Unearthing the origin of incredible acceleration of chemical reaction rates by enzymes has occupied the minds of biochemists and molecular biologists for more than a century (1). The modern paradigm of enzymatic action rests on the ideas presented by Haldane and Pauling, namely, that enzymes selectively stabilize transition states in chemical reactions, thus lowering reaction activation energies (2). The widely held microscopic view of enzymatic catalysis envisions a nearly static enzyme reaction chamber carefully sculpted by evolution to match complementarily the shape and electrostatic surface of the reaction transition state. However, recent NMR experiments have suggested that enzyme active sites are mobile on the microsecond to second time scale, commensurate with the time scales for the corresponding catalytic reactions (3). Single-molecule fluorescent spectroscopy studies also point to nontrivial static and dynamic disorder in enzymatic processes, something that is often masked by ensemble averaging in macroscopic kinetic experiments (4). Even in the light of these findings, the central dogma of modern structural biology has remained largely intact; 3D protein structure determines its function. Indeed, this paradigm has worked wonderfully over many decades to explain numerous biological processes, from enzymatic catalysis to signal transduction. Recent works, however, suggested that from one-sixth to one-third of eukaryotic proteins are either disordered or contain large disordered regions (5, 6), hinting that the traditional interpretation of the structure–function paradigm may be too limiting (6). Disordered proteins regulate many transcriptional and signal transduction processes (7). They even exhibit enzymatic activity, as was recently demonstrated experimentally (8). How this is accomplished in the absence of prearranged 3D structure has not been well understood. In a recent issue of PNAS, Roca et al. (9) have used computer simulations to explain the way a molten globule can accelerate a chemical reaction. Their work, combined with recent advances in understanding the way protein folding and functional landscapes are coupled (6, 10), provide essential groundwork for generalizing the structure–function paradigm by using energy landscape ideas.
How can a disordered protein, which does not adopt any definite 3D structure, carry out specific biological function? In the case of protein–protein interactions, two possibilities come to mind. First, only a small segment of the protein is specifically recognized, based on a signal from a local sequence of amino acids. In a way, a protein segment behaves as a peptide. Alternatively, and quite commonly, a well defined 3D structure is involved in a binding interaction (7). However, without its binding partner, this structure is well disguised among a multitude of other structures that the molten globule ensemble explores, in liquid-like dynamics. This particular structure is nevertheless special. It is low in energy; thus, a funnel is protruded in protein's energy landscape because of structural correlations, similar to the energy landscape of globular proteins with well defined native state (11) (see Fig. 1 Left). In this case, however, the funnel is not deep enough to render the native state the thermodynamic global minimum (6) (see Fig. 1 Center). Instead, additional “pulling down” of the funnel's bottom is needed by specific high-affinity interfacial interactions with protein's binding partner (6). This coupling of folding (3D structure formation) and binding energy landscapes allows molten globules to specifically recognize structural partners (6, 10). Still, a free energy penalty needs to be paid during binding, to overcome the entropy of disordered states, requiring extra stabilization energy at the binding interface. Given this penalty, why would proteins evolve to become disordered? Various hypotheses have been put forward for the ubiquity of the unfolded states: rapid turnover, faster or more specific binding kinetics, and functional promiscuity (7). From a practical viewpoint, it is extremely difficult to either predict or experimentally determine what specific functional role is played by a particular disordered protein, because molten globule dynamics efficiently conceal which particular conformation is the special one. These difficulties are clearly exemplified by current lack of understanding of α-synuclein's function, a disordered protein that is implicated in Parkinson's disease (12).
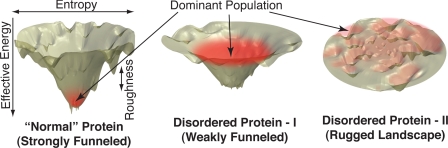
Fig. 1.
The interplay between protein's folding and functional landscapes. (Left) Energy landscapes of globular proteins are thought to be funneled, such that the native state is both a thermodynamic global minimum and is also kinetically accessible (11). It is a common practice to partition the total free energy into protein chain's structural entropy (horizontal axes) and the remaining part, which is often called “effective energy,” or simply energy (vertical axes). Note that the latter also includes entropic contributions, mainly from the solvent. (Center) Energy landscapes of many disordered proteins are likely organized around a special state, characterized by a weakly funneled landscape (6). However, the driving force for folding is too weak, allowing the protein to remain disordered. Interactions with specific targets create additional favorable contacts, deepening the funnel and driving subsequent folding (6). Transient population of catalytically competent states, near the funnel's bottom, may allow for efficient catalysis (9). (Right) A random energy landscape is shown, where search for a specific functionally competent conformation is extremely inefficient.
Could a molten globule also catalyze a chemical reaction? The answer turns out to be affirmative (8). Does a random energy landscape allow structurally specific catalysis? It is very unlikely, because it would take an astronomically long time for the protein to find the catalytically competent conformation, an argument similar to the necessity of having a folding funnel to overcome excessively long folding times (11) (see Fig. 1 Right). Thus, as in the case of coupling of binding and folding landscapes (6), a disordered enzyme's energy landscape must be weakly funneled. Pulling down of the enzyme's funnel upon transition state analogue binding, which is manifest as nudging the molten globule ensemble toward the native state, has been observed both experimentally and computationally (8, 9). On the other hand, because enzyme substrates are typically small, this effect might be less pronounced compared with more extensive protein–protein interfaces. Roca et al. (9) have elucidated the way a molten globule acts as a catalyst for a genetically engineered version of chorismate mutase from Methanococcus jannaschii, transforming chorismate to prephenate (9). Using multiscale modeling, they discovered that activation barriers are low for several conformational basins within the native superbasin, which provide equivalent preorganization of the active site. The reaction barriers were found to be high for nonnative conformations.
For a weakly funneled protein, reaching the catalytically competent conformational manifold may cost some free energy, mainly to overcome excess structural entropy. The probability of observing such a conformation may be so small that these conformations are virtually invisible to spectroscopic interrogation. However, if catalytically induced speed-up for the competent conformation is very large, the molten globule will still exhibit enzymatic activity, perhaps somewhat reduced. In this light, would it make sense for nature to use molten globules for catalysis if it results in a potential slowdown? We can only speculate at this point. For example, extremely efficient control of the catalytic rate may be achieved by posttranslational modifications or single point mutations that effect the native state's metastability by only 1–2 kcal/mol. Furthermore, molten globular proteins are quickly degraded by the proteasome, allowing finer temporal control over the catalytic process. This would be useful if only a short burst of catalytic activity is desired.
Disordered proteins regulate many transcriptional and signal transduction processes.
High-resolution structural biology techniques, such as x-ray crystallography, have taught us a tremendous amount about the mechanisms of enzymatic reactions. It is clear, however, that complementary techniques, both experimental and computational, are needed to study proteins that are not characterized by a well defined structure. Above, I provided an argument that even for many of these proteins there actually exists a special conformation at low energy, which induces weak funneling in the energy landscape. However, if we are given a specific disordered protein with some function, it might be incredibly difficult to determine which conformation is functionally competent among a myriad of other states that the globular dynamics explores. For more ordered, yet still flexible enzymes, it will be interesting to systematically probe their catalytic landscapes (9), perhaps based on a precomputed hierarchy of native-like states (13), to gain deeper insights in to the way enzyme folding (14) and dynamics modulates the kinetics of catalysis. In summary, because disordered proteins play an important role in eukaryotic biology and human health, controlling numerous transcriptional and signal transduction processes, more significant future effort is needed to understand how their energy landscapes determine their function.
Acknowledgments.
My research is supported by National Science Foundation Award CHE-715225, the Arnold and Mabel Beckman Foundation Beckman Young Investigator Award, the Camille and Henry Dreyfus Foundation, and Petroleum Research Fund Award 47593-G6.
Footnotes
The author declares no conflict of interest.
See companion article on page 13877 in issue 37 of volume 105.
References
- 1.Fischer E. Über die optischen Isomeren des Traubenzuckers, der Gluconsäure und der Zuckersäure [Concerning the optical isomers of the grape sugars, gluconic acid and saccharic acid] Ber Dtsch Chem Ges. 1890;23:2611–2620. [Google Scholar]
- 2.Fersht A. Structure and Mechanism in Protein Science. New York: Freeman; 1999. [Google Scholar]
- 3.Henzler-Wildman KA, et al. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 4.Min W, et al. Fluctuating enzymes: Lessons from single-molecule studies. Acc Chem Res. 2005;38:923–931. doi: 10.1021/ar040133f. [DOI] [PubMed] [Google Scholar]
- 5.Romero P, et al. Sequence complexity of disordered proteins. Proteins Struct Funct Genet. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Papoian GA, Wolynes PG. The physics and bioinformatics of binding and folding: An energy landscape perspective. Biopolymers. 2003;68:333–349. doi: 10.1002/bip.10286. [DOI] [PubMed] [Google Scholar]
- 7.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 8.Pervushin K, Vamvaca K, Vgeli B, Hilvert D. Structure and dynamics of a molten globular enzyme. Nat Struct Mol Biol. 2007;14:1202–1206. doi: 10.1038/nsmb1325. [DOI] [PubMed] [Google Scholar]
- 9.Roca M, Messer B, Hilvert D, Warshel A. On the relationship between folding and chemical landscapes in enzyme catalysis. Proc Natl Acad Sci USA. 2008;105:13877–13882. doi: 10.1073/pnas.0803405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy Y, Cho SS, Onuchic JN, Wolynes PG. A survey of flexible protein binding mechanisms and their transition states using native topology-based energy landscapes. J Mol Biol. 2005;346:1121–1145. doi: 10.1016/j.jmb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 11.Wolynes P, Onuchic J, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 12.McNulty BC, Young GB, Pielak GJ. Macromolecular crowding in the Escherichia coli periplasm maintains α-synuclein disorder. J Mol Biol. 2006;355:893–897. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 13.Materese CK, Goldmon CC, Papoian GA. Hierarchical organization of eglin c native state dynamics is shaped by competing direct and water-mediated interactions. Proc Natl Acad Sci USA. 2008;105:10659–10664. doi: 10.1073/pnas.0801850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osvath S, Sabelko JJ, Gruebele M. Tuning the heterogeneous early folding dynamics of phosphoglycerate kinase. J Mol Biol. 2003;333:187–199. doi: 10.1016/j.jmb.2003.08.011. [DOI] [PubMed] [Google Scholar]