84-8 | 15 | C00049 |
| 134.0218 | 1.284 | 521751 | Malic acid | C4H6O5 | 6915-15-7 | 118 | C00149 |
| 136.0412 | 3.607 | 10592900 | Hypoxanthine | C5H4N4O | 68-94-0 | 83 | C00262 |
| 136.0646 | 3.111 | 30958450 | n-Methylnicotinamide | C7H8N2O | 114-33-0 | 3770 | |
| 146.0211 | 1.374 | 59271 | 2-Ketoglutaric acid | C5H6O5 | 328-50-7 | 119 | C00026 |
| 146.0578 | 5.884 | 88422 | Adipic acid | C6H10O4 | 124-04-9 | 115 | |
| 146.1055 | 0.876 | 588966 | Lysine | C6H14N2O2 | 56-87-1 | 25 | C00047 |
| 152.0335 | 4.138 | 329319 | Xanthine | C5H4N4O2 | 69-89-6 | 82 | C00385 |
| 158.0444 | 1.181 | 248520 | Allantoin | C4H6N4O3 | 97-59-6 | 89 | C01551 |
| 160.0736 | 10.375 | 1365197 | 3-Methyladipic acid | C7H12O4 | 1-3-3058 | 3797 | |
| 160.0736 | 11.514 | 1043004 | Pimelic acid | C7H12O4 | 111-16-0 | 3280 | C02656 |
| 164.0480 | 16.733 | 756554 | 4-Hydroxycinnamic acid | C9H8O3 | 6450 | ||
| 166.0633 | 12.054 | 45156 | Phloretic acid | C9H10O3 | 501-97-3 | 4148 | C01744 |
| 168.0289 | 2.978 | 3916 | Uric acid | C5H4N4O3 | 69-93-2 | 88 | C00366 |
| 169.0847 | 0.993 | 189444 | N(pai)-Methyl-L-histidine | C7H11N3O2 | 368-16-1 | 3293 | C01152 |
| 174.0159 | 1.372 | 232344 | Aconitic acid | C6H6O6 | 499-12-7 | 3300 | C00417 |
| 175.0948 | 1.067 | 197980 | Citrulline | C6H13N3O3 | 372-75-8 | 16 | C00327 |
| 176.0323 | 1.388 | 4398341 | Ascorbic acid (vitamin C) | C6H8O6 | 50-81-7 | 249 | |
| 179.0586 | 9.460 | 54173080 | Hippuric acid | C9H9NO3 | 495-69-2 | 1301 | C01586 |
| 191.0583 | 11.753 | 5562405 | 5-Hydroxyindoleacetic acid | C10H9NO3 | 54-16-0 | 2975 | |
| 192.0269 | 1.361 | 1197306 | Isocitric acid | C6H8O7 | 320-77-4 | 3328 | C00311 |
| 194.0720 | 2.799 | 10371060 | Aminohippuric acid | C9H10N2O3 | 61-78-9 | 3927 | |
| 202.1207 | 20.024 | 6810013 | Sebacic acid | C10H18O4 | 111-20-6 | 4240 | C08277 |
| 204.0897 | 5.321 | 30498 | Tryptophan | C11H12N2O2 | 73-22-3 | 33 | C00078 |
| 226.0595 | 4.406 | 22633350 | 3-Nitrotyrosine | C9H10N2O5 | 6383 |
CAS ID, Chemical Abstracts Service identification number; RT, retention time.
Human urine analysis using mass, RT, and MFG
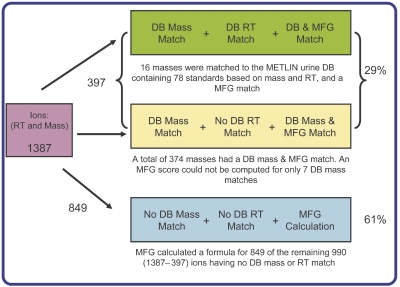
Four replicates, each of two individual human urine samples (A and B), were analyzed by LC/MS and processed in MFE. The resulting data were imported and combined into two projects in MassProfiler software, representing the two urine samples. A total of 1387 features, each having a minimum of at least two isotopes, was found to be present in all replicates in at least one of the two projects. This list of compounds was searched against the METLIN urine database using mass and RT matching. The database search results are summarized in Figure 3. A total of 397 masses (29% of total ions detected) matched the database within the previously specified tolerance windows. Sixteen of these compounds were detected in one of the two human urine samples that matched both the monoisotopic mass and RT of the standards in the database, and had an MFG score of 100 (maximum score is 100) matching the database formula. Another 374 compounds had both a database match and MFG score (50–100) calculated for them; 163 of these had an MFG score of 100, indicating that the mass match from the database correlated well with the isotope patterns for those masses, and hence greater confidence in the molecular formula. Nevertheless, without a RT to match, there is always uncertainty in the chemical identity. An MFG score could not be calculated for only 7 of the 397 masses. For the remaining 990 ions for which there was no mass match to the database, MFG could nevertheless calculate a score for 849 (61%) of them. Overall, MFG computed a score for 90% of the 1387 detected ions. This is encouraging because it implies that as the database is populated with increasing numbers of RTs, there will be this additional parameter, as well as MFG, to indicate how reliable a database match might be.
Figure 3.
A summary of the results for the number of urine metabolite masses detected in both human urine samples A and B that had a METLIN database mass match, RT match, and for which MFG calculation was performed. DB, database; MFG, Molecular Formula Generator; RT, retention time.
To evaluate whether more of the compounds in urine could be matched to the standards, the filtering parameters in MassProfiler were relaxed. This was achieved by: (a) requiring that a mass appear only in at least half (rather than all) the samples in each project, and (b) requiring a minimum of only one isotope for each mass. As expected, the number of compounds matching the standards in the database increased dramatically from 16 to 32. Table 2 shows a list of all compounds from human urine with abundance, mass, RT, and MFG score information that matched the urine standards in terms of mass and RT. Creatinine and uric acid, compounds that one expects to be abundant in urine, were present in both human urine samples A and B with MFG scores of 100.
TABLE 2.
MassProfiler List of Metabolites Detected in Human Urine Samples A and B That Matched the Synthetic Urine Standards in the METLIN Database
| Mass | RT | Name | Formula | Δ Mass (ppm) Urine ΔA | Δ Mass (ppm) Urine ΔB | MFG Score Urine ΔA | MFG Score Urine ΔB |
|---|---|---|---|---|---|---|---|
| 130.0291 | 1.602 | 1,1–Cyclopropanedicarboxylic acid | C5H6O4 | −12.2 | — | 90.2 | — |
| 146.0216 | 1.650 | 2-Ketoglutaric acid | C5H6O5 | 0.4 | 0.8 | 100.0 | 100.0 |
| 191.0579 | 11.837 | 5-Hydroxyi-oleacetic acid | C10H9NO3 | 2.1 | 1.7 | 100.0 | 100.0 |
| 174.0158 | 1.647 | Aconitic acid | C6H6O6 | 3.9 | 3.2 | 100.0 | 100.0 |
| 89.0473 | 1.151 | Alanine | C3H7NO2 | 2.5 | 0.4 | 100.0 | 100.0 |
| 194.0684 | 3.008 | Aminohippuric acid | C9H10N2O3 | 4.2 | 3.0 | 100.0 | 100.0 |
| 132.0524 | 1.376 | Asparagine | C4H8N2O3 | — | 8.2 | — | 100.0 |
| 175.0957 | 1.331 | Citrulline | C6H13N3O3 | 0.1 | 0.3 | 100.0 | 100.0 |
| 131.0704 | 1.150 | Creatine | C4H9N3O2 | −6.0 | −5.7 | 98.3 | 98.7 |
| 113.0590 | 1.161 | Creatinine | C4H7N3O | −1.5 | 1.8 | 100.0 | 100.0 |
| 92.0495 | 1.300 | Glycerol | C3H8O3 | — | −22.4 | — | 69.5 |
| 75.0324 | 1.469 | Glycine | C2H5NO2 | — | −3.9 | — | 100.0 |
| 179.0582 | 9.622 | Hippuric acid | C9H9NO3 | −0.2 | −2.7 | 90.7 | 85.6 |
| 136.0380 | 3.650 | Hypoxanthine | C5H4N4O | 3.0 | 3.1 | 100.0 | 100.0 |
| 213.0089 | 6.186 | I-Oxylsulfuric acid | C8H7NO4S | 3.4 | 3.4 | 59.2 | 59.4 |
| 192.0265 | 1.647 | Isocitric acid | C6H8O7 | 2.6 | 1.1 | 97.4 | 100.0 |
| 146.1050 | 0.888 | Lysine | C6H14N2O2 | 2.4 | 3.0 | 100.0 | 98.1 |
| 182.0788 | 1.041 | Mannitol | C6H14O6 | 0.2 | 0.4 | 100.0 | 100.0 |
| 118.0261 | 3.647 | Methylmalonic acid | C4H6O4 | — | 4.2 | — | 100.0 |
| 169.0849 | 1.022 | N(pai)-Methyl-L-histidine | C7H11N3O2 | 1.2 | 1.2 | 100.0 | 100.0 |
| 189.0626 | 2.211 | N-Acetyl-L-glutamic acid | C7H11NO5 | 5.3 | 2.4 | 81.4 | 95.4 |
| 88.0163 | 1.225 | Pyruvic acid | C3H4O3 | — | −5.1 | — | 100.0 |
| 376.1378 | 14.419 | Riboflavin (vitamin B2) | C17H20N4O6 | 1.6 | — | 88.4 | — |
| 89.0479 | 1.035 | Sarcosine | C3H7NO2 | −2.5 | — | 100.0 | — |
| 118.0269 | 3.653 | Succinic acid | C4H6O4 | 2.2 | — | 100.0 | — |
| 126.0426 | 3.286 | Thymine | C5H6N2O2 | 4.5 | — | 100.0 | — |
| 75.0690 | 1.066 | Trimethylamine N-oxide | C3H9NO | −6.2 | −8.3 | 100.0 | 100.0 |
| 204.0890 | 5.358 | Tryptophan | C11H12N2O2 | 3.7 | 3.4 | 94.0 | 100.0 |
| 181.0725 | 2.326 | Tyrosine | C9H11NO3 | — | 9.1 | — | 100.0 |
| 168.0281 | 2.745 | Uric acid | C5H4N4O3 | 1.6 | 0.0 | 100.0 | 100.0 |
| 138.0415 | 2.415 | Urocanic acid | C6H6N2O2 | 10.4 | 11.3 | 96.6 | 65.9 |
| 152.0334 | 4.146 | Xanthine | C5H4N4O2 | 0.4 | 0.9 | 100.0 | 100.0 |
| 209.0687 | 3.841 | Methylsalicyluric acid | C10H11NO4 | 1.2 | −1.1 | 60.9 | 100 |
| 193.0738 | 10.956 | 2-Methylhippuric acid | C10H11NO3 | −3 | 0.3 | 63.9 | 62.3 |
| 364.2251 | 19.734 | Dihydrocortisol | C21H32O5 | 0.1 | 0 | 68.4 | 77.3 |
| 297.0892 | 11.729 | 5′-Methylthioadenosine | C11H15N5O3S | 0.5 | 1.6 | 66.8 | 64.5 |
In addition to a calculated MFG score, the observed mass (i.e., analyzed by LC/MS) and RT for each metabolite in urine samples A and B, as well as their differences (Δ) between the values for the standards in the METLIN database is shown. RT, retention time.
Although most compounds had an MFG score of 100, a few, such as indoxylsulfuric acid, had low MFG scores. A low MFG score may still be significant, as it is calculated based on mass spectral data for all samples in a project. So, while inspection of a single sample might yield a score of 100, and therefore signify compatibility with the database match, the score can be different when it is calculated for a group of replicate samples (in this case four), where the isotope information is scored differently. In situations where the MFG score is not 100 it is incumbent on the analyst to check the individual spectra to confirm the MFG result.
Human urine analysis using mass and MFG only
Table 2 also includes four examples (at the bottom of the table) of METLIN urine database matches for human urine samples A and B using only mass and MFG scoring (that is, compounds with database matches outside of the synthetic urine standards set). Based on mass information only, mass 209.0687 matched methylsalicyluric acid (molecular formula: C10H11NO4) in the database. Because of no corroborating RT information from a standard for this compound, to verify that methylsalicyluric acid indeed elutes at 3.841 min, we used the MFG calculated score, based on mass spectral data of the isotopes, to assist us in determining the validity of the database match. An MFG score of 100 was calculated for this feature in human sample A, but a score of only 60.9 was calculated for human sample B. Upon closer inspection of the MS spectrum of sample B (graphic zoomed in on the ion 210.07588) for the data at time 3.84 min (Figure 4), the reason for this is quite clear. An isotope distribution calculator for formula C10H12NO4 had predicted that in addition to the first isotope, m/z 210.07660, there exists a second expected isotope of m/z 211.07980 (data not shown). Since the predicted value of the second isotope is much smaller than the observed isotope of m/z 211.09232, it translated to a mass error (Δ ppm), that is greater than the allowable mass error window (> ±7.5 ppm). The software therefore assigned a lower MFG score for the database match (shown in a table as an inset of Figure 4) and also suggested an alternative formula with a higher MFG score. This example is an instance where the MFG score can be a valuable asset in assisting the researcher in determining the confidence to attach to a database match. This is all the more important, as in the case above, where the Δ ppm for the database match for the two urine samples was very good (<1.5 ppm).
Figure 4.
Results of METLIN database mass matching and MFG calculation showing incompatibility for methylsalicyluric acid.
Another example where MFG was useful in the interpretation of the database match was where the mass was found in both human urine samples, but was in disagreement with the database match. For example, Figure 5 shows that mass 364.2251 matched dihydrocortisol in the database to within 0.1 ppm. However, the MFG scores of 68.4 and 77.3 (see Table 2), which incorporate all the spectral data for this mass, indicated that there are uncertainties with this database match. The mass spectrum results at time 19.73 min for urine sample B (Figure 5) revealed an isotope distribution pattern that had a very good mass match to the empirical formula C21H33O5, with the observed errors for the three isotopes from the predicated masses being 0.06, 2.38, and 3.42 ppm respectively. However, the results of the MFG calculation showed that the calculated percent abundances for the second and third isotopes were sufficiently different from the observed data to result in it being ranked lower, despite the fact that all three isotopes had a low mass error. In this case, poorer isotope ratios were due to the weak analyte signal in the TOF detector. In summary, an analyst would likely conclude with a high degree of probability that, having considered the biological source of the samples, the results of the database matches, and MFG scores, dihydrocortisol had indeed been detected, and that injection of an authentic standard to verify the match would be warranted. It should be noted that the differences between MFG and database match will always be subtle, since any differences would have to occur within the user-assigned mass and RT tolerance windows.
Figure 5.
The result of MFG calculation based on the mass spectral data for dihydrocortisol is a ranked list of possible formulas.
CONCLUSIONS
Due to the analytical constraint that mass alone cannot unambiguously assign elemental composition, there is a need to complement database assignment of high mass accuracy data with other techniques such as isotope ratios and RT. Here, we have demonstrated the utility of METLIN Personal Metabolite Database software in assigning the correct elemental compositions for a set of urine metabolite standards. The ability to include RT as a separate, orthogonal variable permits rapid, positive identification of the temporally resolved masses. By also combining MFG capability with mass and RT database matching, the anticipated benefit is to increase the confidence with which both known and unknown compounds are assigned a correct elemental composition.
REFERENCES
- 1.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: A metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen KF, Smedsgaard J. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardised liquid chromatography–UV–mass spectrometry methodology. J Chromatogr A. 2003;1002:111–136. doi: 10.1016/s0021-9673(03)00490-4. [DOI] [PubMed] [Google Scholar]
- 3.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, et al. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 4.Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 5.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: Mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Gao W, Cai J, He S, Zeng R, Chen R. Predicting molecular formulas of fragment ions with isotope patterns in tandem mass spectra. IEEE/ACM Trans Comput Biol Bioinform. 2005;2:217–230. doi: 10.1109/TCBB.2005.43. [DOI] [PubMed] [Google Scholar]
- 9.Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMCBioinformatic s. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]