Abstract
Extensive chemical analyses of spider venoms from many species have revealed complex mixtures of biologically active compounds, of which several have provided important leads for drug development. We have recently shown that NMR spectroscopy can be used advantageously for a direct structural characterization of the small-molecule content of such complex mixtures. Here, we report the application of this strategy to a larger-scale analysis of a collection of spider venoms representing >70 species, which, in combination with mass spectrometric analyses, allowed the identification of a wide range of known, and several previously undescribed, small molecules. These include polyamines, common neurotransmitters, and amino acid derivatives as well as two additional members of a recently discovered family of natural products, the sulfated nucleosides. In the case of the well studied brown recluse spider, Loxosceles reclusa, sulfated guanosine derivatives were found to comprise the major small-molecule components of the venom.
Keywords: chemical prospecting, Loxosceles, metabolomics, natural products, neurotoxins
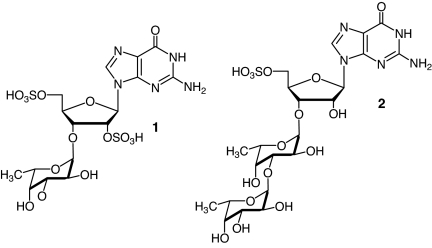
Biological samples commonly contain complex mixtures of small molecules with diverse properties and functions. Their characterization remains a challenge, because many analytical approaches are primarily aimed at identifying specific classes of known compounds. Several recent studies have demonstrated the feasibility of NMR-spectroscopic analyses of complex small-molecule mixtures, including the use of diffusion-ordered spectroscopy (DOSY) (1) or principal component analysis (PCA) in metabolomics (2) as well as the characterization of crude unfractionated natural products extracts using routine 2D NMR spectra (3–5). Compared with MS analyses, 2D NMR spectroscopic investigations of small-molecule mixtures offer the benefit of more detailed structural information, which is of particular relevance for the detection of unanticipated chemotypes. We have recently reported on the utility of direct NMR spectroscopic analysis of unpurified biological extracts as a powerful method for the discovery of natural products (5). Such analyses provide a means for the rapid evaluation of biological samples in an unbiased and nondestructive manner and are particularly useful for the characterization of glandular secretions with specific biological functions. A standard set of spectra [1H, dqfCOSY, heteronuclear multiple quantum correlation (HMQC), heteronuclear multiple bond correlation (HMBC), and NOESY] collected for a crude biological sample will usually permit partial or complete structural characterization of its major components as well as of many minor components. If these “direct” NMR-spectroscopic analyses suggest the presence of compounds of interest, final identification of individual components can be accomplished in a subsequent step, for example by using additional MS analyses, through synthesis of proposed structures, or via NMR-monitored fractionation of the sample aimed at enrichment or isolation of the compounds of interest. In the case of the hobo spider, Tegenaria agrestis, direct NMR spectroscopic analysis facilitated detection and characterization of an entire family of natural products, the sulfated nucleosides, including the sulfated guanosine derivatives 1 and 2 (Fig. 1) (5). Except for a single example (6), sulfated nucleosides had completely escaped detection by conventional isolation and characterization protocols, although spider venoms had been subject to intensive chemical scrutiny that had led to identification of hundreds of proteins, peptides, acylated polyamines, and various small-molecule neurotransmitters (7, 8).
Fig. 1.
Sulfated nucleosides from the hobo spider, Tegenaria agrestis.
The promising results with T. agrestis prompted us to initiate NMR-spectroscopic screening of venom from a large selection of spider species, both to determine the extent to which sulfated nucleosides might occur among the various spider taxa and to obtain an expanded and unbiased view of spider venom composition in general.
Results
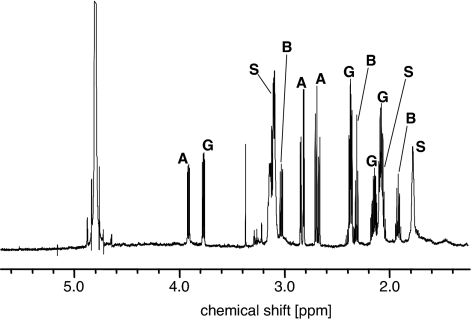
Spider venom samples representing >70 species from 27 families (9, 10) were subjected to direct NMR spectroscopic analysis. For each species, one or more lyophilized samples corresponding to 5–10 μl of crude venom were dissolved, without purification,†† in D2O, and 1H NMR spectra were recorded. These 1H spectra were used to assess the overall composition of nonvolatiles in each venom sample. In a number of instances, the 1H NMR spectrum immediately suggested that simple amino acids and amino acid derivatives typically found in spider venom represented the major components. For example, in the 1H spectrum of Pisaura mirabilis venom, signals representing >90% of the small-molecule content of the venom could be assigned to glutamic acid, aspartic acid, 4-aminobutyric acid, and spermidine (Fig. 2). For venoms of this type, structural assignments were confirmed through NMR-spectroscopic mixing experiments using authentic samples of the identified compounds. The 1H NMR spectra of several other spider venom samples, for example, Dysdera sp., Phyxioshema sp., and Scytodes fusca, showed primarily broad, unresolved peaks characteristic of large peptides and proteins but were virtually devoid of any significant signals representing small molecules. A summary of the small molecules we identified in our venom library is given in Table 1.
Fig. 2.
The 1H NMR spectrum of P. mirabilis venom. Signals corresponding to glutamic acid, aspartic acid, 4-aminobutyric acid, and spermidine are labeled G, A, B, and S, respectively.
Table 1.
Small molecules detected in spider venoms organized by family
| Family | Species | Small molecules | Family | Species | Small molecules |
|---|---|---|---|---|---|
| Agelenidae | Agelena gracilens | Philodromidae | Thanatus striatus | e | |
| Agelena labyrinthica | i | Pisauridae | Dolomedes tenebrosus | b,d,e, | |
| Agelena orientalis | i | Dolomedes gertschi | b,d,e | ||
| Agelenopsis aperta (21, 22) | d,i | Pisaura mirabilis | b,e | ||
| Agelenopsis pennsylvania | Plectreuridae | Plectreurys tristis (23) | e | ||
| Benoitia tadzhika | Salticidae | Phidippus ardenus | |||
| Calilena sp. | Phidippus johnsoni | ||||
| Hololena curta (6, 21) | d,h | Phidippus sp. | d,f | ||
| Loxosceles arizonica | h | Scytodidae | Scytodes fusca | ||
| Loxosceles deserta | h | Segestriidae | Ariadna sp. | c,d | |
| Loxosceles reclusa | h | Sparassidae | Delena cancerides | d,e,g,i | |
| Tegenaria agrestis (5) | d,h | Eusparassus oculatus | a,b,e,f,g,i | ||
| Tegenaria domestica | b,d,g,i | Holconia flindersi | b,d,e | ||
| Amaurobiidae | Coelotes pastoralis | d, h | Isopedella canberrensus | d,e | |
| Paracoelotes birulai (21) | d,i | Isopedella leai | b,d | ||
| Araneidae | Aculepeira sp. | d,i | Micrommata virescens virvirescens | a,ed | |
| Araneus diadematus | c,d,e | Neosparassus diana | b,d,e | ||
| Araneus sp. | e | Olios sp. | e | ||
| Araneus tartaricus | d,e | Theraphosidae | Aphonopelma seemani | a,b,d,i | |
| Argiope lobata | Aphonopelma sp. | b,d,i | |||
| Eriophora edax | d.e | Avicularia avicularis | b,e,i | ||
| Larinioides cornutus | d,e,i | Ceratogyrus marshalli (cornuatus) | b,d,g | ||
| Larinioides sp. | e,i | Chilobrachys sp. | a,d,i | ||
| Clubionidae | Cheiracanthium punctorium punctorium | Citharischius crawshayi | d,e | ||
| Cyrtaucheniidae | Anemesia incana | e | Haplopelma albostriatum | a,d,e,i | |
| Dipluridae | Phyxioshema sp. | Haplopelma lividum | a,d,e,f,i | ||
| Dysderidae | Dysdera sp. | Heteroscodra maculata | B | ||
| Eresidae | Eresus sp. | e | Ornithoctonus huwena | a,b,d,e | |
| Stegodyphus sp. | e | Phormictopus cancerides | b,d | ||
| Filistatidae | Kukulcania sp. | b | Psalmopoeus cambridgei | b,d,g | |
| Gnaphosidae | Drassodes sp. | e | Selenocosmia hainana | a,b,d,e,i | |
| Hexathelidae | Macrothele sp. (24) | b,i | Theridiidae | Latrodectus tadzhicus | i |
| Lycosidae | Rabidosa sp. | d,e | Steatoda giorra | ||
| Hogna sp. | d,e,i | Steatoda paykulliana | b,e,i | ||
| Lycosa proegrandis | d,e,f | Thomisidae | Misumena vatia | b,d,i | |
| Nemesiidae | Raveniola micrura | b,d,e,g,i | Thomisus onustsus | b,i | |
| Oecobiidae | Urotea sp. | Titanoecidae | Titanoeca sp. | d,e | |
| Oxyopidae | Peucetia viridans | e,i | Zodariidae | Lashesana sp. | b,d,e |
| Palpimanidae | Palpimanus sp. | d |
a, acetyl choline; b, γ-aminobutyric acid; c, choline; d, citric acid; e, glutamic acid; f, histamine; g, octopamine; h, sulfated nucleosides; I, other nucleosides.
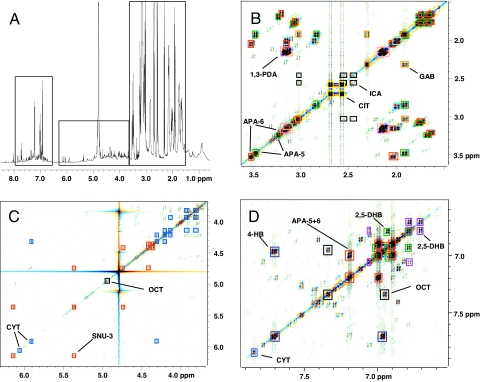
Venom samples of more interesting composition that could not be characterized based solely on their 1H NMR spectra were analyzed further by using 2D NMR spectroscopy, including dqfCOSY, NOESY, HMQC, and HMBC spectra. For the venom samples analyzed in this way, the resulting sets of spectra provided sufficient signal dispersion and connectivity information to assign all major signals to discrete structural fragments, which, in many cases, allowed immediate identification of complete structures. For example, the highly convoluted 1H NMR spectrum of Coelotes pastoralis venom (11) clearly indicates that this venom represents a complex mixture (Fig. 3). However, based on the corresponding dqfCOSY spectrum, structures could be easily proposed for the venom's major components as well as for many minor components. Among the minor components, we identified a sulfated nucleoside, whose presence was suggested by cross-peaks at 5.35 ppm/6.13 ppm, 5.35 ppm/4.74 ppm, and 4.36 ppm/4.42 ppm that appeared to belong to the 2′and 5′ protons of a ribose spin system, which are characteristically shifted downfield in sulfated derivatives (5). Further analysis using HMQC and HMBC spectra, as well as negative-ion electrospray-ionization mass spectrometry (ESI-MS) and UV spectroscopy, confirmed the presence of the previously undescribed disulfated guanosine derivative 3 [Fig. 4 and supporting information (SI) Fig. S1 and Table S1]. In addition, several compounds commonly found in spider venom were identified, including citric acid, isocitric acid, cytosine, γ-amino butyric acid, and octopamine.
Fig. 3.
Detailed NMR-spectroscopic analysis of C. pastoralis venom. (A) The 1H NMR spectrum revealing a highly complex composition. (B–D) Excerpts of the corresponding dqfCOSY spectrum with color-coded annotation of cross-peaks in yellow, citric acid (CIT); orange, γ-aminobutyric acid (GAB); green, acylated polyamine 5 (APA-5); red, acylated polyamine 6 (APA-6) and disulfated guanosine 3 (SNU-3); light blue, other polyamines; black, isocitric acid (ICA) and octopamine (OCT); pink, 1,3-diaminopropylene section of polyamines; and dark purple, 4-hydroxybenzoic acid (4-HB). In addition, D shows evidence for two minor 2,5-dihydroxybenzoic acid derivatives (purple and green, 2,5-DHB).
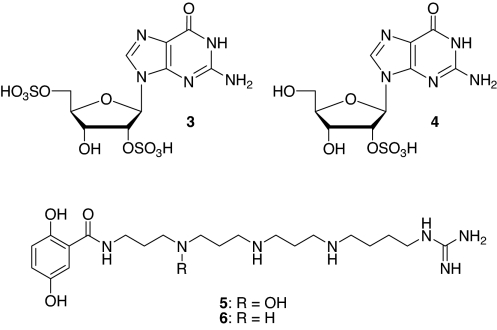
Fig. 4.
New natural products 3-6 identified from C. pastoralis and Loxosceles spp. venom.
Like many of the venoms investigated, C. pastoralis venom was found to contain significant amounts of acylated polyamines, a large family of natural products that has been extensively studied (7). The presence of acyl polyamines in our unpurified venoms was easily recognized based on the characteristic spin systems of their aromatic head groups, whose connections to the polyamine chains were evident from the HMBC spectra. Polyamines frequently occurred as mixtures of several structurally very similar compounds, and as a result, overlap of the corresponding NMR-spectroscopic signals in both 1D and 2D spectra did not always permit complete characterization of individual compounds. Nonetheless, NMR-spectroscopic analyses enabled elucidation of the general structural features of the polyamines present in a sample, which, in cases where the spectra suggested the presence of unusual structural features, were investigated further. In the case of C. pastoralis, two major as well as several minor acylated polyamines were detected in the 1H NMR spectrum. Analysis of the 2D NMR spectra yielded partial structures for the two major polyamines (Tables S2 and S3). Both compounds appeared to possess a 2,5-dihydroxybenzoylaminopropylamine head group and terminate with a 4-guanidinobutylamine (Agmatine) moiety. Complete characterization of the intervening diaminoalkane segments was not possible, however, because of multiple signal overlap. Subsequent analysis of the sample by ESI-MS-MS nevertheless allowed the remainder of both structures, 5 and 6, to be determined (Figs. S2 and S3). For these analyses, the molecular [M + 1]+ peaks corresponding to polyamines 5 and 6 were identified based on the structural characteristics previously identified in the NMR spectra. For example, collisionally induced fragmentation of a terminal agmatine moiety produces a characteristic fragment ion having a mass-to-charge (m/z) ratio of 114 (12). A tandem experiment scanning for those precursor ions giving rise to a fragment of m/z = 114 revealed two major components in the venom sample, whose abundance, elemental composition, and molecular mass indicated that they correspond to the two major polyamines observed in the NMR spectra. Complete structures could then be confirmed based on the predictable fragmentation patterns (12) observed in daughter scans acquired for the selected molecular [M + 1]+ peaks. Aside from these two compounds from C. pastoralis, the major acylated polyamines present in our venom samples appeared to belong to known types and were not characterized in detail.
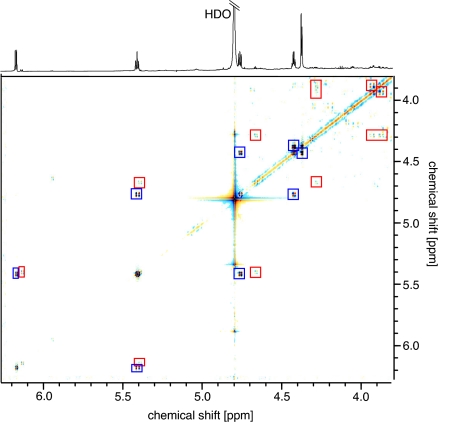
Among the most interesting spiders investigated in the course of this survey were three species from the genus Loxosceles: Loxosceles arizonica (13), Loxosceles deserta (14), and the well known brown recluse, Loxosceles reclusa (13). Loxosceles spiders are notable for the severe necrotic lesions that may result from their bites to humans (15). Surprisingly, we found sulfated nucleosides to comprise by far the most abundant components in the venoms of all three species. Our NMR-spectroscopic analyses revealed that all three Loxosceles species contained 2,5-disulfated guanosine (3) as the major component, in addition to small amounts of what appeared to be 2-sulfated guanosine (4) (Fig. 5 and Table S4). Because the concentrations of the monosulfated derivative 4 in Loxosceles spp. venom were low, and the amounts of venom we had available were limited, we were unable to characterize 4 fully via HSQC and HMBC spectra. Therefore, a sample of 4 was synthesized via nonselective, partial sulfation of a large excess of unprotected guanosine. NMR-spectroscopic analysis of the resulting mixture of monosulfated guanosine derivatives confirmed our structural assignment of 4 as the 2-sulfated derivative. The presence of the guanine moiety in 3 and 4 was further confirmed by UV spectroscopy (Fig. S1) and via negative-ion ESI-MS (Tables S1 and S4), which showed peaks corresponding to [M-1]− at m/z 442.0 and 362.0, respectively.
Fig. 5.
The 1H NMR and dqfCOSY spectra obtained for L. reclusa venom. Cross-peaks corresponding to 2,5-disulfated guanosine (3) and 2-sulfated guanosine (4) are marked blue and red, respectively.
Discussion
A well known limitation of NMR spectroscopy is its relatively poor sensitivity. Despite continuing advances in probe design and available magnet field strength, NMR spectroscopic sensitivity is often orders of magnitude lower than that of MS (16). Nonetheless, our analyses, which in most cases had to rely on samples of only 5–25 μl of crude venom, allowed detailed NMR-spectroscopic characterization of most components accounting for more than ≈1–5% of the small-molecule content of the venom samples. For example, as shown in Fig. 3, most of the signals visible in the dqfCOSY spectrum of C. pastoralis could be assigned to distinct spin systems. The threshold for detection necessarily varied for different compound classes and depended on the size and complexity of the venom samples. For example, whereas 1% of a sulfated ribonucleoside derivative could have been detected even in the most dilute venom samples investigated, the presence of as much as 5% of an unusually functionalized polyamine might not have been detected in venoms containing large amounts of other polyamines.
Our results show that direct NMR-spectroscopic analysis can be used to screen complex biological samples such as spider venom for the presence of small molecules. In combination with mass-spectrometric analyses and synthesis, this methodology enabled us to detect and identify compounds without purification. In the case of the sulfated guanosine derivatives that we identified in venoms obtained from C. pastoralis and Loxosceles spp., complete structures could be proposed based on the NMR data obtained for the entire venom, with subsequent confirmation via independent synthesis. A different situation was presented by the acylated polyamines detected in C. pastoralis, which occur as part of a larger family of structurally similar compounds. Here, signal overlap in the NMR spectra prevented full NMR-spectroscopic characterization of the individual components. However, the NMR spectroscopic analyses revealed important structural features which provided guidance in the rational pursuit of further analyses via MS leading to full identification.
Complementing MS analyses, NMR-spectroscopic screening seems particularly well suited for the survey of arrays of related samples, because the efficiency of analysis inevitably increases as known chemotypes are encountered repeatedly. Quick recognition of the NMR-spectroscopic signals representing such known chemotypes then facilitates detection of unexpected or previously uncharacterized compounds (17). Particularly noteworthy in the present study is the fact that sulfated nucleosides were found to comprise the major components in the venoms of the three Loxosceles species, including the brown recluse spider, L. reclusa. That sulfated nucleosides should comprise the major small-molecule components of this previously well studied venom and yet have remained undetected until now, provides powerful testimony to the effectiveness of primary NMR-spectroscopic screening in natural-products research. Based on the results of this study, it appears that sulfated nucleosides occur in the spider superfamilies Agelenoidea (A. aperta, H. curta), Amaurobioidea (C. pastoralis) and Scytodoidea (Loxosceles spp.). However, given the variability of spider venom composition even among spiders from individual families, it seems likely that similar compounds occur in other superfamilies as well.
The physiological properties of the sulfated nucleosides remain largely unexplored, primarily because their discovery is still very recent and amounts available from their natural sources are insufficient for broad biological screening. We have recently completed the synthesis of a small library of sulfated nucleosides, which is currently being evaluated through the National Institutes of Health's Molecular Libraries Screening Network (MLSCN).
Materials and Methods
Spider Venom.
Venom samples were purchased commercially from Fauna Laboratories, Ltd. and Spider Pharm or were acquired directly from live specimens in our laboratory. In all cases, the venom was milked by using electric stimulation of the venom gland and subsequently lyophilized (18, 19).
Sample Preparation.
Venom was obtained frozen in lyophilized form, generally as a white powder. To each sample was added 0.75 ml (for use of normal 5-mm NMR tubes) or 0.3 ml (for use of Shigemi tubes) of D2O (100 atom % D). The resulting suspension was subjected to sonication for 1 min and then centrifuged for 5 min. The supernatant was removed and analyzed via NMR spectroscopy. After completion of NMR-spectroscopic experiments, select samples were lyophilized and redissolved in water for MS analyses. Authentic samples of common small molecule neurotransmitters, which were used to confirm identities, as well as all NMR solvents, were purchased from Aldrich and Acros Chemicals.
NMR Spectroscopy.
All analyses were carried out by using a Varian INOVA 600-MHz NMR spectrometer, which was equipped with a 5-mm inverse-detection HCN probe. Shigemi tubes were used as needed. NMR spectra were acquired at 25°C by using the standard pulse sequences provided by Varian. Venom samples of complex composition were characterized via a phase-cycled phase-sensitive dqfCOSY spectrum by using the following parameters: 600-ms acquisition time, 250–600 complex increments in F1, and 8, 16, 32, or 64 scans per increment, depending on the concentration of the sample. The numbers of increments (ni) were chosen so that the digital resolution in F1 was roughly the same for all extracts. Generally, 40–80 complex increments per 1 ppm of sweep width in F1 were acquired. For further characterization of selected samples, nongradient HMQC (phase-sensitive), gradient HSQC (phase-sensitive), gradient or phase-cycled HMBC, phase-cycled phase-sensitive NOESY spectra (using a mixing time of 600 ms), or phase-sensitive ROESY spectra (mixing time 300 ms) were acquired.
MS.
MS analyses were carried out as HPLC-MS or via direct injection of the sample via syringe pump by using a Micromass Quattro I tandem MS operated in positive- or negative-ion electrospray mode and an Agilent Series 1100 liquid chromatograph (10 × 250 mm; Supelco 5-μm ODS preparative column eluted at a flow rate of 3.4 ml/min) equipped with a diode array detector. A gradient elution was used, which started with a solvent composition of 5% methanol and 95% water for 3 minutes and then progressed to 100% methanol by 40 min. After UV-detection and before ESI-MS detection, the HPLC effluent was split 40:1. Before injection onto the HPLC, spider venom samples were redissolved in water and filtered over glass wool.
Synthesis.
A stirred suspension of guanosine in dry dimethylformamide (95 mg, 0.36 mmol) under argon was cooled to −10°C, and sulfur trioxide-dimethylformamide complex (75 mg, 0.49 mmol) was added in one portion. The mixture turned clear within 2 min and was allowed to warm to 22°C. After stirring for 2 h, the reaction mixture was cooled to 0°C, and 1 ml of saturated aqueous potassium bicarbonate solution was added with stirring. Subsequently, the mixture was evaporated to dryness, and a portion (10 mg) was analyzed by NMR spectroscopy. The (1H,1H)-dqfCOSY (1H,13C)-HSQC, and (1H,13C)-HMBC spectra revealed a mixture containing 2′,3′,5′-trisulfated, 3′,5′-disulfated, 2′,5′-disulfated, and 5′-sulfated guanosine as the major products, along with smaller amounts of 2′- and 3′-sulfated guanosine. Chemical shift values of the 2′-sulfated and the 2′,5′-disulfated isomers were in agreement with those determined for the natural product. See Tables S1 and S4–S7 for NMR-spectroscopic data.
Supplementary Material
Acknowledgments.
We thank Prof. Andrey Feodorov of Fauna Laboratories, Ltd., for providing spider venoms. This work was supported in part by National Institutes of Health Grant GM 53830.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806840105/DCSupplemental.
In some instances, centrifugation was necessary to remove insoluble material. In addition, it cannot be excluded that in some cases lyophilization caused loss of volatile venom components; however, for the present study, limited availability of nonlyophilized venom samples precluded additional analyses aimed at volatile components, for example, via GC-MS.
References
- 1.Cobas JC, Groves P, Martin-Pastor M, De Capua A. New applications, processing methods and pulse sequences using diffusion NMR. Curr Anal Chem. 2005;1:289–305. [Google Scholar]
- 2.Halouska S, Powers R. Negative impact of noise on the principal component analysis of NMR data. J Magn Res. 2006;178:88–95. doi: 10.1016/j.jmr.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Dossey AT, Walse SS, Rocca JR, Edison AS. Single-insect NMR: A new tool to probe chemical biodiversity. Am Chem Soc Chem Biol. 2006;1:511–514. doi: 10.1021/cb600318u. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F, Dossey AT, Zachariah C, Edison AS, Bruschweiler R. Strategy for automated analysis of dynamic metabolic mixtures by NMR. Application to an insect venom. Anal Chem. 2007;79:7748–7752. doi: 10.1021/ac0711586. [DOI] [PubMed] [Google Scholar]
- 5.Taggi AE, Meinwald J, Schroeder FC. A new approach to natural products discovery exemplified by the identification of sulfated nucleosides in spider venom. J Am Chem Soc. 2004;126:10364–10369. doi: 10.1021/ja047416n. [DOI] [PubMed] [Google Scholar]
- 6.McCormick J, et al. Structure and total synthesis of HF-7, a neuroactive glyconucleoside disulfate from the funnel-web spider Hololena curta. J Am Chem Soc. 1999;121:5661–5665. [Google Scholar]
- 7.McCormick KD, Meinwald J. Neurotoxic acylpolyamines from spider venoms. J Chem Ecol. 1993;19:2411–2451. doi: 10.1007/BF00979674. [DOI] [PubMed] [Google Scholar]
- 8.Schulz S. The chemistry of spider toxins and spider silk. Angew Chem Int Ed Engl. 1997;36:314–326. [Google Scholar]
- 9.Platnick NI. Advances in Spider Taxonomy 1992–1995 with Redescriptions 1940–1980. New York: Am Mus of Nat Hist; 1997. [Google Scholar]
- 10.Platnick NI. The World Spider Catalog, ver 7.5. New York: Am Mus of Nat Hist; 2007. http://research.amnh.org/entomology/spiders/catalog/index.html. [Google Scholar]
- 11.Ovtchinnikov SV. The nominotypical spider subgenus Coelotes. Tethys. Entomol Res. 1841;2:35–48. [Google Scholar]
- 12.Chesnov S, Bigler L, Hesse M. The spider Paracoelotes birulai: Detection and structure elucidation of new acylpolyamines by on-line coupled HPLC-APCI-MS and HPLC-APCI-MS/MS. Helv Chim Acta. 2000;83:3295–3305. [Google Scholar]
- 13.Gertsch WJ, Mulaik S. The spiders of Texas. Bull Am Mus Nat Hist. 1940;77:307–340. [Google Scholar]
- 14.Gertsch WJ, Ennik F. The spider genus Loxosceles in North America, and the West Indies (Araneae, Loxoscelidae) Bull Am Mus Nat Hist. 1983;175:264–360. [Google Scholar]
- 15.Vetter RS, Visscher PK. Bites and stings of medically important venomous arthropods. Int J Dermatol. 1998;37:481–496. doi: 10.1046/j.1365-4362.1998.00455.x. [DOI] [PubMed] [Google Scholar]
- 16.Lacey ME, Subramanian R, Olson DL, Webb AG, Sweedler JV. High-resolution NMR spectroscopy of sample volumes from 1 nl to 10 μl. Chem Rev. 1999;99:3133–3152. doi: 10.1021/cr980140f. [DOI] [PubMed] [Google Scholar]
- 17.Schroeder FC, et al. Differential analysis of two-dimensional NMR spectra: New natural products from a pilot-scale fungal extract library. Angew Chem Int Ed. 2007;46:901–904. doi: 10.1002/anie.200603821. [DOI] [PubMed] [Google Scholar]
- 18.Grant JB, Land B. Transcutaneous amphibian stimulator (TAS): A device for the collection of amphibian skin secretions. Herpetol Rev. 2002;33:38–41. [Google Scholar]
- 19.Norment BR, Smith OE. Effect of Loxosceles reclusa Gertsch and Mulaik venom against hemocytes of Acheta domesticus (Linnaeus) Toxicon. 1968;7:141–144. doi: 10.1016/0041-0101(68)90033-0. [DOI] [PubMed] [Google Scholar]
- 20.Tzouros M, Chesnov S, Bienz S, Hesse M, Bigler L. New linear polyamine derivatives in spider venoms. Toxicon. 2005;46:350–354. doi: 10.1016/j.toxicon.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 21.Jasys VJ, et al. Isolation, structure elucidation, and synthesis of novel hydroxylamine-containing polyamines from the venom of the Agelenopsis aperta spider. J Am Chem Soc. 1990;112:6696–6704. [Google Scholar]
- 22.Quistad GB, Lam WW, Casida JE. Identification of bis(agmatine)oxalamide in venom from the primitive hunting spider, Plectreurys tristis (Simon) Toxicon. 1993;31:920–924. doi: 10.1016/0041-0101(93)90229-c. [DOI] [PubMed] [Google Scholar]
- 23.Yamaji N, et al. Structure and enantioselective synthesis of polyamine toxin MG30 from the venom of the spider Macrothele gigas. Tetrahedron Lett. 2004;45:5371–5373. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.