Abstract
Nanoparticles in a biological fluid (plasma, or otherwise) associate with a range of biopolymers, especially proteins, organized into the “protein corona” that is associated with the nanoparticle and continuously exchanging with the proteins in the environment. Methodologies to determine the corona and to understand its dependence on nanomaterial properties are likely to become important in bionanoscience. Here, we study the long-lived (“hard”) protein corona formed from human plasma for a range of nanoparticles that differ in surface properties and size. Six different polystyrene nanoparticles were studied: three different surface chemistries (plain PS, carboxyl-modified, and amine-modified) and two sizes of each (50 and 100 nm), enabling us to perform systematic studies of the effect of surface properties and size on the detailed protein coronas. Proteins in the corona that are conserved and unique across the nanoparticle types were identified and classified according to the protein functional properties. Remarkably, both size and surface properties were found to play a very significant role in determining the nanoparticle coronas on the different particles of identical materials. We comment on the future need for scientific understanding, characterization, and possibly some additional emphasis on standards for the surfaces of nanoparticles.
Keywords: bionanoscience, mass spectrometry, interactions, proteomics, human plasma
There is a growing appreciation that an understanding of the fundamental interactions of nanoscale objects with living matter will play a central role in nanomedicine, as well as in terms of nanosafety issues. For this reason we believe that accurate and extensive mapping of the biomolecule (e.g., protein) corona around a nanoparticle is emerging as a key objective of bionanoscience. Consequently, we consider that an understanding of the principles governing the formation of the corona—for example, the connection between nanomaterial properties and corona composition—will set new directions for bionanoscience. In biological media (e.g., human plasma) we envisage the particle surface being “dressed” by a corona of biological macromolecules that may be loosely divided into a “soft” component in which rapid dynamical exchange of the biomolecules between medium and particles predominates, and a “hard” corona, whose constituent biological macromolecules have a high affinity for the particle surface. The long residence times of biomolecules (e.g., proteins) in the hard corona make it easier to isolate and identify them (1, 2). This loose division into hard and soft coronas has been illustrated in some examples (1, 2) in which a simple method of “pelleting” (spinning particles and medium, removing the supernatant, and then washing the pellet to discard loosely associated molecules) is found to be consistent with the identification of the hard corona via other techniques. This method must be applied with care (2), because the washing steps can fail to remove excess (abundant) unbound proteins, but it has been applied with some success (3–7). Several investigations of the protein corona around polystyrene particles have used silver-stained 2D PAGE gels to detect the proteins eluted from the particles followed by spot excision, trypsin digestion, and peptide detection with mass spectrometry (3–7). Although our results for the more abundant proteins are similar to those discussed previously, by using 1D gels and mass spectrometry, we show that the hard corona is much more complex than previously considered. This is important, because proteins associated to nanoparticles for sufficient time are likely to be implicated biologically in some transport or other process, and we should by no means expect their abundance to reflect biological impact. Crucially, we also find that, irrespective of the nanoparticle material, both surface properties and particle size influence the composition of the hard corona. This has significant implications for our conception of nanomaterials in a biological environment, because it implies that the bulk characteristics are (much) less important than the surface ones. Indeed, even in framing standards or regulations in nanoscience, one may need to take account of the implications of the research presented here. Systematic studies of polystyrene particles with different sizes and surface charge, combined with a survey of a homologous series of N-isopropylacrylamide (NIPAM)/N-tert-butylacrylamide (BAM) copolymer nanoparticles, is used to illustrate the range of possibilities for hard corona formation.
Results and Discussion
In this study we investigate the effect of nanoparticle size and surface charge on the formation of the protein corona from human plasma. The clotting mechanism is inhibited by EDTA to provide a model system for the biological fluid. Plasma was withdrawn on one occasion from healthy humans (for details see Material and Methods). Highly monodisperse polystyrene nanoparticles (50 nm and 100 nm) that are widely available for future biological studies provide an example of a controlled systematic variation of properties that could affect the protein corona, and where the fate and transport of the particles can be tracked. For each size we select positively charged (by amine modification), nominally “neutral” unmodified (plain) polystyrene particles, and negatively charged (by carboxyl modification) as described in Table 1. The size, zeta potential, and other properties of the particles were determined in the buffering conditions relevant to the plasma corona studies (see Table 1) and, as is usual with commercially supplied samples, significant deviations from nominal specifications were observed. In one case the particles were found (by dynamic light scattering) to disperse poorly, illustrating the extreme care one needs to take with sample characterization before such studies. Thus, the 100-nm amine-modified polystyrene particles did not fulfill the criteria of monodispersity and charge for which they were bought. According to dynamic light-scattering measurements the particles aggregated and zeta-potential measurements in PBS gave a negative surface charge instead of a positive one, as shown in Table 1.
Table 1.
Properties for the 6 polystyrene nanoparticles and data related to proteomics
| 1* | 2* | 3* | 4* | 5* | 6* | |
|---|---|---|---|---|---|---|
| Size†, nm | 100 | 50 | 83 | 37 | 115 | 42 |
| Size in H2O, nm | 4,323 | 57 | 96 | 54 | 121 | 60 |
| Size in buffer‡, nm | 3,293 | 61 | 96 | 53 | 109 | 50 |
| Surface modification | –NH2 | –NH2 | None | None | –COOH | –COOH |
| z-potential in buffer‡, mV | −32 ± 3.6 | +23 ± 1.1 | −40 ± 0.9 | −41 ± 2.4 | −41 ± 2.1 | −42 ± 1.9 |
| Fluorophore | Orange§ | Blue§ | YG§ | YG§ | YG§ | YG§ |
| No. of cut bands | 21 | 25 | 20 | 18 | 19 | 23 |
| No. of identified proteins¶ | 30‖ | 45‖ | 49‖ | 49‖ | 36‖ | 42‖ |
*1, 100-nm amine-modified; 2, 50-nm amine-modified; 3, 100-nm plain; 4, 50-nm plain; 5, 100-nm carboxyl-modified; and 6, 50-nm carboxyl-modified.
†According to the manufacturer.
‡10 mM phosphate, 0.15 M NaCl, 1 mM EDTA, pH 7.5.
§The polystyrene particles were core-labeled with fluorophores: orange for the 100-nm amine-modified, blue for the 50-nm amine-modified, and yellow-green (YG) for the plain and carboxyl-modified particles.
¶Except immunoglobulin.
‖These numbers should not be regarded as absolute numbers; instead, they should be regarded as guidelines to the number of proteins in the hard protein corona.
Particles were incubated with plasma for one hour followed by centrifugation to form pellets of the particles, and extensive washing to remove all of the unbound proteins. Bound proteins—the hard corona—were eluted from the particles and separated by 1D PAGE. Selected bands were excised and the proteins trypsin digested before detection by mass spectrometry, that is, it is peptides from the protein amino acid sequence that are detected. Sequence coverage refers to the degree to which the whole-protein amino acid sequence has been covered by the peptides detected by mass spectrometry.
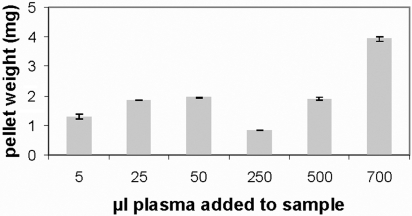
Different ratios between plasma concentration and total particle surface area were studied, keeping the ratios the same regardless of the particle size, and we report detailed results for conditions under which the pellet size is maximized. Fig. 1 shows the pellet sizes for the 50-nm carboxyl-modified particles as an example. The difference in pellet sizes is also reflected in the intensity of the bands in the 1D PAGE gels, see supporting information (SI) Fig. S1. As can be seen in Fig. 1, the pellet size first increases with increasing plasma concentration. However, a subsequent decrease of the pellet size can be observed for all particles (see Fig. S1), except the 100-nm amine-modified particles, for a certain region of plasma concentration. If the plasma concentration is elevated over this region the pellet size starts to increase again. The amine-modified 100-nm particles did not show this drop in pellet size with increased plasma concentration; instead, the pellet size increased with increasing plasma concentration. The different behavior for the amine-modified 100-nm particles compared with the other ones could be an effect of aggregation of the amine-modified 100-nm particles (see Table 1). Again, one sees the consequences of (often unnoticed) deviations in nanoparticle properties from nominal specifications. The result for a “true” 100-nm positively charged polystyrene particle will probably be different from results reported in this article. These observations are intriguing and may reveal important observations in more full investigations. In essence, for the moment, we see the maximum as a reference point for ourselves and other researchers at which the corona can be determined and compared for different systems. The effect itself is possibly due to complex associative fluid phenomena (such as those that drive phase separations) controlling effective interparticle interactions, which vary with the nature of the added plasma. It could also be a matter of practical importance whether the mechanism of protein corona formation around nano- and microparticles is affected by this (although, as yet, our investigations reveal no such effects).
Fig. 1.
Dry pellet formed for carboxyl-modified polystyrene 50-nm particles incubated with increasing plasma concentrations. The error bars represent the difference from two separate experiments. SDS/PAGE for the samples is shown in Table S3.
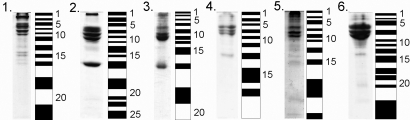
Fig. 2 shows how the lanes from the 1D gels were cut into individual samples that were then trypsin digested and analyzed by mass spectrometry. The full 1D gel lanes were analyzed, with all visible bands being cut individually, and the spaces between the bands also being analyzed. Table 1 shows the number of bands cut for each particle reported. The full list of detected proteins and the proteomics data can be found in Table S3. For the ‘neutral’ particles, the major part of the “hard” corona consists of fibrinogen, IgG, albumin, and inter-alpha-trypsin inhibitor heavy chain, as shown previously (3–7). However, there are quite significant groups of proteins identified, even for the neutral particles. Table 1 shows the total number of identified proteins on the six different particles (except IgG, which will be discussed separately), and it is significant that some tens of proteins are reliably detected for all of the nanoparticles studied.
Fig. 2.
Illustration of SDS/PAGE gels and bands excised. Lanes: 1, 100-nm amine-modified; 2, 50-nm amine-modified; 3, 100-nm plain; 4, 50-nm plain; 5, 100-nm carboxyl-modified; and 6, 50-nm carboxyl-modified. The selected lanes for the different particle types were cut according to the pattern shown to the right of the gels. The numbers to the right of the pattern show the numbering for the proteomic data found in Table S3. The plasma concentrations for the different samples were: 1, 2.8; 2, 0.56; 3, 0.28; 4, 0.56; 5, 0.56; and 6, 5.6 ml of plasma per m2 particle surface.
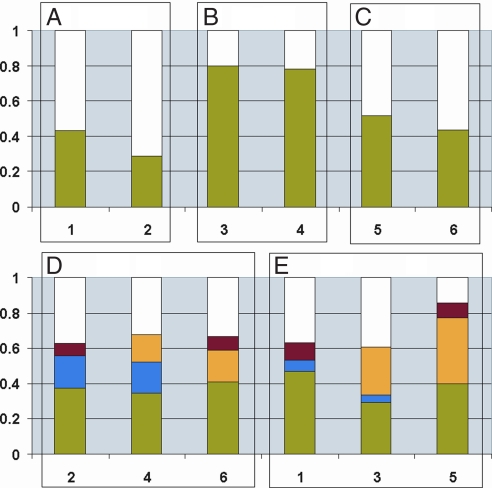
A general comparison of the degree of similarity of the protein coronas around the different nanoparticles is presented in Fig. 3. We emphasize that, although this is a useful manner in which to understand the relationship of physiochemical properties to overall corona structure, even minor compositional changes (such as those shown below to arise from nanoparticle size alone in the case of “neutral” polystyrene nanoparticles) are sufficient to cause a change in the (potential) biological impact. Fig. 3 illustrates the fraction of proteins that is unique for each particle type, as well as the fractions detected on two or more particles. Fig. 3 A–C illustrates the overall effects of particle size. Thus, the coronas around two different sized neutral polystyrene particles (Fig. 3B) are very similar, with ≈80% homology between the two coronas, suggesting that the molecular (e.g., hydrophobic) properties are more important than size for that case. In contrast, a size effect is observed for both the amine- (Fig. 3A) and carboxyl- (Fig. 3C) modified particles. There are differences for both the amine- and carboxyl-modified particle pairs that can be interpreted as a size effect. Interpretation of the differences in the composition of the protein coronas observed for the two amine-modified particles is hampered by the difficulties with the dispersion and zeta characterization of the 100-nm particles (see Table 1). As a result the observed differences could result from one or a combination of the following properties: size, charge, and/or changed surface properties because of different fluorophores. Still, we retain this result, in part, to illustrate the current situation for commonly available reference nanoparticles.
Fig. 3.
Comparison of the overlap in the protein coronas of the different polystyrene particles. Graphs: 1, 100-nm amine-modified; 2, 50-nm amine-modified; 3, 100-nm plain; 4, 50-nm plain; 5, 100-nm carboxyl-modified; and 6, 50-nm carboxyl-modified. The fractions shown are calculated without including different Ig chains. (A–C) Comparison of the similarity between the coronas around different size particles with similar surface properties: fraction of proteins found on both particles (green), and fraction of proteins that is found on one size but not the other (white). (D and E) Comparison of the similarities of the corona for particles of the same size but different surface charges: fraction of proteins found on all three particles (green), fraction of proteins found on the amine-modified and plain particles (blue), fraction of proteins found on the plain and carboxyl-modified particles (yellow), fraction of proteins found on the amine- and carboxyl-modified particles (red), and fraction of proteins found on just one specific particle surface (white).
The homology between the coronas for the carboxyl-modified particles is only ≈50%. This is a striking confirmation that the corona around nanoparticles will be size-dependent, and one may therefore expect different biological impacts, not just from the direct consequence of size, but from the implicit interactions from the corona. This is an important point that must in future be appreciated in in vitro and in vivo studies in nanomedicine and nanosafety.
Polystyrene particles are slightly negatively charged as a result of surface charges, which arise from fragments of the initiator used to start the polymerization reaction. However, by convention, these are considered to be plain or neutral particles, because they are not specifically functionalized. In this work, the polystyrene particles used have positive (amine-modified), neutral (the plain unmodified particles), or negative (carboxyl-modified) charges, and can be classified according to their surface chemistry or zeta potential. We discuss the overall dependence of the resultant protein corona on surface charge and zeta potential and identify adsorption patterns and similarities. The interpretation of these data is further complicated by the fact that the nanoparticles are labeled with different fluorophores, as result of the fact that they are commercially sourced and therefore dependent on what is available. The amine-modified particles are labeled with a blue fluorophore (50-nm particles) and an orange fluorophore (100-nm particles), whereas the carboxyl-modified and plain particles are labeled with a yellow-green fluorophore. Müller et al. (7) have shown that fluorophore labeling of manufactured polystyrene particles affects the distribution of the most common proteins adsorbed to the particles compared with unlabeled manufactured polystyrene particles. However, all particles used in this study are fluorophore-labeled and the fluorophores are expected to be buried in the core of the particles. Furthermore, the four plain and carboxyl-modified particles are all labeled with the same fluorophore, making it possible to compare the coronas formed and to draw some general conclusions about how different surface charges can affect the hard corona composition. It transpires that protein coronas for the same particle size but different surface charge (and surface chemistry) exhibit a rich range of behaviors (see Fig. 3 D and E).
For the 50-nm particles (Fig. 3D), ≈35–40% of the proteins are common to all three coronas (green), whereas ≈35% of the proteins are unique to nanoparticles of a given surface composition (white). The remaining ≈25–30% of the corona for the carboxyl-modified (yellow) and amine-modified (blue) particles is composed of different (≈15–20% for each type) proteins that are also found in the plain corona. A small component (≈5–10%, red) that is common for the two charged particles is not found in the protein corona around the plain (neutral) particle.
The picture is different when the coronas around the three different 100-nm particle types are compared (Fig. 3E). For the carboxyl-modified particles only a small fraction (≈15%) of proteins (white) are unique, whereas the plain and amine-modified particles (which were not 100 nm but were aggregated with a size on the order of micrometers) again each have ≈35% proteins (white) that are unique to them. The surface-modified particles have ≈50% proteins in common, and ≈35% in common with the plain particles (green). The plain and carboxyl-modified particles share a large fraction of proteins, ≈25% and ≈35%, respectively (yellow), whereas only a small fraction, ≈5%, of the proteins are shared only between the plain and amine-modified particles (blue). As was also observed for the 50-nm particles, in the 100-nm particles case, just a few proteins (<10%) are shared only between the charged particles (red).
Bearing in mind that the nanoparticle material, for the plain and carboxyl- or amine-modified particles, is (in the interior) chemically identical, the fact that surface properties so significantly affect the global composition of the hard protein corona is striking. It emphasizes the need for careful controls in future experimental design, not just of nanoparticle size, but particularly of surface properties, if one seeks to carry out systematic and reproducible bionanoscience.
Table 2 shows a selection of proteins, grouped according to their function, derived from Table S2, which is derived from the complete proteomics data presented in Table S1.
Table 2.
Selection of identified proteins grouped according to their function
| 1* | 2* | 3* | 4* | 5* | 6* | |
|---|---|---|---|---|---|---|
| Immunoglobulin | ||||||
| Ig alpha-1 chain C region | X | X | X | X | ||
| Ig alpha-2 chain C region | X | |||||
| Ig gamma-1 chain C region | X | X | X | X | X | |
| Ig gamma-2 chain C region | X | X | X | X | ||
| Ig gamma-3 chain C region | X | X | ||||
| Ig gamma-4 chain C region | X | X | X | |||
| Ig kappa chain C region | X | X | X | X | X | X |
| Ig lambda chain C regions | X | X | X | X | X | X |
| Ig mu chain C region | X | X | X | X | ||
| Immunoglobulin J chain | X | X | ||||
| Ig kappa chain V-I region | X | X | X | |||
| Ig kappa chain V-II region | X | X | X | X | ||
| Ig heavy chain V-III region | X | X | ||||
| Ig kappa chain V-IV region | X | X | ||||
| Lipoproteins | ||||||
| Apolipoprotein A-I | X | X | X | X | X | X |
| Apolipoprotein A-II | X | X | ||||
| Apolipoprotein A-IV | X | X | X | X | X | |
| Apolipoprotein B-100 | X | X | X | X | ||
| Apolipoprotein C-I | X | X | X | X | ||
| Apolipoprotein C-III | X | X | X | |||
| Apolipoprotein D | X | X | X | |||
| Apolipoprotein E | X | X | X | X | ||
| Apolipoprotein F | X | |||||
| Apolipoprotein L1 | X | X | ||||
| Beta-2-glycoprotein 1 (apolipoprotein H) | X | X | X | X | X | |
| Clusterin (apolipoprotein J) | X | X | X | X | X | X |
| Complement pathways | ||||||
| Complement C1r | X | |||||
| Complement C1 s | X | X | X | |||
| Complement C1q | X | X | X | X | ||
| Complement C2 | ||||||
| Complement C3 | X | X | X | X | ||
| Complement C4 | X | X | X | X | ||
| Complement C5 | X | X | ||||
| Complement C6 | X | X | ||||
| Complement C8 | X | X | ||||
| Complement C9 | X | |||||
| Complement factor B | X | X | X | |||
| Complement factor H | X | X | X | X | X | X |
| Complement factor I | X | |||||
| Complement C4b-binding protein | X | X | X | X | X | |
| Plasma protease C1 inhibitor | X | X | X | |||
| Acute-phase protein | ||||||
| Mannose-binding protein | X | |||||
| Alpha 1-antitrypsin | X | X | X | |||
| Alpha 1-antichymotrypsin | X | X | X | X | ||
| Fibrinogen | X | X | X | X | X | X |
| Plasminogen | X | X | X | X | ||
| Complement factors (see above) | X | X | X | X | X | X |
| Serum amyloid P component | X | X | ||||
| Serum amyloid A | X | |||||
| Transthyretin | X | X | X | |||
| Coagulation factors | ||||||
| Fibrinogen (factor I) | X | X | X | X | X | X |
| V | X | |||||
| XI (plasma thromboplastin antecedent) | X | X | ||||
| Prekallikrein | X | |||||
| Kininogen | X | X | X | X | X | |
| Fibronectin | X | X | X | X | X | |
| antithrombin III | X | X | ||||
| Vitamin K-dependent protein S | X | X | X | X | ||
| Plasminogen | X | X | X | X | ||
| Alpha 2-antiplasmin | X | X | ||||
*1, 100-nm amine-modified; 2, 50-nm amine-modified; 3, 100-nm plain; 4, 50-nm plain; 5, 100-nm carboxyl-modified; and 6, 50-nm carboxyl-modified.
The first group of proteins in Table 2 lists Ig fractions that have been identified as part of the hard-protein corona around the different particles. The differences in the coronas are significant, with different forms of the constant regions and the variable regions of immunoglobulins interacting preferentially with the plain particles. Interestingly, the 100-nm carboxyl-modified particles also have quite a large number of immunoglobulins in their corona, whereas the 50-nm carboxyl- and amine-modified particles have remarkably little. The great variability due to size and charge is intriguing. Immunoglobulins (IgGs) of the type identified on the particles are involved in many processes from immunity response to allergic reaction and anaphylactic shock. IgG is involved in transport across the placenta, as well as the general process of opsonization for presentation to macrophage, for example (8).
The second group of proteins, apolipoproteins, are involved in the transportation of lipids and cholesterol in the bloodstream (9) and, as such, are expected to greatly affect the intracellular trafficking, fate, and transport of nanoparticles in cells and animals.
We have previously pointed out that nanoparticles of different materials and different sizes associate with apolipoproteins derived from HDL and chylomicrons (1). The identities of the proteins attached to HDL, LDL, and VLDL have recently been analyzed using proteomics (10–16). In particular, the list of proteins believed to be associated with HDL particles has grown significantly in recent years (17).
The 50-nm amine-modified particles have the highest number of detected apolipoproteins, 9 of the 10 identified being associated with HDL, suggesting that 50-nm amine-modified particles associate with HDL. Apolipoprotein B-100 (the major protein in VLDL and LDL particles, but not HDL) (16) is associated with all 50-nm particles, suggesting that the 50-nm particles associate to some degree with VLDL and LDL.
The two differently sized plain polystyrene particles show less significant differences in the general composition of the hard-protein corona. However, apolipoprotein B-100 is detected throughout the gel for the 100-nm particles, meaning that it is represented in high concentration in the sample, whereas it is not detected at all on the 50-nm particles. This indicates that the 100-nm plain particles may have a preferential interaction with LDL and/or VLDL compared with the 50-nm plain particles. The carboxyl-modified particles also show this size difference in the presence of apolipoprotein B-100.
However, apolipoproteins are, in general, less frequently detected for the carboxyl-modified particles than for the plain particles. For example, apolipoprotein B-100 is detected in 7 of the bands cut for the 100-nm plain particles, whereas it is detected in only 1 of the bands cut for the 100-nm carboxyl-modified particles with sequence coverage of 2%. The apolipoproteins are also less frequently detected for the 100-nm amine-modified particles, with apolipoprotein A-I detected in only one of the bands cut with a sequence coverage of 12% compared with the best sequence coverage for apolipoprotein A-I of 78% for the 50-nm amine-modified particles.
These preferential interactions between different lipoproteins and nanoparticles are worthy of further investigation. To what degree, for example, the 50-nm amine particles follow characteristic fate pathways of HDL, and the 100-nm particles act on LDL receptors, is a topic of considerable future interest.
Protein group three in Table 2 lists proteins in the complement pathways, a key part of the innate immune response. As for immunoglobulins, the plain particles bind most proteins related to the complement pathways, closely followed by the 100-nm carboxyl-modified particles. The cell membrane-penetrating complex, complement C9, is detected on the 100-nm plain particles. The three 50-nm nanoparticles have relatively few complement-pathway proteins detected in their coronas. This variations in which complement-pathway proteins bind as a function of nanoparticle charge and size might suggest that different particles activate different complement pathways to differing extents, and again this is a topic of considerable interest for future investigations.
Group four in Table 2 shows acute-phase proteins in the different coronas, and closely follows the results for the complement-pathway proteins. The plain and carboxyl-modified 100-nm particles again have quite a number of different acute-phase proteins in their coronas. A difference can be observed for the 50-nm amine-modified particles. These also have many acute-phase proteins identified as part of the protein corona, whereas only few complement-pathway proteins could be found compared with the plain particles. Again, these observations might suggest the activation of differing inflammatory responses by the particles of different size and surface charge.
The last group of proteins listed in Table 2 concerns proteins involved in the coagulation process. As can be seen in the table, the plain, the carboxyl-modified, and the 50-nm amine-modified polystyrene nanoparticles have almost the same coagulation-related proteins present in their coronas. The 100-nm amine-modified particles have a slightly different corona content when it comes to proteins involved in the coagulation process. However, there are several proteins from the coagulation process detected in all of the particle coronas. To what degree this reflects the hard protein corona in vivo or is an artifact of the use of EDTA in the experimental buffer (to prevent the coagulation cascade) has yet to be investigated. As they stand, the results are suggestive of different coagulation and platelet response profiles.
Proteins that are not listed in Table 2 can be found in Table S2. In all six coronas inter-alpha-trypsin inhibitors, serum albumin, clusterin, and vitronectin are detected. In the coronas around the plain particles, some proteins related to iron, heme, and hemoglobin transport were detected but are absent in the coronas around the 100-nm modified particles.
The idea that the pronounced charge dependence of the corona truly is a surface effect may be further supported by results (Fig. S3) from the study of a series of positively and negatively charged 50:50 NIPAM/BAM copolymer nanoparticles in which the charges are spread throughout the particle body (rather than being only at the particle surface as in the polystyrene particles). Thus, random copolymer nanoparticles of NIPAM:BAM with increasing amounts of either acrylic acid (negative charge) or N-N′-N″-dimethylaminopropyl acrylamide (positive charge) were synthesized, and their zeta potentials determined as before (see in Table S3). The number of charges is estimated from the initial composition of monomers, but the fact that the zeta potential is largely unaffected, whereas the swelling temperature is shifted strongly at high charge density, confirms the idea that the surface charge is not much affected, with the majority of the changes residing in the interior (bulk) of the particles. The fact that the gel band pattern is independent of the type and amount of charge in the investigated range (the gels are strikingly similar to ref. 1) suggests that differences in protein corona might be driven by surface-charge properties, rather than the overall force derived from the core of the nanoparticle.
Conclusions
As interest in understanding the principles governing bionanointeractions grows we can expect the quest for the fundamental interacting unit in biological media to accelerate. In this article we have shown that, even though many of the major highly abundant proteins (these being somewhat featureless from a biological activity point of view) in the corona are independent of size and surface charge, a whole range of proteins (many of them having quite distinct biological roles relevant to nanomedicine and nanosafety) form part of the corona. Bearing in mind that, in such well known cases of biological nanoparticles as the LDL and HDL particles themselves, where single or few copies of surface-expressed proteins dominate the biological impacts, we believe that the presence of these proteins could be highly significant. In turn, the nature of the proteins in the corona is (as expected) determined by the local chemical property of the nanomaterial. However, even for a fixed material type, the size of the particle, and its surface modification are able to entirely change the nature of the biologically active proteins in the corona, and thereby possibly also the biological impacts.
There remains, of course, the fundamental question as to the format of presentation of these proteins, and to what degree they are still able to present native epitopes when embedded in the corona. The answer to such questions will likely determine the future roadmap of nanomedicine, nanosafety, and perhaps impact the overall context of nanoscience itself.
Materials and Methods
Polystyrene Nanoparticles.
Polystyrene latex beads were purchased from Sigma (amine-modified 50-nm and 100-nm labeled with blue and orange fluorophores, respectively) and from Polysciences [both unmodified (plain) and carboxyl-modified 50-nm and 100-nm, labeled with yellow-green fluorophore]. All nanoparticles were used as received.
Size and Zeta-Potential Determination for the Particles.
The size and zeta-potential for the polystyrene particles were determined with a Malvern Zetasizer 3000HSa. Polystyrene particles were diluted with either water or PBS before measurement. The measurements were conducted at 25°C by using 75 μg/ml and 15–25 μg/ml concentration of the 50-nm and 100-nm particles, respectively.
Human Plasma.
Blood was taken from 10 different seemingly healthy donors. Each donor donated blood for 10 × 3-ml tubes containing EDTA to prevent blood clotting. The blood donation was arranged such that the blood samples were labeled anonymously. They could not be traced back to a specific donor, however, it was possible to use plasma from just one of the donors for a specific experiment. The tubes were centrifuged, for 5 min at 800 RCF to pellet the red and white blood cells. The supernatant (the plasma) was transferred to labeled tubes and stored at −80°C until used. On thawing the plasma was centrifuged again for 2 min at 16.1 kRCF to further reduce the presence of red and white blood cells.
Polystyrene Nanoparticle Incubation with Plasma.
All experiments were conducted at least twice to ensure reproducibility of the particle–protein complex pellet sizes, general pattern, and band intensities on the 1D gels. Particle suspensions of 1.66 mg/ml were incubated with different concentrations of human blood plasma in 10 mM phosphate, 0.15M NaCl, 1 mM EDTA, pH7.5, for 1 h (total volume, 750 μl). The ratio of total particle-surface area to plasma concentration was kept the same for the two different particle sizes to ensure comparability between the results. The samples were centrifuged to pellet the particle–protein complexes. The pellet was resuspended in PBS, transferred to a new vial, and centrifuged again to pellet the particle–protein complexes; this procedure was repeated three times. After the third washing step the supernatant did not contain any detectable amount of proteins. The proteins were eluted from the particles by adding SDS-sample buffer to the pellet and boiling the solution. The proteins were separated by 12% SDS/PAGE 1D gels.
Determination of Pellet Weight.
Polystyrene nanoparticles were mixed with different amounts of human plasma as previously described. The samples were centrifuged to pellet the particle–protein complexes and the supernatants were discarded. The pellets were dried overnight at 60°C and then weighed with a Sartorius SE2 microbalance (Sartorius). The results represent the average over two individual series and the error bars represent the standard deviation.
Protein identification by Mass Spectrometry.
Bands of interest from SDS/PAGE gels (12%) were excised and digested in-gel with trypsin according to the method of Shevchenko et al. (18). The resulting peptide mixtures were resuspended in 0.1% formic acid and analyzed by electrospray liquid chromatography mass spectrometry (LC MS/MS). An HPLC (Surveyor, ThermoFinnigan) was interfaced with an LTQ ion trap mass spectrometer (ThermoFinnigan). Chromatography buffer solutions (Buffer A, 0.1% formic acid; Buffer B, 100% acetonitrile and 0.1% formic acid) were used to deliver a 72-min gradient (5 min sample loading, 32 min to 40% Buffer B, 2 min to 80%, hold 11 min, 1 min to 0%, hold for 20 min, 1 min flow adjusting). A flow rate of 150 μl/min was used at the electrospray source. Spectra were searched by using the SEQUEST algorithm (19) against the indexed UniProtKB/Swiss-Prot database (http://www.expasy.org, release July 3, 2007) (see Table S1). The probability-based evaluation program Bioworks Browser was used for filtering identifications; proteins with Xcorr (1, 2, 3) = (1.90, 2.00, 2.50) and a peptide probability of 1e−5 or better were accepted.
Supplementary Material
Acknowledgments.
This work was supported by European Union Sixth Framework Program project NanoInteract Grant NMP4-CT-2006-033231, Irish Research Council for Science, Engineering and Technology (M.L.), and European Science Foundation EpitopeMap.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805135105/DCSupplemental.
References
- 1.Cedervall T, et al. Detailed Identification of plasma proteins adsorbed on copolymer nanoparticles. Angew Chem Int Ed. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- 2.Cedervall T, et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci USA. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gessner A, Lieske A, Paulke BR, Müller RH. Influence of surface charge density on protein adsorption on polymeric nanoparticles: Analysis by two-dimensional electrophoresis. Eur J Pharm Biopharm. 2002;54:165–170. doi: 10.1016/s0939-6411(02)00081-4. [DOI] [PubMed] [Google Scholar]
- 4.Gessner A, Lieske A, Paulke BR, Müller RH. Functional groups on polystyrene model nanoparticles: Influence on protein adsorption. J Biomed Mater Res A. 2003;65A:319–326. doi: 10.1002/jbm.a.10371. [DOI] [PubMed] [Google Scholar]
- 5.Gessner A, et al. Nanoparticles with decreasing surface hydrophobicities: Influence on plasma protein adsorption. Int J Pharm. 2000;196:245–249. doi: 10.1016/s0378-5173(99)00432-9. [DOI] [PubMed] [Google Scholar]
- 6.Lück M, Paulke B-R, Schröder W, Blunk T, Müller RH. Analysis of plasma protein adsorption on polymeric nanoparticles with different surface characteristics. J Biomed Mater Res. 1998;39:478–485. doi: 10.1002/(sici)1097-4636(19980305)39:3<478::aid-jbm19>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Müller RH, Rühl D, Lück M, Paulke BR. Influence of fluorescent labelling of polystyrene particles on phagocytic uptake, surface hydrophobicity, and plasma protein adsorption. Pharm Res. 1997;14:18–24. doi: 10.1023/a:1012043131081. [DOI] [PubMed] [Google Scholar]
- 8.Jefferis R, Kumararatne DS. Selective Igg subclass deficiency—Quantification and clinical relevance. Clin Exp Immunol. 1990;81:357–367. doi: 10.1111/j.1365-2249.1990.tb05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson NL, Anderson NG. The human plasma proteome—History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 10.Heller M, et al. Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins. Proteomics. 2005;5:2619–2630. doi: 10.1002/pmic.200401233. [DOI] [PubMed] [Google Scholar]
- 11.Hortin GL, Shen R-F, Martin BM, Remaley AT. Diverse range of small peptides associated with high-density lipoprotein. Biochem Biophys Res Commun. 2006;340:909–915. doi: 10.1016/j.bbrc.2005.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics II: Mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005;5:1431–1445. doi: 10.1002/pmic.200401010. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson H, Leanderson P, Tagesson C, Lindahl M. Lipoproteomics I: Mapping of proteins in low-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005;5:551–565. doi: 10.1002/pmic.200300938. [DOI] [PubMed] [Google Scholar]
- 14.Mancone C, et al. Proteomic analysis of human very low-density lipoprotein by two-dimensional gel electrophoresis and MALDI-TOF/TOF. Proteomics. 2007;7:143–154. doi: 10.1002/pmic.200600339. [DOI] [PubMed] [Google Scholar]
- 15.Rezaee F, Casetta B, Levels JHM, Speijer D, Meijers JCM. Proteomic analysis of high-density lipoprotein. Proteomics. 2006;6:721–730. doi: 10.1002/pmic.200500191. [DOI] [PubMed] [Google Scholar]
- 16.Vaisar T, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reilly MP, Tall AR. HDL proteomics: Pot of gold or Pandora's box? J Clin Invest. 2007;117:595–598. doi: 10.1172/JCI31608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 19.Yates JR, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.