Abstract
Phylogenetic reconstructions are often plagued by difficulties in distinguishing phylogenetic signal (due to shared ancestry) from phylogenetic noise or homoplasy (due to character-state convergences or reversals). We use a new interpretive hypothesis, termed hemiplasy, to show how random lineage sorting might account for specific instances of seeming “phylogenetic discordance” among different chromosomal traits, or between karyotypic features and probable species phylogenies. We posit that hemiplasy is generally less likely for underdominant chromosomal polymorphisms (i.e., those with heterozygous disadvantage) than for neutral polymorphisms or especially for overdominant rearrangements (which should tend to be longer-lived), and we illustrate this concept by using examples from chiropterans and afrotherians. Chromosomal states are especially powerful in phylogenetic reconstructions because they offer strong signatures of common ancestry, but their evolutionary interpretations remain fully subject to the principles of cladistics and the potential complications of hemiplasy.
Keywords: cladistics, gene trees, lineage sorting, phylogeny, species trees
In phylogenetic analyses, systematists routinely strive to distinguish homology (trait similarity due to shared ancestry) from homoplasy (trait similarity arising from evolutionary convergence, parallelism, or character-state reversals). Homology can offer valid phylogenetic signal, whereas homoplasy is regarded as evolutionary noise that, if not properly accommodated, jeopardizes phylogenetic reconstructions. Homology itself has distinct components, as first emphasized by Hennig (1) in his insightful distinction between symplesiomorphies (traits showing shared-ancestral homology) and synapomorphies (traits with shared-derived homology). From a Hennigian perspective, only valid synapomorphies properly earmark clades.
The critical distinctions between homoplasy and homology and between different kinds of homology have served the field of systematics well. However, a difficulty arises when a shared-derived genetic trait that from mechanistic considerations should be homoplasy-free nonetheless recurs in two or more taxa that seem to be unrelated. For example, suppose that a derived chromosomal inversion with presumably unique (monophyletic) endpoint breaks is present in two or more species that belong to disparate clades. Under the traditional interpretive framework outlined above, this phylogenetic dilemma could only be resolved in either of two ways: by supposing that the inversion has evolved multiple times independently, notwithstanding mechanistic karyotypic arguments to the contrary; or by supposing that the shared trait does earmark a bona-fide organismal clade, notwithstanding independent phylogenetic evidence to the contrary.
Here, we raise another potential explanation for this kind of phylogenetic enigma, and illustrate its application to karyotypic data involving chromosomal syntenies in mammals. Each synteny is a large conserved block of DNA, i.e., a linked assemblage of ordered loci. Numerous syntenies have been revealed in various mammals through chromosomal painting, principally by using flow-sorted whole human chromosomes as genetic probes (2). These syntenic blocks, sometimes shared across even distantly related species, may involve entire chromosomes, chromosomal arms, or chromosomal segments. Based on the premise that each syntenic assemblage in extant species is of monophyletic origin, researchers have reconstructed phylogenies and ancestral karyotypes for numerous mammalian taxa (ref. 3 and references therein). Normally, the phylogenetic inferences from these cladistic appraisals are self-consistent (across syntenic blocks) and taxonomically reasonable, and they have helped greatly to identify particular mammalian clades. However, in a few cases problematic phylogenetic patterns of the sort described above have emerged.
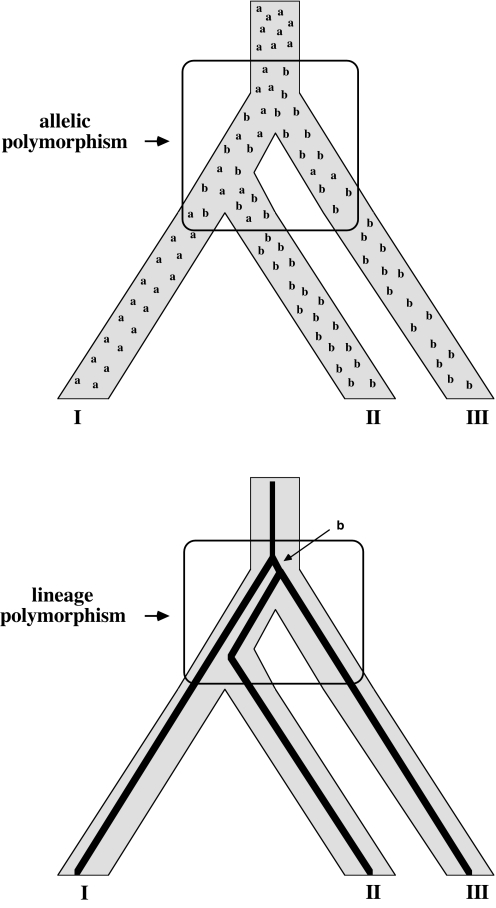
We recently introduced the term hemiplasy (3), which we defined as any topological discordance between a gene tree and a species tree attributable to the phylogenetic sorting of genetic polymorphisms across successive nodes in a species tree. Two fundamentally equivalent diagrammatic representations of hemiplasy are shown in Fig. 1. Therefore, hemiplasy is a genuine form of trait homology (orthology, either of alleles or of genealogical lineages) that gives the illusion of homoplasy in an organismal tree, but nonetheless is not homoplasy at the gene-tree level. Hemiplasy is also to be distinguished from other potential sources of gene-tree/species-tree discordance, including introgression, genetic transformation, or viral-mediated DNA transfer (which can be important in some biological settings, but which we do not consider further in this article). Here, we raise the possibility that hemiplasy might account for some of the phylogenetic anomalies that seem to be present in the taxonomic distributions of particular syntenic blocks of genes within mammalian karyotypes.
Fig. 1.
Schematic representations of hemiplasy. Shown are the distributions of a genic or chromosomal polymorphism (Upper) and a set of genealogical lineages (Lower) that traversed successive speciation nodes in an organismal phylogeny (broad branches) only to become fixed, by lineage sorting, in descendant species in a pattern that appears at face value to be discordant with the species phylogeny. In these diagrams, species II and III both have the homologous and derived character “b,” so the gene tree gives the impression that species II and III are the more closely related. However, in truth species I and II are sister taxa, despite the fact that species I alone retains the ancestral genetic condition “a.”
Background
Theory.
Nei (ref. 4, pp. 401–403) quantified the theoretical probability of what we would now term hemiplasy for the simplest possible evolutionary case: neutral alleles in three related and geographically unstructured species. Under that scenario, the probability of a gene-tree/species-tree discordance is (2/3)e−T/2N where T is the number of generations between the successive speciation events, N is the effective population size, and e is the base of the natural logarithms. According to this formula, hemiplasy is likely under some realistic evolutionary parameters. For example, the single-locus probability of hemiplasy (or the expected percentage of loci displaying hemiplasy) is ≈50% when T/2N = 0.3, as would be true, for example, if two speciation nodes were ≈1.0 million generations apart and the effective population size between nodes was ≈1.7 million individuals. Similar theory applied to more complex phylogenetic settings can be found in refs. 5 and 6.
SINE Data.
Short-interspersed elements (SINEs) have long been touted as powerful phylogenetic markers, for several reasons (7): each SINE is a retropseudogene that resides at a specific chromosomal location; each occupied site is thought to represent a single (monophyletic) insertion event; SINEs are stable once inserted into a genome [in large part because no known mechanism exists for excision of an element, such that SINE absence at a chromosomal site presumably reflects the ancestral state (8)]; and large numbers of independent SINEs are dispersed in the genomes of most eukaryotic organisms. For these reasons, any SINE shared by different species was initially assumed to be definitive in marking an organismal clade. As phrased by Nikaido et al. (9), with respect to SINEs “the probability that homoplasy will obscure phylogenetic relationships is, for all practical purposes, zero.”
However, it soon became clear that different SINEs sometimes (albeit rarely) disagree in the organismal clades they presumably delineate. This and other evidence led Hillis (10) to note that lineage sorting can introduce homoplasy-like outcomes (that we would now term hemiplasy) when a polymorphism becomes fixed in some but not all of the descendants of a polymorphic ancestor. Researchers quickly acknowledged this possibility for some of the occasional character conflicts among SINES. For example, Shedlock and Okada (11) wrote that “if the time to fixation transcends species boundaries formed in rapid succession, such as can occur during explosive radiations, inconsistent patterns of SINE insertions may be observed because of ancestral polymorphism.” Similar statements now appear as standard caveats for published phylogenies on SINEs (12), and there has emerged a widespread recognition that SINES are merely “nearly perfect” (13) rather than perfect phylogenetic markers.
Karyotypic Hemiplasy.
Syntenic blocks involving entire chromosomes, chromosomal arms, or large chromosomal segments are sometimes shared across even distantly-related mammalian species. Standard wisdom is that each such syntenic block is of monophyletic origin, because the independent assembly of a shared synteny in different lineages seems mechanistically highly implausible (14). In other words, shared syntenic associations are much more useful in defining clades than chromosomal reorganizations that disrupt an ancestral synteny, because the latter might recur by means of breakpoint reuse (15–18), especially in regions of segmental duplications (19), high concentrations of repetitive elements, and fragile sites (18, 20, 21). With respect to monophyletic origin (but not with respect to the probability of later evolutionary loss), the potential phylogenetic utility of syntenic blocks thus bears a considerable analogy to the phylogenetic utility of SINE elements, both of which are viewed as rare genomic changes (22).
During the course of comparing previously identified syntenic blocks in eutherian mammals against outgroup taxa (23), we noticed at least two candidate examples of hemiplasy (involving chiropterans and afrotherians) that have motivated this report. In what follows, each syntenic block in a given mammalian species is named according to the human chromosome or chromosomal arm to which its linked loci are apparently homologous, as judged by cross-species chromosome painting (CP) by using human chromosomes as genetic probes. These data are sometimes used to construct phylogenetic trees or, more frequently, the chromosomal characters themselves are mapped onto a consensus sequence-based tree.
Chiroptera.
By using similar types of chromosomal analyses, cross-species CP coupled with chromosomal mapping to a consensus sequence-based tree derived from (24–26), Mao et al. (27) identified 10 presumed instances of homoplasy (convergent evolution in this case) in Chiroptera that they regarded as weakening the reliability of chromosomal characters for resolving interfamily relationships of bats. For example, a presumably homoplasic synteny (chromosomal block HSA 1/6/5) was shared by particular species representing two rather distinct bat families, Pteropodidae and Megadermatidae (Fig. 2). However, an alternative possibility is that this and other such cases might be due to hemiplasy.
Fig. 2.
Phylogenetic relationships among bat species representing four taxonomic families (redrawn from ref. 27). Also shown is the presence of the chromosomal synteny 1/6/5, which formerly was interpreted to be homoplasic (27) but might instead be an example of hemiplasy.
By using calibrations from a relaxed Bayesian molecular clock (28), Eick et al. (24) estimated the following divergence times for pertinent nodes in the chiropteran tree (Fig. 2): 41 mya for the split of Rhinolophidae from Hipposideridae; 54 mya for the split of the Megadermatidae from Hipposideridae plus Rhinolophidae; and 58 mya for the divergence of Pteropodidae from Rhinolophidae plus Hipposideridae plus Megadermatidae. These estimates provide a temporal framework for the required persistence times (under the hemiplasy scenario) of chromosomal polymorphisms across successive speciation nodes. If the syntenic block HSA 1/6/5 was part of a polymorphism in the ancestral lineage leading to Pteropodidae and Megadermatidae, this polymorphism must have persisted for ≈4 million years before sorting eventually into the descendant lineages where the different chromosomal arrangements today are housed. Similar reasoning can be applied to the other “homoplasic” karyotypic states identified by Mao et al. (27).
Most of the rearrangements responsible for repatterning chiropteran genomes involve centric or Robertsonian fusions (29, 30), which, unless in monobrachial combinations (31), are perceived as not being particularly underdominant (i.e., possessing heterozygous disadvantage). So, perhaps such rearrangements occasionally do survive as polymorphic states for considerable lengths of time, although this would also depend on historical variables including effective population size and spatial population structure. Robertsonian polymorphisms, such as the 2n = 56, 58, 62 series documented in Rhinolophus hipposideros from different geographic areas and the 2n = 42 − 44 variation in Rhinolophus pearsoni from various provinces in China (32), are well known in bats, and in some other mammalian groups including rodents (33, 34), bovids (35), insectivores (36), and primates (37). In some cases (such as in the 44-chromosome gibbons), multiple related species share polymorphic chromosomal conditions, indicating that the polymorphisms have survived speciation events (38).
Afrotheria.
This well supported mammalian clade of Afro-Arabian origin is comprised of elephants, sireneans, elephant shrews, golden moles, tenrecs, and the aardvark. One of the most enduring phylogenetic problems within the group concerns the position of aardvark and elephant shrews with respect to other afrotherians (39). Strong support exists for a clade (the Afroinsectiphillia) comprising elephant shrews, aardvarks, tenrecs, and golden moles (40–42), but sister group relationships within the Afroinsectiphillia remain vague. For example, Amrine-Madsen et al. (40) and Murphy et al. (42), among others, find evidence for a phylogenetic association of elephant shrew, tenrec, and golden mole to the exclusion of aardvark (Fig. 3A), whereas Waddell and Shelley (41) conclude from an analysis of a different concatenation of nuclear and mitochondrial sequences that an association exists of aardvark, tenrec, and golden mole to the exclusion of elephant shrew (Fig. 3B). To further complicate matters, cytogenetic data (43) seem at face value to unite elephant shrew and aardvark to the exclusion of golden mole (Fig. 3C). Underpinning this latter suggestion are two syntenic blocks (HSA 10q/17 and HSA 3/20) that are present in elephant shrew (43, 44) and aardvark (45) but absent in golden mole (43).
Fig. 3.
Competing phylogenetic hypotheses for species comprising the Afroinsectiphillia clade (aardvark, elephant shrew, golden mole, and tenrec). (A) Clade comprising the elephant shrew, tenrec, and golden mole to the exclusion of aardvark based on concatenations of nuclear and mitochondrial DNA sequences (40, 42). (B) Clade composed of aardvark plus tenrec and a weaker grouping of these taxa and the golden mole to the exclusion of elephant shrew based on a different concatenation (41). (C) Clade composed of aardvark and elephant shrew to the exclusion of golden mole based on cross-species CP (43).
To reconcile these seemingly incompatible phylogenetic interpretations, at least three possibilities exist (apart from independent convergent evolution of the syntenic blocks, which seems highly unlikely on mechanistic grounds). First, perhaps the phylogeny in Fig. 3A is correct, in which case 10q/17 and 3/20 could be long-retained symplesiomorphies (stemming from the ancestor of Afroinsectiphillia) in the extant elephant shrew and aardvark lineages. In principle, hemiplasy for 10q/17 and 3/20, as diagrammed in Fig. 1, could also account for this phylogeny. Second, perhaps the phylogeny in Fig. 3C is correct, but this would contradict much other phylogenetic evidence. Last, perhaps Fig. 3B is correct, in which case the 10q/17 and 3/20 syntenies must have been polymorphic in the ancestor to Afroinsectiphillia and the polymorphisms were later lineage-sorted in a way that produced the gene tree/species tree discordances. This latter possibility would be an example of hemiplasy in a phylogeny for which the alternative explanation of symplesiomorphy (as traditionally defined) would not apply.
Both the 10q/17 and 3/20 syntenies in the aardvark and elephant shrew appear to result from Robertsonian fusions (43–45) that probably arose ≈75 mya in a common ancestor to Afroinsectiphillia (these syntenies are not present in the closest outgroups, elephant and manatee; refs. 45–47). Likely origination dates for the elephant shrew lineage and for the tenrec/golden mole/elephant shrew clade (Fig. 3) are reportedly ≈73 mya and ≈65 mya, respectively (42). Thus, if either of the hypothetical scenarios involving lineage sorting and hemiplasy is correct, then these derived chromosomal syntenies must have persisted as polymorphisms (with their respective counterpart arrangements) for at least ≈2 million years, and maximally for more than ≈10 million years to temporally encompass the relevant speciation nodes.
Discussion
Distinguishing phylogenetic signal (due to shared ancestry) from phylogenetic noise or homoplasy (e.g., due to character-state convergence) is the perennial challenge in systematics. The complications of homoplasy are widely appreciated in DNA sequence analyses (as well as in morphology-based systematics), but they can also attend efforts to reconstruct phylogenies from other categories of data including chromosomal characters (48–51). Another type of potential phylogenetic complication is hemiplasy, as defined in Background and Fig. 1. Although the distinction between gene trees and species phylogenies has long been appreciated (52–56), the phylogenetic ramifications of hemiplasy (in contrast with those of homoplasy) have often been overlooked.
Hemiplasy as a theoretically plausible evolutionary phenomenon is not in doubt. It is a logical consequence of several biological realities: lineage turnover by means of differential organismal reproduction, the fact that species are composed of populations of individuals, the fact that every new mutation originates in one individual or family, and, therefore, the fact that any evolutionary change of state necessitates a transitional phase of genetic polymorphism at the population level. Hemiplasy can also affect any and all classes of phylogenetic marker, regardless of their mechanistic susceptibilities to homoplasy. The outstanding question is whether hemiplasy materially affects real datasets.
Some of the most favorable opportunities for identifying instances of hemiplasy involve datasets for which homoplasy can be essentially disregarded as a complicating evolutionary factor. For example, all extant occurrences of a SINE element at a particular chromosomal site presumably trace back to a single (monophyletic) insertion event, so convergent evolution can often be quite safely eliminated as an explanation for why particular species might share a particular SINE; and this feature in turn has also helped researchers identify some probable (but formerly overlooked) instances of hemiplasy in SINE data (see Background).
Here, we have adopted a similar rationale to address the possibility that some chromosomal syntenies, each thought to be of monophyletic origin, might likewise be present in seemingly unrelated species because of lineage sorting from polymorphic conditions. The term hemiplasy formalizes and extends some of the pioneering cytogenetic observations by Dutrillaux and coworkers involving idiosyncratic lineage sorting in primates (57), and it offers an alternative explanation for some chromosomal states that conventionally were interpreted to have arisen convergently in different lineages, or were subject to evolutionary reversals each requiring the precise disruption of two adjacent syntenies.
The phenomenon of hemiplasy is most plausible when the internodal distances in a phylogenetic tree are short (relative to effective population sizes) and/or when the persistence time of a polymorphism is long. The latter, in turn, is more likely for neutral polymorphisms (such as some Robertsonian fusions that have little or no impact on fertility) than it is for polymorphisms that are underdominant, and it is especially likely for balanced polymorphisms. Hemiplasy is also more likely in species that are subdivided geographically, such that the multiple genetic elements of a collective polymorphism are buffered against extinction by virtue of being housed and perhaps fixed in different populations. Conversely, hemiplasy is least likely under circumstances where the fixation of new genetic variants occurs rapidly in each nonstructured species through genetic drift, inbreeding, selection in favor of homozygotes, or meiotic drive.
Even in genuinely homoplasy-free datasets, the distinction between a gene tree and species tree and the possibility of hemiplasy mean that no single genetic character can be deemed definitive in earmarking a clade. In recognition of this fact, Waddell et al. (58) proposed a statistical framework for testing clades by using data from SINEs. The particular tests apply to any rooted tree, with three taxa, under a Wright–Fisher coalescent model and assumptions of panmixia, nonoverlapping generations, and constant population size. From their analyses (which in effect take lineage sorting and the possibility of hemiplasy into account), at least three SINE (or chromosomal) characters (none being contradictory) would be required to reject alternative phylogenetic groupings at the 95% confidence level.
In practice, even this small number of synapomorphic traits might be difficult to attain in various types of karyotypic data. Consider, for example, the Afroinsectiphillia, where only two shared syntenies (HSA 10q/17 and HSA 3/20) currently support a sister association between elephant shrew and aardvark (the probability of this outcome by chance lineage sorting is 0.11; see table 3 in ref. 58). Thus, strict rejection of the alternate hypotheses [(aardvark plus golden mole) plus elephant shrew, or (elephant shrew plus golden mole) plus aardvark] is not yet possible. Such considerations highlight a general caveat about the use of otherwise powerful chromosomal rearrangements in constructing phylogenies: the number of informative characters at a particular node must be above the critical threshold required for statistical support (given the reality of the distinction between gene trees and a species tree).
In closing, we do not claim to have proven any instances of hemiplasy in the current karyotypic datasets on syntenic blocks. Rather, our intent has been to raise consciousness about the hemiplasy phenomenon in the field of chromosomal research, and thereby stimulate further discussion along these lines. The impact of hemiplasy is a function of several historical variables (including population demographics, selection mode and intensity, and internodal times) that traditionally have been almost entirely neglected in phylogenetic reconstructions based on chromosomal characters. It would be useful to evaluate the possibility of karyotypic hemiplasy in a wide variety of phylogenetic settings.
Acknowledgments.
We thank Peter Waddell for drawing our attention to the applicability of statistical analyses developed for SINEs in testing of the validity of phylogenetic affinities based on chromosomal data. Gauthier Dobigny, Lutz Froenicke, and Darren Griffin provided insightful comments on various drafts of this manuscript. T.J.R. was supported by the National Research Foundation (South Africa), and A.R.-H. was supported by a Spanish Ministry of Education and Science (MEC) Postdoctoral Fellowship. J.C.A. was supported by funds from the University of California at Irvine.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hennig W. Grundzüge einer Theorie der Phylogenetischen Systematik. Berlin: Deutscher Zentralverlag; 1950. (1996 Phylogenetic Systematics (University of Illinois Press, Urbana, Illinois) (English) [Google Scholar]
- 2.Ferguson-Smith MA, Trifonov V. Mammalian karyotype evolution. Nat Rev Genet. 2007;8:950–962. doi: 10.1038/nrg2199. [DOI] [PubMed] [Google Scholar]
- 3.Avise JC, Robinson TJ. Hemiplasy: A new term in the lexicon of phylogenetics. Syst Biol. 2008;57:503–507. doi: 10.1080/10635150802164587. [DOI] [PubMed] [Google Scholar]
- 4.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ Press; 1987. [Google Scholar]
- 5.Rosenberg NA. The probability of topological concordance of gene trees and species trees. Theor Pop Biol. 2002;61:225–247. doi: 10.1006/tpbi.2001.1568. [DOI] [PubMed] [Google Scholar]
- 6.Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006;2:e68. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shedlock AM, Okada N. SINE insertions: Powerful tools for molecular systematics. BioEssays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 8.Nishihara H, et al. A retroposon analysis of Afrotherian phylogeny. Mol Biol Evol. 2005;22:1823–1833. doi: 10.1093/molbev/msi179. [DOI] [PubMed] [Google Scholar]
- 9.Nikaido M, Rooney AP, Okada N. Phylogenetic relationships among cetartiodactyls based on evidence from insertions of SINEs and LINEs: Hippopotamuses are the closest extant relatives of whales. Proc Natl Acad Sci USA. 1999;96:10261–10266. doi: 10.1073/pnas.96.18.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillis DM. SINEs of the perfect character. Proc Natl Acad Sci USA. 1999;96:9979–9981. doi: 10.1073/pnas.96.18.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shedlock AM, Milinkovitch MC, Okada N. SINE evolution, missing data, and the origin of whales. Syst Biol. 2000;49:808–817. doi: 10.1080/106351500750049851. [DOI] [PubMed] [Google Scholar]
- 12.Shedlock AM, Takahashi K, Okada N. SINEs of speciation: Tracking lineages with retroposons. Trends Ecol Evol. 2004;19:545–553. doi: 10.1016/j.tree.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Ray DA, Xing J, Salem A-H, Batzer MA. SINEs of a nearly perfect character. Syst Biol. 2006;55:928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- 14.Müller S, et al. Molecular dissection of human chromosome 3 and 21 evolution. Proc Natl Acad Sci USA. 2000;97:206–211. doi: 10.1073/pnas.97.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pevzner PA, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci USA. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao S, et al. Human, mouse, and rat genome large-scale rearrangements: Stability versus speciation. Genome Res. 2004;14:1851–1860. doi: 10.1101/gr.2663304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy WJ, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Herrera A, Castresana J, Robinson TJ. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 2006;7:R115. doi: 10.1186/gb-2006-7-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehrer-Sawatzki H, Cooper DN. Molecular mechanisms of chromosomal rearrangement during primate evolution. Chromosome Res. 2008;16:41–56. doi: 10.1007/s10577-007-1207-1. [DOI] [PubMed] [Google Scholar]
- 20.Robinson TJ, Ruiz-Herrera A, Froenicke L. Dissecting the mammalian genome: New insights into chromosomal evolution. Trends Genet. 2006;22:297–301. doi: 10.1016/j.tig.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Herrera A, et al. Evolutionary conserved chromosomal segments in the human karyotype are bounded by unstable chromosome bands. Cytogenet Genome Res. 2005;108:161–174. doi: 10.1159/000080812. [DOI] [PubMed] [Google Scholar]
- 22.Rokas A, Holland PW. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 23.Robinson TJ, Ruiz-Herrera A. Defining the ancestral eutherian karyotype: A cladistic interpretation of chromosome painting and genome sequence assembly data. Chromosome Res. 2008 doi: 10.1007/s10577-008-1264-0. in press. [DOI] [PubMed] [Google Scholar]
- 24.Eick G, Jacobs DS, Matthee CA. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera) Mol Biol Evol. 2005;22:1869–1886. doi: 10.1093/molbev/msi180. [DOI] [PubMed] [Google Scholar]
- 25.Teeling EC, et al. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 26.Jones G, Teeling EC. The evolution of echolocation in bats. Trends Ecol Evol. 2006;21:149–156. doi: 10.1016/j.tree.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Mao X, et al. Comparative cytogenetics of bats (Chiroptera): The prevalence of Robertsonian translocations limits the power of chromosomal characters in resolving interfamily phylogenetic relationships. Chromosome Res. 2008;16:115–170. doi: 10.1007/s10577-007-1206-2. [DOI] [PubMed] [Google Scholar]
- 28.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 29.Baker RJ, Bickham JW, Arnold ML. Chromosomal evolution in. Rhogeessa (Chiroptera: Vespertilionidae): Possible speciation by centric fusions. Evolution. 1985;39:233–243. doi: 10.1111/j.1558-5646.1985.tb05663.x. [DOI] [PubMed] [Google Scholar]
- 30.Ao L, et al. Karyotypic evolution and phylogenetic relationships in the order Chiroptera as revealed by G-banding comparisons and chromosome painting. Chromosome Res. 2007;15:257–267. doi: 10.1007/s10577-007-1120-7. [DOI] [PubMed] [Google Scholar]
- 31.Baker RJ, Bickham JW. Speciation by monobrachial centric fusion. Proc Natl Acad Sci USA. 1986;83:8245–8248. doi: 10.1073/pnas.83.21.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao X, et al. Karyotype evolution in Rhinolophus bats (Rhinolophidae, Chiroptera) illuminated by cross-species chromosome painting and G-band comparison. Chromosome Res. 2007;15:835–848. doi: 10.1007/s10577-007-1167-5. [DOI] [PubMed] [Google Scholar]
- 33.Koop BF, Baker RJ, Genoways HH. Numerous chromosomal polymorphisms in a natural population of rice rats (Oryzomys, Cricetidae) Cytogenet Cell Genet. 1983;35:131–135. doi: 10.1159/000131854. [DOI] [PubMed] [Google Scholar]
- 34.Capanna E, Castiglia R. Chromosomes and speciation in Mus musculus domesticus. Cytogenet Genome Res. 2004;105:375–384. doi: 10.1159/000078210. [DOI] [PubMed] [Google Scholar]
- 35.Gallagher DS, Womack JE. Chromosome conservation in the Bovidae. J Hered. 1992;83:287–298. doi: 10.1093/oxfordjournals.jhered.a111215. [DOI] [PubMed] [Google Scholar]
- 36.Searle JB, Wójcik JM. In: Evolution of Shrews. Wójcik JM, Wolsan M, editors. Bialowieza, Poland: Mammal Research Institute; 1998. pp. 219–268. [Google Scholar]
- 37.Torres OM, Enciso S, Ruiz F, Silva E, Yunis I. Chromosome diversity of the genus Aotus from Colombia. Am J Primatol. 1998;44:255–275. doi: 10.1002/(SICI)1098-2345(1998)44:4<255::AID-AJP2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 38.Jauch A, et al. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc Natl Acad Sci USA. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson TJ, Seiffert E. Afrotherian origins and interrelationships: New views and future prospects. Curr Top Dev Biol. 2004;63:37–60. doi: 10.1016/S0070-2153(04)63002-X. [DOI] [PubMed] [Google Scholar]
- 40.Amrine-Madsen H, Koepfli K-P, Wayne RK, Springer MS. A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships. Mol Phylogenet Evol. 2003;28:225–240. doi: 10.1016/s1055-7903(03)00118-0. [DOI] [PubMed] [Google Scholar]
- 41.Waddell PJ, Shelley S. Evaluating placental inter-ordinal phylogenies with novel sequences including RAG1, gamma-fibrinogen, ND6, and mt-tRNA, plus MCMC-driven nucleotide, amino acid, and codon models. Mol Phylogenet Evol. 2003;28:197–224. doi: 10.1016/s1055-7903(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 42.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–2041. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson TJ, Fu B, Ferguson-Smith MA, Yang F. Cross-species chromosome painting in the golden mole and elephant shrew: Support for the mammalian clades Afrotheria and Afroinsectiphillia but not Afroinsectivora. Proc R Soc London Ser B. 2004;108:348–354. doi: 10.1098/rspb.2004.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svartman M, Stone G, Page JE, Stanyon R. A chromosome painting test of the basal eutherian karyotype. Chromosome Res. 2004;12:45–53. doi: 10.1023/b:chro.0000009294.18760.e4. [DOI] [PubMed] [Google Scholar]
- 45.Yang F, et al. Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ancestral karyotype. Proc Natl Acad Sci USA. 2003;100:1062–1066. doi: 10.1073/pnas.0335540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frönicke L, Wienberg J, Stone G, Adams L, Stanyon R. Towards the delineation of the ancestral eutherian genome organization: Comparative genome maps of human and the African elephant (Loxodonta africana) generated by chromosome painting. Proc R Soc London Ser B. 2003;270:1331–1340. doi: 10.1098/rspb.2003.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kellogg ME, et al. Chromosome painting in the manatee supports Afrotheria and Paenungulata. BMC Evol Biol. 2007;7:6. doi: 10.1186/1471-2148-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consigliere S, Stanyon R, Koehler U, Agoramoorthy G, Wienberg J. Chromosome painting defines genomic rearrangements between red howler monkey subspecies. Chromosome Res. 1996;4:264–270. doi: 10.1007/BF02263675. [DOI] [PubMed] [Google Scholar]
- 49.Neusser M, Wienberg J, Stanyon R, Müller S. Molecular cytotaxonomy of Platyrrhini: Comparative analysis of five species by multicolour reciprocal chromosome painting gives evidence for a classification of Callimico goeldii with Callitrichidae. Cytogenet Cell Genet. 2001;94:206–215. doi: 10.1159/000048818. [DOI] [PubMed] [Google Scholar]
- 50.de Oliveira EH, et al. The phylogeny of howler monkeys (Alouatta, Platyrrhini): Reconstruction by multicolor cross-species chromosome painting. Chromosome Res. 2002;10:669–683. doi: 10.1023/a:1021520529952. [DOI] [PubMed] [Google Scholar]
- 51.Rens W, et al. Reversal and convergence in marsupial chromosome evolution. Cytogenet Genome Res. 2003;102:282–290. doi: 10.1159/000075764. [DOI] [PubMed] [Google Scholar]
- 52.Hudson RR. Testing the constant-rate neutral allele model with protein sequence data. Evolution. 1983;37:203–217. doi: 10.1111/j.1558-5646.1983.tb05528.x. [DOI] [PubMed] [Google Scholar]
- 53.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahata N, Nei M. Gene genealogy and variance of interpopulational nucleotide differences. Genetics. 1985;110:325–344. doi: 10.1093/genetics/110.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neigel JE, Avise JC. In: Evolutionary Processes and Theory. Nevo E, Karlin S, editors. New York: Academic; 1986. pp. 515–534. [Google Scholar]
- 56.Maddison WP. Gene trees in species trees. Syst Biol. 1997;56:504–514. [Google Scholar]
- 57.Dutrillaux B, Couturier J. The ancestral karyotype of platyrrhine monkeys. Cytogenet Cell Genet. 1981;30:232–242. doi: 10.1159/000131614. [DOI] [PubMed] [Google Scholar]
- 58.Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]