Abstract
The topology of metabolic networks may provide important insights not only into the metabolic capacity of species, but also into the habitats in which they evolved. Here we introduce the concept of a metabolic network's “seed set”—the set of compounds that, based on the network topology, are exogenously acquired—and provide a methodological framework to computationally infer the seed set of a given network. Such seed sets form ecological “interfaces” between metabolic networks and their surroundings, approximating the effective biochemical environment of each species. Analyzing the metabolic networks of 478 species and identifying the seed set of each species, we present a comprehensive large-scale reconstruction of such predicted metabolic environments. The seed sets' composition significantly correlates with several basic properties characterizing the species' environments and agrees with biological observations concerning major adaptations. Species whose environments are highly predictable (e.g., obligate parasites) tend to have smaller seed sets than species living in variable environments. Phylogenetic analysis of the seed sets reveals the complex dynamics governing gain and loss of seeds across the phylogenetic tree and the process of transition between seed and non-seed compounds. Our findings suggest that the seed state is transient and that seeds tend either to be dropped completely from the network or to become non-seed compounds relatively fast. The seed sets also permit a successful reconstruction of a phylogenetic tree of life. The “reverse ecology” approach presented lays the foundations for studying the evolutionary interplay between organisms and their habitats on a large scale.
Keywords: growth environments, metabolic networks, seed compounds, reverse ecology
Numerous biological systems can be represented as networks, encapsulating many of their essential properties (1). The structure and topology of these biological networks are not merely abstract descriptions of the complex interactions in a given system, but are also major determinants of the system's function and dynamics. In particular, a wide range of analytical approaches has been used to study topological characteristics of metabolic networks and their bearings on various metabolic functional properties, including scaling (2), regulation (3), universality (4), and robustness to metabolic gene knockouts (5, 6). Furthermore, as metabolic networks function within the context of biochemical environments and interact with these environments by taking up or secreting various organic and inorganic compounds, previous studies have also addressed the effect that these environmental interactions have on the metabolic process, as manifested in, for example, the distribution of metabolic fluxes within the network (7) or the organism's growth rate (8).
However, as the interactions with the environment must themselves be reflected in the structure of the evolved metabolic networks, these networks can be used not only to infer metabolic function but also to obtain insights into the growth environments in which the species evolved. Specifically, by analyzing the topology of a given metabolic network, we show that the set of compounds that are acquired exogenously (termed “seed set”; see also refs. 9 and 10) can be identified. Assuming that the environment of a species determines the metabolites it extracts from its surroundings to a considerable extent, the seed set can serve as a good proxy for its environment. This “reverse ecology” approach thus goes beyond previous research on the evolution of metabolic networks (11, 12) and metabolic scope analysis (9, 10, 13, 14) in enabling the evolutionary history of both metabolic networks and metabolic growth environments to be traced.
In view of this approach, in this article we first introduce the concept of metabolic networks' seed sets and provide a formal methodology to computationally infer the seed set of a given network. We next integrate this methodology with large-scale metabolic data to compile a comprehensive large-scale dataset describing the seed sets of hundreds of species. The predicted seed sets are shown to accord with biological observations across compounds and across species, validating the potential and relevance of our computational framework and compiled dataset. This dataset is then analyzed to obtain novel insights into the evolutionary dynamics of metabolic networks and the determinants that affect their interfaces with the environment.
Results
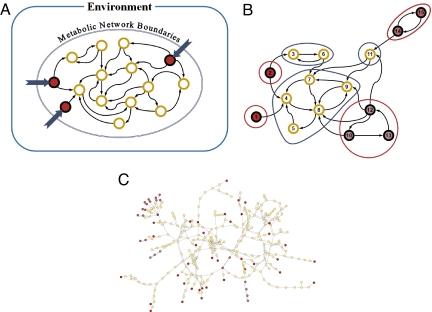
We represent the metabolic network of a given species as a directed graph whose nodes represent compounds and whose edges represent reactions linking substrates to products [Materials and Methods and supporting information (SI) Materials and Methods]. This graph-based representation of metabolic reactions is a common tool in analyzing and studying metabolic networks (1, 2) and can be obtained from large-scale, cross-species databases [e.g., KEGG (15)]. It should be noted, however, that such directed graphs are simplifications of the actual underlying metabolic networks, ignoring, for example, reaction stoichiometry (see Discussion). Compounds that appear in the network are referred to as occurring compounds. Formally, we define the seed set of the network (9, 10) as the minimal subset of the occurring compounds that cannot be synthesized from other compounds in the network (and hence are exogenously acquired) and whose existence permits the production of all other compounds in the network (Fig. 1A).
Fig. 1.
Identifying seed compounds in metabolic networks. (A) A schematic representation of the interaction of a metabolic network with its environment. Seed compounds must be externally acquired from the environment and are highlighted in red. (B) The procedure for identifying seed compounds is illustrated in a simple synthetic network. The network is decomposed into its SCC (illustrated as contour lines surrounding sets of nodes) using Kosaraju's algorithm (19). SCC decomposition reduces the seed detection problem to the simpler problem of detecting source components (i.e., components with no incoming edges) in a directed acyclic graph, where each source component forms a collection of candidate seed compounds. The source components are highlighted in red. The color saturation of the original nodes denotes the seed's confidence level, C (Materials and Methods), with a darker red indicating a higher confidence level. Although some of the seed compounds are easily identified (e.g., those forming the first step of an isolated and directed metabolic pathway), in a complex network the full set of seed compounds cannot easily be found without such a graph-theory algorithm. (C) The metabolic network of Buchnera with the seed compounds highlighted in red as in B. The seed set in this organism (which possesses a metabolic network of 314 occurring compounds) is composed of 61 chemicals (of these, only 38 have a confidence level C = 1). See also Table S2.
Our definition of seed compounds differs from that of essential compounds in that we require the production of all compounds in the network (and the potential activation of all of the metabolic pathways), regardless of their actual dynamic activation in a given environmental condition. In practice, organisms can survive in a wide range of environmental conditions and in each environment may activate only a subset of the pathways in the network, using a different set of exogenously acquired compounds (7, 16). Accordingly, the seed set can be conceived as the union of the essential sets required in all of these environments. Assuming that various alternative pathways have evolved and are retained because of their adaptive value in some environment (17), the seed set represents the overall, static metabolic “interface” (or metabolic “potential”; see also ref. 18) of each organism (and may serve as a characterizing proxy for its effective biochemical habitat).
We developed a graph-based algorithm to detect the seed set of a given network (see Materials and Methods and Fig. 1B for more details). This algorithm is based on a fast method for strongly connected components (SCC) decomposition (19) and can therefore easily scale up and be applied to large-scale network data. Next we constructed the metabolic networks of 478 species (Table S1) using data from a large-scale metabolic reactions database (Materials and Methods) and applied the seed set detection algorithm to identify the seed compounds in each network (Dataset S1). This compilation results in a comprehensive large-scale dataset of predicted biochemical environments of hundreds of species and facilitates a cross-species comparison of such seed sets.
Clearly, large-scale metabolic data are usually based on genome annotation, largely from automated, comparison-based methods (15), and as such are bound to be incomplete and inaccurate (20). This can potentially have a marked effect on the composition of the inferred seed sets. However, examining the effect of missing or erroneous data (SI Text and Fig. S1), we find that the identified seed sets are fairly robust to perturbations of the raw metabolic data. Still, considering the inherent noise and incompleteness associated with these data, we focus here mostly on the identification of significant large-scale statistical signals and phylogenetic patterns characterizing the seed set composition across the tree of life.
To exemplify the composition of a seed set obtained by our analysis and its alignment with known findings concerning the organism's environment, we focus first on a single species whose habitat is simple and well characterized. The obligate endocellular symbiont Buchnera aphidicola has lost many biosynthetic genes and demonstrates extremely successful symbiosis with its aphid host; it provides the host with amino acids that are essential for aphids (i.e., that aphids cannot synthesize) and relies on the host for nutrients it cannot synthesize (such as certain amino acids that are nonessential for aphids) (21–23). Buchnera has retained substrate-specific transporters only for glucose and mannitol (21) and is responsible for sulfate assimilation, a capability not possessed by the aphid (24). Finally, it lacks all of the TCA cycle genes except for those coding for the 2-oxoglutarate dehydrogenase complex (21). The composition of the B. aphidicola seed set obtained by our analysis (Fig. 1C and Table S2) is in clear agreement with the above observations. It contains the most abundant nonessential amino acids for aphids, glutamate and glutamine [which Buchnera uses as a substrate for the synthesis of other, essential amino acids (23)], and is devoid of all of the host essential amino acids. The seed set also includes glucose and mannitol (as the only carbon sources), 2-oxoglutarate, and sulfate, as well as thiamine (vitamin B1) and spermine (an essential growth factor).
To further confirm that the composition of the seed sets obtained by our analysis is consistent with known large-scale biological observations concerning the compounds that various species extract from their environments, we consider several key compounds and examine their presence and absence pattern in the occurring compound sets (phyletic occurrence pattern) and seed sets (phyletic seed pattern) across all of the species in our analysis (SI Materials and Methods and Fig. S2). For example, while many species can synthesize all of the amino acids they require, animals have lost their ability to make some amino acids (referred to as essential amino acids) and acquire them through their diet. Conversely, some obligate intracellular parasites have lost the ability to produce the nonessential amino acids and rely on their host for exogenous provision of these amino acids (21, 25). Comparing the resulting phyletic pattern for phenylalanine (an essential amino acid) with that obtained for glutamate (a nonessential amino acid), we find these patterns to be in complete accordance with the above observations (Fig. S3). Another example is biotin (vitamin B7), an essential cofactor in carboxylation reactions. Of the 42 species that were reported in a recent comparative genomics study (26) to synthesize biotin (and hence do not require biotin uptake from the environment), indeed, 40 have biotin as an occurring compound but not as a seed compound. Of the 24 species that were reported as lacking this capacity, 20 do have biotin as a seed. Interestingly, the four species that seem to lack the capacity to synthesize biotin and do not have biotin in their seed sets all have as a seed the same biotin biosynthetic precursor, dethiobiotin, which has been shown to allow various biotin auxotrophic bacteria to grow in the absence of biotin (27). Additional examples and details are presented in SI Text and SI Materials and Methods. Identified seeds are also correlated with global topological features and are enriched in certain metabolic pathways (SI Text).
Further validation, spanning both multiple species and multiple compounds, builds on data concerning the biosynthetic capacity of several agents of human ehrlichiosis (an emerging infectious disease, primarily transmitted by ticks or trematodes). These intracellular vector-borne pathogens are of particular interest in the context of our work, because their life cycle involves both vertebrate and invertebrate hosts and thus requires metabolic adaptation to very different environments (see also ref. 28). A recent comparative genomics study explored the ability of these newly sequenced Anaplasmataceae (as well as that of other species from the Rickettsias order and other insect symbionts) to synthesize amino acids and major vitamins and cofactors (ref. 29 and table 5 therein). Comparing our identified seed sets with these data (spanning nine species and 30 compounds; Table S3), we examined whether compounds reported in ref. 29 not to be synthesized in a specific species (and therefore to be exogenously acquired) are correctly classified by our algorithm as seeds, and whether compounds that can be synthesized are correctly classified as non-seeds (SI Materials and Methods). We find an overall strong agreement between the two datasets, with a classification accuracy of 79% [P < 10−6; and accuracy of 93% (P < 10−5) for the 10 cofactors alone; SI Materials and Methods]. Moreover, focusing only on our ability to correctly predict exogenously acquired seed compounds, we reach 95% precision (percent of correctly identified seeds out of all predicted seeds; SI Materials and Methods) and 67% recall (percent of correctly identified seeds of all exogenously acquired compounds). Considering the inherent noise in the underlying metabolic data (see SI Text), these scores attest to the ability of our method to successfully identify exogenously acquired compounds.
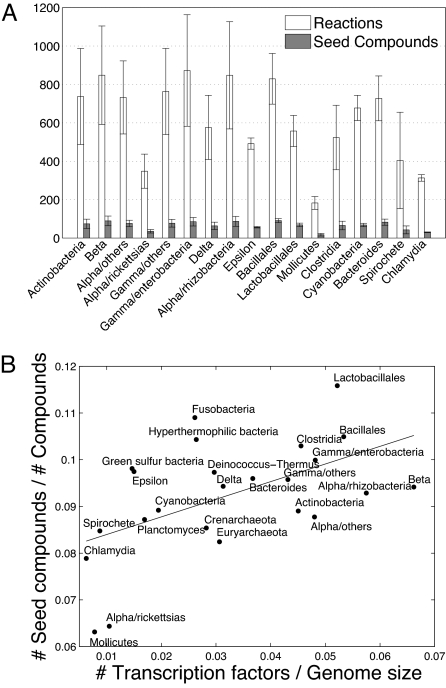
Having demonstrated that the obtained seed set data agree with various biological observations both across compounds and across species, we now turn to identify large-scale relations between the size and composition of the identified seed sets and the species' environments. Here we limit our analysis only to the prokaryotic species, for which large-scale environmental data can be obtained (Materials and Methods). Furthermore, prokaryotes facilitate a comparison between different habitats without the complications associated with tissue-specific metabolism or varying trophic levels. We find that organisms that live in extreme and narrowly defined habitats (e.g., Archaea) tend to have smaller networks and smaller seed sets (see also ref. 13 and Fig. S4). This strong correlation between the organism's environment and the network's structure and organization is particularly apparent for the bacterial phyla; the phyla with the smallest metabolic networks and smallest seed sets are Rickettsias, Mollicutes, Spirochete, and Chlamydia, which are mostly obligate intracellular parasites, inhabiting well defined and predictable environments (Fig. 2A). Moreover, specialized species living in a highly predictable environment (e.g., marine thermal vents) not only tend to have fewer occurring compounds in their networks than those living in multiple habitats, but also require a smaller fraction of these compounds as seeds (P < 3 × 10−4 and P < 0.03, respectively; Wilcoxon rank sum test). The positive correlation between variable environments and larger seed sets is further confirmed by a marked statistical correlation between the fraction of the occurring compounds included in the seed set (a normalized measure of seed sets' size, controlling for network size variation) and an index of environmental variability (0.27, P < 0.004; Spearman rank correlation; Materials and Methods). Moreover, we also find a high correlation between the fraction of the compounds in the seed set and the ratio between the number of transcription factors and genome size (0.4, P < 2 × 10−7; Spearman rank correlation; Materials and Methods and Fig. 2B), the latter of which is known to correlate with habitat variability (30). These results suggest that although species that rely on a highly predictable environment can take up from it many compounds (rather than synthesizing them), overall they still extract significantly fewer compounds than those organisms that have to survive in a wide range of environmental niches. These findings also support our intuition above; the seed set is a union of the various essential sets that correspond to the different environmental conditions in which the species can survive.
Fig. 2.
The size of seed sets across different phyla. (A) The average number of reactions and seed compounds across different bacterial taxonomic phyla. The number of seed compounds is estimated by the number of source components. Evidently, phyla that include mostly obligate intracellular parasites have, on average, the smallest metabolic networks and smallest seed sets. (B) The fraction of the occurring compounds included in the seed set as a function of the ratio between the number of transcription factors and the genome size, across prokaryotic phyla. Again, phyla of intracellular parasites (e.g., Rickettsias, Mollicutes, Spirochete, and Chlamydia) inhabiting well defined and predictable environments have small seed sets (even when normalized by the number of compounds in the network) and a small number of transcription factors. The solid line represents a linear regression. The strong correlation attests to the alignment between the size of the seed set and habitat variability.
Using a covariation correlation assay (Materials and Methods) we further confirm that the growth environment of the various prokaryotic species correlates not only with the size of the seed sets but also with their composition. Data concerning the growth environment of each species are represented as a vector of four attributes (salinity, oxygen requirements, temperature range, and habitat) using discrete categories to describe each attribute (Materials and Methods, SI Materials and Methods, and SI Text). Considering the 446 bacterial and archaeal taxa for which environmental data can be obtained, a significant correlation of 0.25 (Pearson correlation test, P < 10−3; Materials and Methods) is found between the environmental “signature” and the seed composition of a species. This correlation is in fact higher than that found between the environmental signature and the occurring compounds composition (0.21, Pearson correlation test, P < 10−3) despite the fact that occurring sets are markedly bigger than seed sets and potentially carry more information. For certain environmental attributes, species that share the same attribute value also tend to have similar seed sets. Specifically, species with microaerophilic, facultative, and aerobic oxygen requirements, multiple habitat, and mesophilic temperature range have significantly more similar seed sets than expected by chance (P < 0.05 after multiple testing correction; Materials and Methods).
Next, we characterize the evolutionary dynamics governing the gain and loss of seed and non-seed occurring compounds across a phylogenetic tree of life and the transitions between seed and non-seed states. To this end we analyzed the seed and occurring sets using methods borrowed from molecular evolution analysis (31, 32) and gene conservation analysis (33) and applying several phylogenetic analysis approaches (including both maximum parsimony algorithms and maximum likelihood models; Materials and Methods). Specifically, we considered a reference sequence-based tree and estimated the rate at which various compounds are integrated into (and lost from) evolving metabolic networks, and the rates at which seed compounds become non-seed occurring compounds and vice versa (Materials and Methods; Table 1). These estimates control for phylogenetic relations and can separate speciation dynamics from transition events.
Table 1.
The relative frequencies of transitions across the phylogenetic tree between the various states of a compound
| Original state | New state |
||
|---|---|---|---|
| Absent | Non-seed | Seed | |
| Absent | — | 10.2058 | 7.2822 |
| Non-seed | 20.5747 | — | 8.0365 |
| Seed | 31.9681 | 21.9327 | — |
These frequencies describe the expected number of transitions from one state to the other among 100 transition events in a random set of compounds with an equal number in each state. The values presented are based on ancestral network reconstruction (Materials and Methods, second assay). The transition rates that were obtained based on the compounds' phyletic patterns (first assay) and on maximum likelihood estimates (third assay) demonstrate qualitatively similar trends and can be found in Tables S4 and S5. Note that this measure controls for the different frequencies of the various states in the data and hence is not biased by the smaller number of seed states.
Our findings suggest that novel compounds are integrated into metabolic networks as either seed or non-seed compounds, where integration events of non-seeds are ≈1.5–2 times more frequent than integration of seeds (Table 1). Yet seed compounds have a higher tendency than non-seed compounds to be dropped completely from the network. Moreover, the rate of transition of a seed compound into a non-seed occurring compound is higher than that of the reverse process (Table 1). These findings suggest that, in general, the seed status is a transient phase in the “life” of a compound and that seed compounds tend to either be completely dropped from the network or change into non-seed compounds relatively fast. We further calculate several conservation measures (Materials and Methods) to estimate the expected evolutionary “lifespan” of the seed and non-seed states. We find that the seed state is significantly less conserved than the non-seed state (loss rates of 0.0523 and 0.0378, respectively; P < 10−12, Wilcoxon rank sum test), confirming again the transient nature of the seed status.
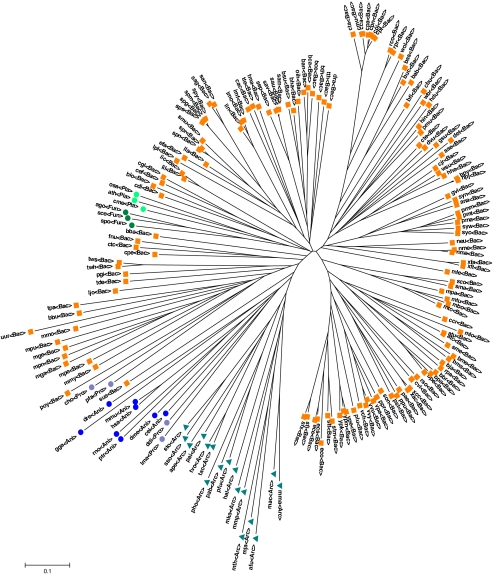
Finally, the seed content of the various species included in our analysis was used to reconstruct a phylogenetic tree de novo (SI Materials and Methods) in a manner analogous to gene content-based phylogenies. Remarkably, this tree of life not only successfully clusters most of the taxonomic groups (Fig. 3) but is just as accurate (measured by its distance from a reference sequence-based tree; SI Materials and Methods) as a tree based on the entire set of occurring compounds (Table 2), despite being based on a significantly smaller number of compounds (seed compounds account, on average, for only 10.8% of the occurring compounds). This accurate reconstruction of a phylogenetic tree is not expected by chance for such small subsets of the occurring compounds, as demonstrated by the markedly less accurate trees obtained with random compound sets (Table 2), suggesting that the identified seed sets are a significant and fundamental characteristic of each species and its evolutionary history. A principal component analysis further demonstrates that the composition of the seed sets can be used to partition the major taxonomic groups (SI Text).
Fig. 3.
Phylogenetic tree based on seed compounds content. 〉Bac〈, Bacteria (orange squares); 〉Arc〈, Archaea (cyan triangles); 〉Pla〈, plants (light green circles); 〉Ani〈, animals (blue circles); 〉Fun〈, fungi (dark green circles); 〉Pro〈, protists (purple circles).
Table 2.
The distances of phylogenetic trees based on seed compounds sets, occurring compounds sets, and random compound sets from a sequence-based reference tree
| Reconstruction method | Distance measure | Seed sets | Occurring sets | Random sets |
|---|---|---|---|---|
| NJ | BSD | 3.22 | 3.23 | 5.61 (0.01) |
| NJ | SD | 224 | 214 | 341.2 (4.2) |
| FM | BSD | 3.23 | 3.25 | 5.64 (0.01) |
| FM | SD | 216 | 228 | 339.8 (4.6) |
Random compound sets are of the same size as the real seed sets. Results for the random sets show average value and standard deviation. Trees are reconstructed and compared by using various common algorithms (SI Materials and Methods). NJ, neighbor-joining; FM, Fitch–Margoliash; BSD, branch score distance; SD, symmetric difference.
Discussion
This study introduces a large-scale reconstruction of metabolic environments using a cross-species analysis of 478 metabolic networks (and >2,200 metabolic compounds) to infer the set of compounds that each species extracts from its environment. Our analysis is based on network topology alone and ignores many other properties of metabolic reactions such as stoichiometry (the quantitative relationships between the reactants and the products of each reaction), rate, and dynamics. The network representation also weighs all pathways equally, ignoring the important distinction between catabolic and anabolic pathways. Incorporating these properties into the metabolic network model and applying a more involved analysis (such as constraint-based stoichiometric modeling) can potentially yield more accurate results (34) (see also SI Text on the potential effect of topology-based analysis on seed identification in autocatalytic cycles). The seed sets obtained with our simplified network model may thus suggest large-scale patterns in the metabolic data rather than reflect accurate stoichiometric constraints. Yet, despite these shortcomings, topological analysis has several important advantages: Most importantly and essential to the kind of analysis presented here, metabolic network topologies can readily be obtained for hundreds of species (unlike stoichiometric and kinetic models that are available only for a very small number of species), allowing a phylogenetic, large-scale analysis (30). Topology-based models also lend themselves to methods and algorithms (mostly borrowed from graph theory or complex network analysis), facilitating analyses that may not be tractable in other, more complicated models. Specifically, seed set identification—for which we have introduced a fast and relatively simple algorithm in the graph representation—is an extremely challenging task in stoichiometric networks, demanding complex optimization schemes (such as mixed-integer programming) that do not scale up to real-life size networks. Lastly, the identified seed compounds dataset was shown to agree with biological observations across species and across compounds and facilitated the characterization of various environmental factors that affect the seed sets and the dynamics by which metabolic networks evolve.
Seed sets' size was shown to correlate strongly with environmental variability, and their composition was shown to covary with several environmental features. The estimated transition rates between seeds and non-seeds and the gain and loss rate of compounds provide a detailed characterization of the overall patterns governing network evolution and suggest a complex dynamic process. The seed status of a compound appears to be relatively transient, whereas such compounds tend to be rapidly lost or convert to non-seed compounds (probably as adaptation occurs and the synthesis of these compounds from new seeds evolves). These dynamics echo those revealed in studies of horizontal gene transfer that have shown that such transfer is more likely to occur in peripheral reactions involved in nutrient uptake or first metabolic steps (11). The transition rate of seeds to non-seeds, which was found to be higher than the transition rate of the reverse process, is also in agreement with the retrograde model of network evolution (35) (positing a substrate-driven process where metabolic pathways are assembled “backwards” in an opposite direction to the flow of the metabolic pathway). However, the higher overall rate of non-seed compounds' integration and the still relatively high rate of transition of non-seeds into seeds (representing the evolution of strategies for externally acquiring previously produced compounds) suggest that other processes [e.g., the patchwork model (36)] may play an important role in the evolution of metabolic networks and that these processes are not mutually exclusive (see also ref. 37). The remarkable capacity for adaptation to a wide range of environmental niches is further exemplified by the successful reconstruction of a seed-based tree of life. The seed set analysis presented in this paper and the above findings illustrate the enormous potential of the “reverse ecology” approach (30) and facilitate further large-scale, cross-species studies concerning the evolutionary forces that shape the interplay between living organisms and their habitats.
Materials and Methods
Metabolic Networks and Relevant Data.
Metabolic networks data were downloaded from the KEGG database (15), version 41.1 (February 2007). In total, the metabolic networks of 558 species (Table S1), covering all taxonomic groups, were reconstructed (SI Materials and Methods). Draft genomes and EST contigs (KEGG organism codes with prefix “d” or “e”) were excluded from the analysis. We also discarded species that have <100 reactions, leaving a total of 478 species. An additional network, composed of the union of the reaction lists of all species, was also reconstructed and is referred to as the global network. Data concerning bacterial and archaeal environments were obtained from the prokaryotic attributes table of the NCBI Genome Project (www.ncbi.nlm.nih.gov/genomes/lproks.cgi). Each species is represented as a vector of four attributes denoting its salinity requirements (nonhalophilic, mesophilic, moderate halophile, or extreme halophile), its oxygen requirements (aerobic, microaerophilic, facultative, or anaerobic), its temperature range (cryophilic, psychrophilic, mesophilic, thermophilic, or hyperthermophilic), and its habitats (host-associated, aquatic, terrestrial, specialized, or multiple). See SI Materials and Methods for a detailed description. Further environmental data, based on a manually curated version of the above table and ranking the environmental variability of 117 bacterial species, were obtained from ref. 30. We also obtained data concerning the number of transcription factors and genome sizes of 159 bacterial species from ref. 38 and used the ratio of these two values as an additional, quantitative measure of environmental variability (30).
Identifying Seed Compounds.
Each network was decomposed into its strongly connected components using Kosaraju's algorithm (19) (SI Materials and Methods and Fig. S5). A strongly connected component is a maximal set of nodes such that for every pair of nodes u and v there is a path from u to v and a path from v to u. The strongly connected components form a directed acyclic graph whose nodes are the components and whose edges are the original edges in the graph that connect nodes in two different components. In this graph, each component without incoming edges and at least one outgoing edge is defined as a source component. Each source component in the SCC decomposition forms a collection of candidate seed compounds. The set of seed compounds must include exactly one compound from each source component and should not include any other compound. In the following, we briefly provide the intuition (based on a graph-based representation of the network; see also the discussion above concerning reaction stoichiometry): First, it should be noted that every strongly connected component is an equivalence class; if one of the compounds in the component can be produced then all others can be produced as well. Second, because source components do not have any incoming edges, if none of the compounds in a source component is present in the seed set, none of the compounds in this component can be produced. Finally, if at least one compound from each source component is included in the seed set, a path from a seed compound to any other compound in the network can be found and hence all compounds in the network can be produced. Because all of the compounds in a source component are equally likely to be included in the seed set, each of these compounds was assigned a confidence level, C = 1/(component size), denoting the compound's probability of being a seed. We used a threshold of C ≥ 0.2 to determine whether a compound should be regarded as a seed or not (including all compounds that are part of source components of size 5 or less). With this threshold value we discarded on average only 3.3% of the seeds. Using other threshold values (specifically, C ≥ 0.1 or C ≥ 0.01) did not significantly change any of our results. Dataset S1 describes the composition of the seed set in each species (with the associated C values). See also Fig. S6, illustrating the metabolic network of yeast with the seed compounds highlighted.
Covariation Correlation Assay.
To examine the correlation between seed set composition and environmental attributes across all bacterial species, we applied an assay similar to the one used in ref. 39. Given N species, two N × N distance matrices, Ss and Sh, were constructed. Ss represents the pairwise Jaccard distance (40) between the seed sets of the various species. Sh represents the pairwise Hamming distance between the vectors of attributes describing the environments of these species. The Pearson correlation between the (n2 − n)/2 entries forming the lower triangle of Ss and Sh was calculated. Statistical significance of the resulting correlation was computed by shuffling the species' labels 1,000 times and calculating the probability to achieve an equal or higher absolute value correlation score by chance. An additional assay examines the similarity among seed sets of various species with a certain environmental attribute value. The average pairwise distance between the seed sets of all species with that specific attribute value was calculated and compared with the average distances obtained for 100,000 random collections of species (of the same size) to determine its statistical significance. The resulting P values were corrected for multiple testing via the false discovery rate procedure (41).
Phylogenetic Analysis, Transition Rates, and Conservation.
We consider a well established, sequence similarity-based tree as a reference phylogeny (42). This tree is based on 31 orthologs and includes a relatively large number of species, covering most of the taxonomic groups for which metabolic data are available. Our phylogenetic-based analyses were restricted to the species that could be matched to those included in the reference tree, resulting in a total of 178 species. A given compound in each species (extant or ancestral) can take one of three distinct states: absent (completely absent from the occurring compounds set), non-seed (an occurring compound that is not part of the seed set), or seed (an occurring compound that is part of the seed set). Our seed set analysis determines the state of each compound in the extant species. To calculate the evolutionary transition rate between the different states across the phylogenetic tree, we applied three assays analogous to those used in nucleotide substitution analysis (see also SI Materials and Methods). In the first assay, the compounds' states in each internal node of the phylogenetic tree (representing ancestral species) were predicted, using Fitch's small parsimony algorithm (43). Fitch's algorithm finds the most parsimonious state assignment for all of the internal nodes of a phylogenetic tree, given a phyletic pattern that assigns states to the terminal, current species nodes. We then calculate the relative frequencies of the substitution between the different states following the Tamura and Nei approach (31), a commonly applied method for substitution rate estimation (SI Materials and Methods). In the second assay, the state of each compound in the internal nodes of the tree is estimated, but this time based on the reconstruction of the metabolic network in each ancestral species, following Kreimer et al. (28). The seed set detection algorithm is then applied to each ancestral network to obtain the set of occurring and seed compounds in the internal nodes. Tamura and Nei's method is used again to estimate the relative transition rates. In the third assay, a maximum likelihood approach is applied to the phyletic patterns of all of the compounds in our analysis to obtain a maximum likelihood estimate of the substitution rates. This is computed with the PAML package (32) using the UNREST model. Two additional conservation measures, propensity for gene loss (PGL; a maximum parsimony measure) (44) and gene loss rate (GLR; a maximum likelihood measure) (33), were also applied to the phyletic patterns of the various compounds to compute the tendency of a compound to lose its state as a seed compound (or its state as a non-seed compound) during the evolutionary process. With these measures we do not distinguish between cases where the compound was completely dropped from the network and cases where its state converted to another state, but rather aim to characterize the level of conservation of each state. These conservation measures can therefore be conceived as representing the average “lifespan” of the state. The results obtained for the PGL measure were qualitatively similar to those obtained with the GLR measure and are not presented.
Supplementary Material
Acknowledgments.
We thank the anonymous reviewers and the editor for their helpful comments. We are grateful to Tomer Shlomi for numerous valuable suggestions (including the use of SCC decomposition). We thank Ruth Hershberg, Laurent Lehmann, DT Levy, and Jeremy Van Cleve for their comments. E.B.'s research is supported by the Yeshaya Horowitz Association through the Center for Complexity Science, the Morrison Institute for Population and Resource Studies, and by a grant to the Santa Fe Institute from the James S. McDonnell Foundation 21st Century Collaborative Award Studying Complex Systems. This research is supported in part by National Institutes of Health Grant GM28016 (to M.W.F.). M.K.'s research is supported by grants from the Israel Science Foundation and the Israeli Ministry of Science and Technology. E.R.'s research is supported by grants from the Israel Science Foundation, the German–Israeli Fund, the Yishayahu Horowitz Center for Complexity Science, and the Tauber Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806162105/DCSupplemental.
References
- 1.Alon U. Biological networks: The tinkerer as an engineer. Science. 2003;301:1866–1867. doi: 10.1126/science.1089072. [DOI] [PubMed] [Google Scholar]
- 2.Jeong H, Tombor B, Albert R, Oltvai Z, Barabasi A. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 3.Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles E. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–193. doi: 10.1038/nature01166. [DOI] [PubMed] [Google Scholar]
- 4.Smith E, Morowitz H. Universality in intermediary metabolism. Proc Natl Acad Sci USA. 2004;101:13168–13173. doi: 10.1073/pnas.0404922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papp B, Pal C, Hurst L. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature. 2004;429:661–664. doi: 10.1038/nature02636. [DOI] [PubMed] [Google Scholar]
- 6.Deutscher D, Meilijson I, Kupiec M, Ruppin E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat Genet. 2006;38:993–998. doi: 10.1038/ng1856. [DOI] [PubMed] [Google Scholar]
- 7.Almaas E, Kovacs B, Vicsek T, Oltvai Z, Barabási A. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427:839–843. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- 8.Ibarra R, Edwards JS, Palsson B. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- 9.Handorf T, Ebenhoh O, Heinrich R. Expanding metabolic networks: Scopes of compounds, robustness, and evolution. J Mol Evol. 2005;61:498–512. doi: 10.1007/s00239-005-0027-1. [DOI] [PubMed] [Google Scholar]
- 10.Raymond J, Segre D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 11.Pal C, Papp B, Lercher M. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 12.Yamada T, Kanehisa M, Goto S. Extraction of phylogenetic network modules from the metabolic network. BMC Bioinformatics. 2006;7:130. doi: 10.1186/1471-2105-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebenhoh O, Handorf T, Heinrich R. A cross species comparison of metabolic network functions. Genome Inform. 2005;16:203–213. [PubMed] [Google Scholar]
- 14.Ebenhoh O, Handorf T, Kahn D. Evolutionary changes of metabolic networks and their biosynthetic capacities. IEE Proc Syst Biol. 2006;153:354–358. doi: 10.1049/ip-syb:20060014. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, et al. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klausmeier C, Litchman E, Levin S. A model of flexible uptake of two essential resources. J Theor Biol. 2007;246:278–289. doi: 10.1016/j.jtbi.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 17.Ancel Meyers L, Bull J. Fighting change with change: Adaptive variation in an uncertain world. Trends Ecol Evol. 2002;17:551–557. [Google Scholar]
- 18.Palumbo M, Colosimo A, Giuliani A, Farina L. Functional essentiality from topology features in metabolic networks: A case study in yeast. FEBS Lett. 2005;579:4642–4646. doi: 10.1016/j.febslet.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 19.Aho A, Hopcroft J, Ullman J. The Design and Analysis of Computer Algorithms. Reading, MA: Addison-Wesley; 1974. [Google Scholar]
- 20.Green M, Karp P. The outcomes of pathway database computations depend on pathway ontology. Nucleic Acids Res. 2006;34:3687–3697. doi: 10.1093/nar/gkl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 22.Moran N, Mira A. The process of genome shrinkage in the obligate symbiont buchnera aphidicola. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-12-research0054. :research0054.1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran N, Degnan P. Functional genomics of buchnera and the ecology of aphid hosts. Mol Ecol. 2006;15:1251–1261. doi: 10.1111/j.1365-294X.2005.02744.x. [DOI] [PubMed] [Google Scholar]
- 24.Douglas A. Sulphate utilization in an aphid symbiosis. Insect Biochem. 1998;18:599–605. [Google Scholar]
- 25.Stephens R, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 26.Hebbeln P, Rodionov D, Alfandega A, Eitinger T. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci USA. 2007;104:2909–2914. doi: 10.1073/pnas.0609905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowman W, DeMoll E. Biosynthesis of biotin from dethiobiotin by the biotin auxotroph lactobacillus plantarum. J Bacteriol. 1993;175:7702–7704. doi: 10.1128/jb.175.23.7702-7704.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreimer A, Borenstein E, Gophna U, Ruppin E. The evolution of modularity in bacterial metabolic networks. Proc Natl Acad Sci USA. 2008;105:6976–6981. doi: 10.1073/pnas.0712149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunning Hotopp J, et al. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parter M, Kashtan N, Alon U. Environmental variability and modularity of bacterial metabolic networks. BMC Evol Biol. 2007;7:169. doi: 10.1186/1471-2148-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1951. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 33.Borenstein E, Shlomi T, Ruppin E, Sharan R. Gene loss rate: A probabilistic measure for the conservation of eukaryotic genes. Nucleic Acids Res. 2007;35:e7. doi: 10.1093/nar/gkl792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rokhlenko O, Shlomi T, Sharan R, Ruppin E, Pinter R. Constraint-based functional similarity of metabolic genes: Going beyond network topology. Bioinformatics. 2007;23:2139–2146. doi: 10.1093/bioinformatics/btm319. [DOI] [PubMed] [Google Scholar]
- 35.Horowitz H. On the evolution of biochemical synthesis. Proc Natl Acad Sci USA. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazcano A, Miller S. The origin and early evolution of life: Prebiotic chemistry, the pre-RNA world, and time. Cell. 1996;85:793–798. doi: 10.1016/s0092-8674(00)81263-5. [DOI] [PubMed] [Google Scholar]
- 37.Rison S, Thornton J. Pathway evolution, structurally speaking. Curr Opin Struct Biol. 2002;12:374–382. doi: 10.1016/s0959-440x(02)00331-7. [DOI] [PubMed] [Google Scholar]
- 38.Babu M, Teichmann S, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J Mol Biol. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 39.Kaufman A, Dror G, Meilijson I, Ruppin E. Gene expression of Caenorhabditis elegans neurons carries information on their synaptic connectivity. PLoS Comput Biol. 2006;2:e167. doi: 10.1371/journal.pcbi.0020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vaudoise Sci Nat. 1908;44:223–270. [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 42.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 43.Fitch W. Towards defining the course of evolution: Minimum change for a specific tree topology. Syst Zool. 1971;20:406–416. [Google Scholar]
- 44.Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.