Abstract
Bacterial flagella contain membrane-embedded stators, Mot complexes, that harness the energy of either transmembrane proton or sodium ion gradients to power motility. Use of sodium ion gradients is associated with elevated pH and sodium concentrations. The Mot complexes studied to date contain channels that use either protons or sodium ions, with some bacteria having only one type and others having two distinct Mot types with different ion-coupling. Here, alkaliphilic Bacillus clausii KSM-K16 was shown to be motile in a pH range from 7 to 11 although its genome encodes only one Mot (BCl-MotAB). Assays of swimming as a function of pH, sodium concentration, and ion-selective motility inhibitors showed that BCl-MotAB couples motility to sodium at the high end of its pH range but uses protons at lower pH. This pattern was confirmed in swimming assays of a statorless Bacillus subtilis mutant expressing either BCl-MotAB or one of the two B. subtilis stators, sodium-coupled Bs-MotPS or proton-coupled Bs-MotAB. Pairs of mutations in BCl-MotB were identified that converted the naturally bifunctional BCl-MotAB to stators that preferentially use either protons or sodium ions across the full pH range. We then identified trios of mutations that added a capacity for dual-ion coupling on the distinct B. subtilis Bs-MotAB and Bs-MotPS motors. Determinants that alter the specificity of bifunctional and single-coupled flagellar stators add to insights from studies of other ion-translocating transporters that use both protons and sodium ions.
Keywords: alkaliphile, MotAB, MotPS, motility, bacteria
The bacterial flagella that support swimming in liquid and on surfaces are rigid, propeller-like structures. Their rotation is powered by electrochemical gradients of either protons or sodium ions across the cytoplasmic membrane (1, 2). Ion channels are part of membrane-embedded stator structures at the base of the flagella. They play a critical role in the transduction of chemiosmotic energy into mechanical energy. The stators of the flagellar motor are most often called Mot complexes. Mot complexes are thought to contain four MotA-like proteins, each of which has four transmembrane segments (TMS), and two MotB-like proteins, each of which has a single TMS (1–3). The MotB-like proteins have a major role in determining ion-coupling specificity (4, 5). Initially, only proton-coupled motility was documented in bacteria, e.g., in Escherichia coli (1, 2). However, work of Imae and colleagues in the 1980s showed that extremely alkaliphilic Bacillus species use only sodium ion gradients for motility (6, 7). This was followed by evidence that Vibrio alginolyticus and Bacillus subtilis possess two distinct stators with different ion specificities that make contributions to swimming under particular physical–chemical conditions of pH, salinity, and viscosity (8–12).To date, only a limited number of additional flagellar stators have been directly probed for a capacity to use sodium as the coupling ion. The MotAB-like system of alkaliphilic Bacillus clausii KSM-K16 that is described here is an example of a single stator that can use both protons and sodium ions.
B. clausii strains are a group of alkaliphilic bacteria that have received attention because of their use in probiotic formulations (13) and their production of industrially useful enzymes (14, 15). The genome sequence of B. clausii KSM-K16 revealed a single set of genes encoding a MotAB-like pair of proteins (GenBank accession no. AP006627). We use the designation BCl-MotAB for this Mot stator. The BCl-MotA and MotB proteins were closely related to the Bs-MotA and MotB that constitute the proton-coupled Mot in B. subtilis (identity: 52% and 45%, similarity: 73% and 64%, respectively). They less closely resembled the Bs-MotP and MotS that constitute a sodium-coupled Mot in B. subtilis (identity: 32% and 28%, similarity: 57% and 55%, respectively). Motility at very alkaline pH values is expected to require use of a sodium-coupled Mot (6, 16, 17). The presence of the single BCl-MotAB species in B. clausii KSM-K16 therefore raised the possibility that either the motility of this alkaliphile strain was lost in the upper pH range for growth or the BCl-MotAB is capable of using both protons and sodium ions so that its coupling pattern changes over its pH range as the relative magnitudes of the transmembrane sodium and proton motive forces change. Preliminary assays showed that B. clausii KSM-K16 was motile at pH values from 7 to 11, so we probed the effects of added sodium and ion-specific motility inhibitors on swimming. Because B. clausii KSM-K16 is not genetically accessible, we used a statorless (ΔmotABΔmotPS) mutant strain of B. subtilis as the host for experiments in which the functional properties of BCl-MotAB could be compared side by side with those of bona fide proton- and sodium-coupled Mot stators, Bs-MotAB and Bs-MotPS, respectively. The results supported the hypothesis that BCl-AB is bifunctional with respect to ion-coupling capacity. We then identified mutational changes in BCl-MotB that converted the bifunctional BCl-MotAB stator one that prefers either protons or sodium ions at both neutral and highly-alkaline pH and applied an analogous strategy to confer a bifunctional capacity on the ion-specific Bs-MotAB and Bs-MotPS of B. subtilis.
Results and Discussion
B. clausii Motility Responds to Added Sodium, Cyanide m-Chlorophenyl Hydrazone (CCCP), and 5-(N-Ethyl-N-Isopropyl)-Amiloride (EIPA) Differently at Neutral and Alkaline pH.
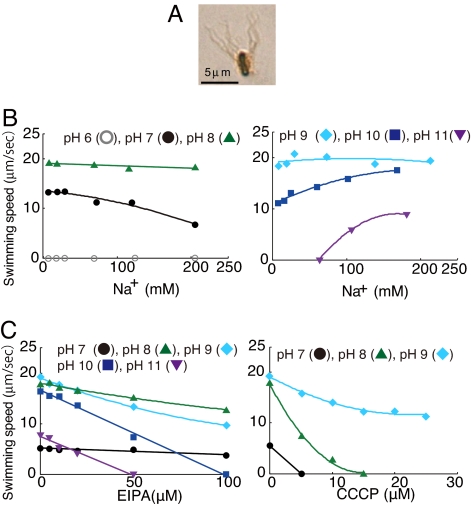
Cells of B. clausii KSM-K16 had an average of 12 flagellae per cell, with an average length of 6 μm, when grown under several different conditions of pH and sodium concentration [Fig. 1A and supporting information (SI) Fig. S1]. Swimming was observed over a pH range from 7.0 to 11.0 (Fig. 1B). The dependence of swimming on the concentration of sodium ions was examined in alkaline complex medium. This medium contained 6 mM sodium. Stimulation of swimming by higher sodium concentrations was observed only at pH 10 and above (Fig. 1B). At pH 10, stimulation by sodium was modest, but at pH 11, swimming of B. clausii depended on added sodium ions and required a threshold sodium concentration of ≈70 mM. Threshold concentrations at different levels were also observed for sodium-dependent Bs-MotPS at pH 7.0 (5) and pH 8.5 (shown in later figures). Such thresholds have been observed when freely swimming cells of other bacteria were assayed, but their basis is incompletely understood (1).
Fig. 1.
Properties of the flagella and swimming of B. clausii KSM-K16. Cells were grown at 30°C in alkaline complex medium (pH 9.0 and 230 mM Na+). (A) Stained flagellae of wild-type B. clausii KSM-K16. Cells were stained with staining solution that contained 5% (wt/vol) tannic acid as described in Materials and Methods. (B and C) The pH values used in the particular assay are designated by symbols that are indicated. (B) The swimming speed of wild-type B. clausii in alkaline complex medium as a function of the sodium concentration and pH. The swimming speed was assayed at various concentrations of sodium as described in Materials and Methods. (C) Effect of EIPA and CCCP on the swimming speed of wild-type B. clausii. Swimming speeds were measured at various concentration of EIPA (Left) or CCCP (Right) in alkaline complex medium containing 230 mM sodium.
The sodium channel inhibitor EIPA, an amiloride analogue, did not affect swimming of B. clausii at pH 7.0 and affected swimming only modestly at pH 8.0 at inhibitor concentrations up to 100 μM (Fig. 1C Left). Complete inhibition of swimming at pH 10.0 and 11.0 was mediated by EIPA at concentrations of 100 and 50 μM, respectively. Conversely, addition of the protonophore CCCP, which dissipates electrochemical proton gradients, completely inhibited swimming at pH 7.0 and 8.0, whereas swimming at pH 9.0 was not inhibited greatly by uncoupler even at CCCP concentrations up to 25 μM (Fig. 1C Right). At the same pH, CCCP at 20 μM completely inhibited swimming powered by proton-coupled Bs-MotAB (shown in a later figure). Therefore, the modest inhibition of BCl-MotAB by 20 μM CCCP at pH 9.0 was not due to loss of the capacity of CCCP to uncouple at pH 9.0.
The results suggested that the BCl-MotAB stator complex used protons with high preference at pH 7.0 and used sodium ions at pH 11. The modest inhibition of swimming by high concentrations of sodium at pH 7.0 (Fig. 1B) was observed previously with proton-powered motility of hybrid MotPB stators from B. subtilis. Elevated cytosolic sodium was hypothesized to mediate this inhibition via a cytoplasmic binding site for sodium (5). At pH values from 8.0 to 10.0, robust swimming across a range of sodium concentrations suggested that both protons and sodium ions could be used to power motility.
Properties of BCl-MotAB Resemble Those of Bs-MotAB at pH ≤ 8.5 and Resemble Those of Bs-MotPS at pH > 8.5.
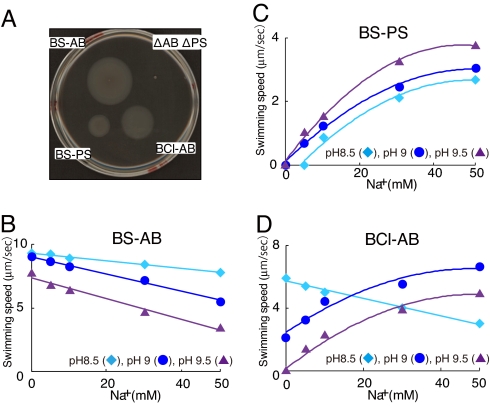
To further probe the ion-coupling preferences of BCl-MotAB at different pH values, we directly compared the swimming properties conferred by BCl-MotAB with those conferred by proton-coupled Bs-MotAB and sodium-coupled Bs-MotPS. BCl-motAB was introduced into the chromosome a B. subtilis strain (designated ΔABΔPS) from which both native Bs-motAB and Bs-motPS were deleted. This produced the mutant strain named BCl-AB that expressed BCl-motAB under control of the B. subtilis motAB promoter. The B. subtilis motAB and motPS were each expressed in the same manner in strains of ΔABΔPS designated as BS-AB and BS-PS, respectively. All three stators restored motility to the nonmotile ΔABΔPS strain on soft-agar plates (Fig. 2A). Swimming-speed assays in liquid were conducted in tryptone-yeast extract medium (TY) that contained 15 mM sodium and in TY/20 medium that was TY diluted 1:20 with water and contained micromolar concentrations of sodium (5). The richer TY medium supported faster swimming speeds than TY/20. As observed earlier (5, 9), the BS-AB strain expressing proton-coupled Bs-MotAB consistently swam at faster speeds than the BS-PS strain expressing sodium-coupled Bs-MotPS in BS-PS. The swimming speed of BCl-AB was between that of the other two strains. Sodium-coupled swimming in some bacteria occurs with speeds higher than observed for proton-coupled swimming, although this is not the case with the two B. subtilis stators (1, 18). We have hypothesized that in laboratory strains of B. subtilis, the interactions of Bs-MotAB are optimized for interactions with the single FliG rotor protein relative to those of Bs-MotPS (5, 9).
Fig. 2.
The motility of a statorless B. subtilis mutant (ΔABΔPS) expressing B. subtilis motAB (BS-AB), B. subtilis motPS (BS-PS), or B. clausii motAB (BCl-AB). (A) Motility in a soft-agar plate assay. Fresh cells were inoculated into LB medium (22) (pH 7.0) soft agar and incubated at 37°C for 16 h. (B–D) Motility in liquid medium. BS-AB (B), BS-PS (C), and BCl-AB (D) cells were grown at 37°C in LB medium (pH 7.0), and the swimming speed was measured at various concentrations of sodium in 1/20 tryptone yeast extract (TY) medium (5) as described in Materials and Methods. The pH values of TY/20 medium used in each assay are designated by symbols as indicated.
The swimming speeds of the B. subtilis strains were first studied at pH values from 6.0 to 9.5 in TY medium, in which the BS-AB and BS-PS strains exhibited distinct responses to changes in both pH and sodium ion concentration (5). Overall, the responses of BCl-AB strain resembled those of BS-AB in the pH range from 6 to 8.5 and those of BS-PS at pH 9 and 9.5. However, the sodium content of TY medium supported significant swimming of the completely sodium-coupled BS-PS strain in the absence of added sodium (Fig. S2). Therefore, detailed comparisons of the sodium-dependence of swimming were conducted on the BS-AB, BS-PS, and BCl-AB strains in TY/20 medium at pH ≥8.5. At this pH, sodium appeared to be the preferred ion for powering the BCl-MotAB stator. In the TY/20 medium, the BS-AB strain exhibited the fastest swimming speed at either pH 8.5 or 9.0. Slower swimming was observed at pH 9.5 and at elevated sodium concentrations (Fig. 2B). The basis for inhibitory effects of sodium on the proton-coupled stator has not been investigated. In contrast to the BS-AB swimming pattern, the BS-PS strain exhibited no swimming in the absence of added sodium at either pH 8.5, 9.0 or 9.5, and the fastest swimming was at pH 9.5 (Fig. 2C). Swimming of the BCl-AB strain was inhibited rather than stimulated by sodium at pH 8.5, as seen with the proton-coupled BS-AB control strain, but BCl-AB exhibited a sodium-dependent pattern like the sodium-coupled BS-PS strain at pH 9.5 (Fig. 2D). At pH 9.0, BCl-AB exhibited evident stimulation by increasing sodium concentrations while still swimming in the absence of added sodium. This suggested that BCl-MotAB uses both protons and sodium ions at pH 9.0 (Fig. 2D).
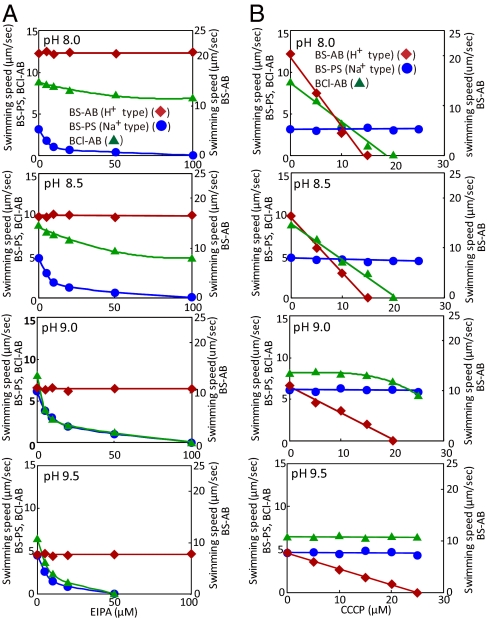
We then examined the effects of added EIPA and CCCP on the swimming speed of the B. subtilis strains with the different stators in TY medium containing 63 mM sodium over a range of pH from 8.0 to 9.5 (Fig. 3). The two B. subtilis stators functioned as expected (5). The swimming speeds of the sodium-coupled BS-PS strain were extremely sensitive to inhibition by EIPA but insensitive to inhibition by CCCP at every pH value. Swimming of the BS-AB strain, on the other hand, was sensitive to inhibition by CCCP but not by EIPA at every pH value. The BCl-AB strain exhibited a lower swimming speed than BS-AB, but its inhibition pattern resembled that of BS-AB at pH 8.0 and 8.5. Swimming of the BCl-AB strain differed from that of BS-AB in showing modest inhibition by EIPA, as opposed to none, and in requiring a higher concentration of CCCP for full inhibition of swimming. At pH 9.0 and 9.5, the swimming speed of the BCl-AB strain was as sensitive to EIPA inhibition as that of sodium-coupled BS-PS (Fig. 3A, bottom two images). Still, there was an indication of continued proton use by BCl-MotAB stator in the BCl-AB strain at pH 9.0. At pH 9.0, swimming of the BCl-AB strain was inhibited 28% by 25 μM CCCP, whereas swimming of the sodium-coupled BS-PS strain was not inhibited at all (Fig. 3B, third image in the column). At pH 9.5, the inhibition pattern of the BCl-AB strains was completely comparable with that of BS-PS (Fig. 3 A and B bottom images). This was consistent with the complete dependence of swimming by the BCl-AB strain on added sodium (Fig. 2D).
Fig. 3.
Effect of EIPA and CCCP on swimming of B. subtilis expressing different stators. The swimming speed of BS-AB (filled diamond), BS-PS (filled circle), and BCl-AB (filled triangle) in TY medium containing 63 mM sodium was examined at various concentrations of EIPA (A) or CCCP (B). The pH values of the TY medium used for the assays were pH 8.0 (First), 8.5 (Second), 9.0 (Third), and 9.5 (Fourth). The swimming speed was analyzed as described in Materials and Methods.
The comparative swimming assays of BCl-MotAB and control stators of known ion-coupling specificity in the B. subtilis setting further supported the conclusion that the MotAB flagellar stator of B. clausii is bifunctional. BCl-MotAB primarily uses proton coupling for swimming at pH ≤8.5, primarily sodium coupling for swimming at pH 9.0 and exclusively uses sodium coupling at pH 9.5 in the B. subtilis host. The pH at which complete sodium-dependence is observed is lower in the B. subtilis host (pH 9.5) than in the native B. clausii setting (pH 11), although in both settings, BCl-MotAB increases its use of sodium as the pH becomes increasingly alkaline. Perhaps the different pH for transition to sodium dependence relates to differences in membrane properties or bioenergetic parameters that impact the functional properties of the stator.
Mutations in BCl-MotB Change the Profile of the Bifunctional BCl-MotAB Stator to a Proton-Coupled or Sodium-Coupled Stator Profile.
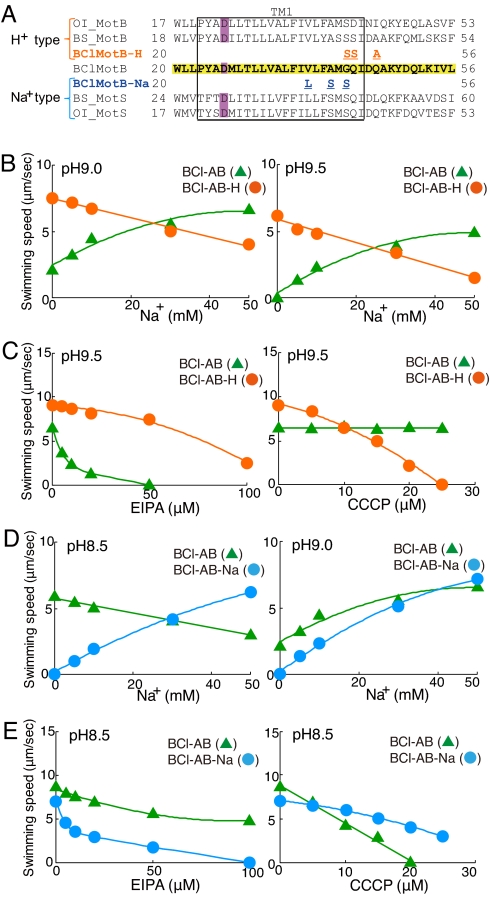
The next challenge was to identify determinants that could change the bifunctional B. clausii MotAB system to one that primarily uses either protons or sodium at both neutral and alkaline pH. The MotB-like proteins are major determinants of the ion specificity of the B. subtilis MotAB vs. MotPS systems (5). To choose mutations, we focused on the alignment of the BCl-MotB with MotB and MotS from B. subtilis and from modestly alkaliphilic Oceanobacillus iheyensis HTE831, each of which possesses both stator types (Fig. 4A). We noted that membrane protein segments of alkaliphiles that are exposed outside the surface of the cytoplasmic membrane generally have greatly reduced abundance of basic amino acids and increased acidic residues relative to the same segments of neutrophile homologues (17). We expected that a conserved ion-selective domain would be located in or on the surface of the more highly conserved transmembrane segment rather than in these poorly conserved hydrophilic segments further outside the surface.
Fig. 4.
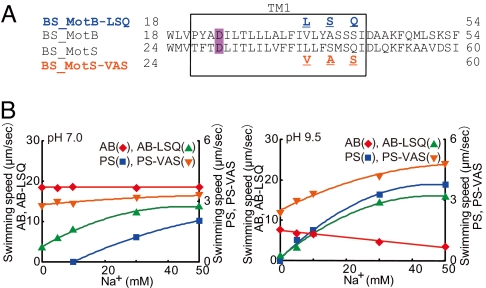
Effect of EIPA and CCCP on swimming of B. subtilis expressing wild-type BCI-AB or mutant B. clausii stators. (A) Multiple alignment of the region containing the single transmembrane segment of Oceanobacillus iheyensis MotB (OI_MotB) and MotS (OI_MotS), B. subtilis MotB (BS_MotB), MotS (BS_MotS), and B. clausii MotB (BClMotB). The sequence of B. clausii MotB is highlighted in yellow. The red letters show locations of point mutations in B. clausii MotB mimicking BS_MotB to yield BClMotB-H. The blue letters show locations of point mutations in B. clausii MotB mimicking BS_MotS to yield BClMotB-Na. D26 is critical for flagellar rotation and is highlighted with violet (2). (B) The swimming speeds of strains BCl-AB (filled triangles) and BCl-AB-H (filled circles) were measured in TY/20 medium [pH 9.0 (Left) and pH 9.5 (Right)] at various concentrations of sodium. (C) Effect of EIPA and CCCP on swimming speeds of strains BCl-AB (filled triangles) and BCl-AB-H (filled circles). The swimming speeds were measured in TY medium (pH 9.5, 63 mM Na+) at various concentrations of EIPA (Left) or CCCP (Right). (D) The swimming speeds of strains BCl-AB (filled triangles) and BCl-AB-Na (filled circles) were measured in TY/20 medium [pH 8.5 (Left) and pH 9.0 (Right)] at various concentrations of sodium. (E) Effect of EIPA and CCCP on the swimming speeds of strains BCl-AB (filled triangles) and BCl-AB-Na (filled circles). The swimming speeds were measured in TY medium (pH 8.5, 63 mM Na+) at various concentrations of EIPA (Left) or CCCP (Right).
For construction of a proton-coupled stator (BCl-MotAB-H) and a sodium-coupled stator (BCl-MotAB-Na) from bifunctional BCl-MotAB, we initially made two distinct triple mutants in which one of the mutations was common to both triple mutants. We hypothesized that replacing the nonconsensus G42 with the conserved serine might reduce flexibility that contributes to conformational changes in support of the switching of ion-coupling preference. Therefore, the G42S change was introduced into both triple mutant forms of BCl-MotB. For production of a BCl-MotB-H candidate, the other two mutations were Q43S and Q46A, changing two glutamines that are not present in the MotB of either B. subtilis or O. iheyensis to the sequence found in B. subtilis MotB (Fig. 4A). For BCl-MotB-Na, the two mutations made in addition to G42S were V37L and A40S, using the amino acids found in B. subtilis MotS to replace the B. clausii residues that are not found in those positions of the two homologues (Fig. 4A). Introduction of two hydroxyamino acids to foster sodium coupling was consistent with participation of such residues in the sodium-binding sites of rotor rings of two ATPases (19, 20). In addition, a larger alignment (Fig. S3) showed that valine was conserved among stators predicted to be MotAB types, whereas leucine was conserved among MotS or PomB types, several of which had been shown to use sodium coupling. Interestingly, the backbone carbonyl oxygen of a valine is involved in sodium ion coordination in the Ilyobacter tartaricus sodium-ATPase that can also use protons (19, 21) and that of leucine is involved in sodium coordination of the sodium-ATPase from Enterococcus hirae (20).
The two triple-mutant BCl-MotB forms were introduced into BCl-MotAB. The properties of the mutant stators BCl-MotAB-H and BCl-MotAB-Na were then studied in B. subtilis ΔABΔPS (strains BCl-AB-H and BCl-AB-Na, respectively) in comparison with the wild-type B. clausii stator expressed in strain BCl-AB. The BCl-AB-H strain exhibited a BS-AB type of pattern: In TY/20 medium, swimming was best at the lowest sodium concentration at pH 9.0 and 9.5 (Fig. 4B); in TY medium with 63 mM sodium at pH 9.5, swimming by the mutant was only partially inhibited by 100 μM EIPA, whereas swimming supported by the parent stator in BCl-AB was completely inhibited by 50 μM EIPA (Fig. 4C Left); and conversely, swimming by the BCl-AB strain was not inhibited by CCCP up to 25 μM, whereas swimming supported by the triple-mutant BCl-MotAB-H stator in strain BCl-AB-H was completely inhibited (Fig. 4C Right). These results showed that the BCl-MotAB-H stator used protons rather than sodium as the major coupling ion for flagellar rotation at pH 9.0 and 9.5. The BCl-AB-Na strain exhibited a BS-PS type of pattern: Swimming of BCl-AB-Na was sodium dependent at pH 8.5 and 9.0 in TY/20 medium (Fig. 4D); swimming at pH 8.5 was completely inhibited at by 100 μM EIPA, whereas the parent BCl-AB was only partially inhibited; and swimming of the BCl-AB-Na strain was more resistant to inhibition by CCCP than swimming of BCl-AB (Fig. 4E). We also conducted a set of experiments on the full panel of single-stator strains at pH 7.0 and examined the swimming speed as a function of sodium, EIPA, or CCCP concentration. Again, the pattern of responses of BCl-MotAB-H most closely resembled that of Bs-MotAB, and that of BCl-MotAB-Na most closely resembled that of Bs-MotPS in these assays (Fig. S4). The change in ion-coupling profiles of the two BCl-MotAB mutant forms suggested that the mutational changes in BCl-MotB optimized the binding and/or translocation of one of the coupling ions (e.g., protons for BCl-MotAB-H) so that the other cation (e.g., sodium for BCl-MotAB-H) was no longer used preferentially at pH values where it is favored for coupling of the parent BCl-MotAB.
To investigate the responsible amino acid change(s) in the two triple mutants, each of the mutations of the triple mutants was introduced into BCl-MotB alone or together with one of the other mutations for BCl-MotAB-H or BCl-MotAB-Na and expressed in strain ΔABΔPS. The effect of sodium on swimming at pH 9.0 was tested as an index of sodium-coupling retention in the BCl-MotAB-H stator. The analyses showed that the BCl-MotB-Q43S mutation was critical for loss of sodium coupling and that a combination of the Q43S mutation plus either the G42S or Q46A change was required to achieve loss of sodium use (Fig. S5). Q43 is predicted to be located at the border of the TMS and outer surface by the secondary structure prediction programs TMHMM (www.cbs.dtu.dk/services/TMHMM/) and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/) (Fig. S6). For assessment of the BCl-MotAB-Na type, the single- and double-mutant forms in ΔABΔPS were tested for sodium-dependent swimming at pH 8.5. The results showed that the BCl-MotB-V37L mutation was critical and that a combination of V37L and either the A40S or the G42S mutation was required for the conversion to sodium coupling at low pH (Fig. S5). V37 is predicted to be located in the middle of the single TMS of BCl-MotB on the same face as the conserved aspartic acid residue, D26, that is required for flagellar rotation (Fig. S6) (2). Thus the mutation in G42 was not necessary for conversion of the bifunctional B. clausii to either single-coupling form. There was a critical residue for generating each form, Leu37 and Val37, respectively, for proton and sodium preference. In addition there was a requirement for one additional change for which there was some flexibility.
Triple Mutants of Proton-Specific Bs-MotAB and Sodium-Specific Bs-MotPS Gain a Bifunctional Capacity.
We were interested in testing whether the general strategy that converted the native bifunctional BCl-MotAB to more single ion-coupled forms could confer a bifunctional capacity on the two native single ion-coupled stators of B. subtilis. Based on the alignment of Bs-MotB and Bs-MotS (Fig. 5A), we constructed two mutants. In mutant Bs-MotAB-LSQ, the crucial Val of Bs-MotB was mutated to Leu together with A → S and S → Q changes. All three of the changes in Bs-MotB introduced residues corresponding to the MotS sequence in those positions. In mutant Bs-MotPS-VAS, the crucial Leu of Bs-MotS was mutated to Val together with S → A and Q → S changes that changed Bs-MotS residues to the MotB sequence (Fig. 5A). When compared with the parent Bs-MotAB in strain BS-AB at pH 7.0, swimming of mutant BS-AB-LSQ at pH 7.0 was clearly stimulated by sodium, although it was not completely dependent as was BS-PS (Fig. 5B Left). At pH 9.5, swimming of BS-AB-LSQ depended on sodium and was almost as robust as that of BS-PS (Fig. 5B Right). When compared with parent Bs-MotPS in strain BS-PS at pH 7.0, swimming of mutant BS-PS-VAS exhibited very little stimulation by sodium and was more like BS-AB in its response to sodium (Fig. 5B Left). At pH 9.5, BS-PS-VAS, unlike the BS-PS strain with the parent stator, exhibited a significant capacity to swim without added sodium but still exhibited stimulation by sodium (Fig. 5B Right). The results suggested that the two mutant forms retained their original ion-coupling capacity but gained a capacity to couple swimming to a new cation. The patterns of inhibition by EIPA and CCCP supported this conclusion (Fig. S7).
Fig. 5.
Mutations that affect the ion-coupling capacities of Bs-MotAB and Bs-MotPS. (A) An alignment of a region of Bs-MotB and Bs-MotS (central two lines). The mutational changes introduced into the MotB of Bs-MotAB-LSQ are shown in the top line and those introduced into the MotS of Bs-MotPS are shown in the bottom line. (B) Swimming speed of Bs-MotAB, Bs-MotAB-LSQ, Bs-MotPS, and Bs-MotPS-VAS, as designated by symbols, was assayed as a function of sodium concentration at pH 7.0 (Left) and 9.5 (Right) in TY/20.
A major finding of the current study was the demonstration that a single bacterial flagellar stator of B. clausii can couple motility to either protons or sodium ions. This finding may facilitate identification of additional examples of bifunctional cation-coupling capacity among the rapidly growing number of bacteria that exhibit motility in neutral to alkaline pH range and for which genomic sequence is available. We also identified mutations that can convert the bifunctional stator to more specific types and mutations that confer bifunctionality on the single ion-coupled B. subtilis stators. Further mutagenesis studies and, ultimately, the correlation of these types of data with high-resolution structural data on these and other transporters that catalyze both proton- and sodium-coupled bioenergetic work will inform detailed models of the cation-binding sites.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
The strains and plasmids used in this study are shown in Table S1. B. subtilis 168 strain BR151MA (wild type) and its derivatives were grown at 37°C in LB medium (22). When appropriate, media were supplemented with erythromycin (0.3 μg/ml), neomycin (7.5 g/ml), or chloramphenicol (5 μg/ml). LB medium containing 0.3% (wt/vol) Noble agar (Difco) was used in assays of motility on soft-agar plates. For assays of swimming speed in liquid, TY medium [1% (wt/vol) Bacto Tryptone and 0.5% (wt/vol) yeast extract] or a 1:20 dilution of TY with water (TY/20) was used at the indicated values of pH and sodium, EIPA, or CCCP (23). The actual final sodium concentration of each assay medium was determined with a flame photometer (Model AMA-175; Tokyo Koden) calibrated with standard sodium solutions of known concentrations. B. clausii KSM-K16 cells were grown at 30°C in alkaline complex medium (pH 9.0 and 230 mM sodium) containing 15.5 g of K2HPO4, 4.5 g of KH2PO4, 0.05 g of MgSO4·7H2O, 0.34 g of citric acid, 5 g of peptone, 2 g of yeast extract, 5 g of glucose, and 10.6 g of Na2CO3 per liter. For swimming speed assays with inhibitors, cells were grown in this medium and then resuspended in fresh medium at the indicated values of pH and sodium content for the assays. For swimming assays over a range of sodium, the cells were grown and harvested as above and then washed and resuspended in alkaline complex medium in which the Na2CO3 was replaced by K2CO3. Sodium was added as indicated, and adjustments in the potassium content were made to maintain constant ionic strength. The sodium concentration of the medium was determined as described above. E. coli cells were grown at 37°C in LB medium. In experiments in which sodium concentrations were varied, constant ionic strength was maintained in the media by adjusting the potassium content of the medium.
Construction of Plasmid Coding B. clausii motAB Under B. subtilis PmotAB Promoter and Its Point Mutations.
All recombinant DNA manipulations were carried out by standard methods. The primers used in this study are shown in Table S2. For cloning of the B. clausii motAB gene under the B. subtilis PmotAB promoter, we performed gene splicing by the overlap extension (gene SOEing) method based on PCR (24). The primers were synthesized based on sequences from B. subtilis and B. clausii. The B. subtilis PmotAB promoter was amplified by PCR using B. subtilis chromosomal DNA as the template with the set of primers, BSmotAB-BamHI-F designed with a BamHI site and BSPmotAB-BclmotAB-R in which a sequence of N-terminal region of the B. clausii motAB gene was introduced. The B. clausii motAB gene was amplified by PCR using B. clausii chromosomal DNA as the template with the set of primers BclmotAB-4 and BSPmotAB-BclmotAB-F in which the promoter sequence of B. subtilis PmotAB promoter was introduced. We performed PCR with these two product mixtures as the template to join the B. subtilis PmotAB promoter and the B. clausii motAB gene. The amplified fragment was digested by BamHI and SphI and cloned into BamHI- and SphI-digested pDR67, yielding pDR-AB. To introduce amino acid substitutions into the B. clausii motB gene, we used the gene SOEing method. We synthesized pairs of mutant primers to the sense strand of the B. clausii motB gene, with a mismatch at the mutation site. We amplified the B. clausii motAB under a B. subtilis PmotAB promoter by PCR with mutant primers and then cloned into BamHI- and SphI-digested pDR67. Mutations that resulted in amino acid substitutions were introduced into the B. subtilis motB and motS genes by using the same method. We amplified the B. subtilis motAB and motPS under a B. subtilis PmotAB promoter by PCR with mutant primers and then cloned the fragment into BamHI- and SphI-digested pDR67. The presence of these mutations was confirmed by DNA sequencing. Each plasmid was integrated into the amyE locus of ΔABΔPS B. subtilis host. Recombinant transformants were selected by conventional techniques. The presence of the insert with the correct sequence was confirmed.
Measurement of Swimming Speed.
B. subtilis cells were cultured at 37°C for 6 h (to an A600 nm of 0.5–0.6) in LB medium (pH 7.0) and diluted 50-fold into TY or TY/20 medium at the pH and with additions that are specified for each experiment. In B. clausii, alkaline complex medium was used for culture and swimming assays. Cell motility was observed under a dark-field microscope using a Leica DMRE microscope and Progressive 3CCD color video camera system (Model DXC-9000; Sony) and recorded on videotape. Swimming speed was determined by direct tracing of the moving cells on the video monitor. All results shown are the averages of three independent experiments in which the speed of 20 different cells was measured.
Visualization of Flagella.
Flagellar staining was carried out as described by Aono et al. (25). B. subtilis cells were cultured at 37°C for 5 h in LB medium and transferred gently to a microscope slide. The sample was air-dried and treated for 3 min with staining solution containing 5% (wt/vol) tannic acid, 0.75% (wt/vol) FeCl2, and 0.01% NaOH, followed by ammoniac silver nitrate for 1 min. Observations of flagella were made by using a Leica DMLB100 bright-field microscope and Leica DC300F camera, Leica IM50 version 1.20 software (Leica Geosystems). The number of flagellae per cell and flagellar length were measured for 20 cells by using NIH ImageJ version 1.36b software.
Supplementary Material
Acknowledgments.
We thank Kao Corporation for providing B. clausii KSM-K16 and Mr. N. Morishita for technical assistance. This work was supported by National Institutes of Health Research Grant GM28454 (to T.A.K.), grants from the Kurata Memorial Foundation for Promoting Science and Special Research, from Toyo University, from the 21st Century Center of Excellence Program of the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M.I.), and a Japan Society for the Promotion of Science Research Fellowships for Young Scientists (to N.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802106105/DCSupplemental.
References
- 1.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Kojima S, Blair DF. The bacterial flagellar motor: Structure and function of a complex molecular machine. Int Rev Cytol. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- 3.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 4.Asai Y, Kawagishi I, Sockett RE, Homma M. Coupling ion specificity of chimeras between H+- and Na+-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J. 2000;19:3639–3648. doi: 10.1093/emboj/19.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito M, Terahara N, Fujinami S, Krulwich TA. Properties of motility in Bacillus subtilis powered by the H+-coupled MotAB flagellar stator, Na+-coupled MotPS or hybrid stators MotAS or MotPB. J Mol Biol. 2005;352:396–408. doi: 10.1016/j.jmb.2005.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirota N, Imae Y. Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J Biol Chem. 1983;258:10577–10581. [PubMed] [Google Scholar]
- 7.Sugiyama S. Na+-driven flagellar motors as a likely Na+ re-entry pathway in alkaliphilic bacteria. Mol Microbiol. 1995;15:592. doi: 10.1111/j.1365-2958.1995.tb02273.x. [DOI] [PubMed] [Google Scholar]
- 8.Atsumi T, et al. Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus. J Bacteriol. 1996;178:5024–5026. doi: 10.1128/jb.178.16.5024-5026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito M, et al. MotPS is the stator-force generator for motility of alkaliphilic Bacillus and its homologue is a second functional Mot in Bacillus subtilis. Mol Microbiol. 2004;53:1035–1049. doi: 10.1111/j.1365-2958.2004.04173.x. [DOI] [PubMed] [Google Scholar]
- 10.Kawagishi I, et al. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J Bacteriol. 1995;177:5158–5160. doi: 10.1128/jb.177.17.5158-5160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarter LL. Multiple modes of motility: A second flagellar system in Escherichia coli. J Bacteriol. 2005;187:1207–1209. doi: 10.1128/JB.187.4.1207-1209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terahara N, et al. An intergenic stem-loop mutation in the Bacillus subtilis ccpA-motPS operon increases motPS transcription and the MotPS contribution to motility. J Bacteriol. 2006;188:2701–2705. doi: 10.1128/JB.188.7.2701-2705.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senesi S, Celandroni F, Tavanti A, Ghelardi E. Molecular characterization and identification of Bacillus clausii strains marketed for use in oral bacteriotherapy. Appl Environ Microbiol. 2001;67:834–839. doi: 10.1128/AEM.67.2.834-839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kageyama Y, et al. Intragenomic diversity of the V1 regions of 16S rRNA genes in high-alkaline protease-producing Bacillus clausii spp. Extremophiles. 2007;11:597–603. doi: 10.1007/s00792-007-0074-1. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, et al. Purification and properties of an alkaline protease from alkalophilic Bacillus sp. KSM-K16. Appl Microbiol Biotechnol. 1995;43:473–481. doi: 10.1007/BF00218452. [DOI] [PubMed] [Google Scholar]
- 16.Fujinami S, Terahara N, Lee S, Ito M. Na+ and flagella-dependent swimming of alkaliphilic Bacillus pseudofirmus OF4: A basis for poor motility at low pH and enhancement in viscous media in an “up-motile” variant. Arch Microbiol. 2006;187:239–247. doi: 10.1007/s00203-006-0192-7. [DOI] [PubMed] [Google Scholar]
- 17.Krulwich TA, Hicks DB, Swartz TH, Ito M. In: Physiology and Biochemistry of Extremophiles. Gerday C, Glansdorff N, editors. Washington, DC: Am Soc Microbiol Press; 2007. pp. 311–329. [Google Scholar]
- 18.Magariyama Y, et al. Very fast flagellar rotation. Nature. 1994;371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 19.Meier T, et al. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 20.Murata T, et al. Structure of the rotor of the V-Type Na+-ATPase from Enterococcus hirae. Science. 2005;308:654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- 21.Neumann S, Matthey U, Kaim G, Dimroth P. Purification and properties of the F1F0 ATPase of Ilyobacter tartaricus, a sodium ion pump. J Bacteriol. 1998;180:3312–3316. doi: 10.1128/jb.180.13.3312-3316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Frisch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 23.Aagaard A, Brzezinski P. Zinc ions inhibit oxidation of cytochrome c oxidase by oxygen. FEBS Lett. 2001;494:157–160. doi: 10.1016/s0014-5793(01)02299-2. [DOI] [PubMed] [Google Scholar]
- 24.Horton RM. In vitro recombination and mutagenesis of DNA. Methods Mol Biol. 1996;67:141–149. doi: 10.1385/0-89603-483-6:141. [DOI] [PubMed] [Google Scholar]
- 25.Aono R, Ogino H, Horikoshi K. pH-dependent flagella formation by facultative alkaliphilic Bacillus sp C-125. Biosci Biotechnol Biochem. 1992;56:48–53. doi: 10.1271/bbb.56.48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.