Abstract
Plant RNA silencing machinery enlists four primary classes of proteins to achieve sequence-specific regulation of gene expression and mount an antiviral defense. These include Dicer-like ribonucleases (DCLs), Argonaute proteins (AGOs), dsRNA-binding proteins (DRBs), and RNA-dependent RNA polymerases (RDRs). Although at least four distinct endogenous RNA silencing pathways have been thoroughly characterized, a detailed understanding of the antiviral RNA silencing pathway is just emerging. In this report, we have examined the role of four DCLs, two AGOs, one DRB, and one RDR in controlling viral RNA accumulation in infected Arabidopsis plants by using a mutant virus lacking its silencing suppressor. Our results show that all four DCLs contribute to antiviral RNA silencing. We confirm previous reports implicating both DCL4 and DCL2 in this process and establish a minor role for DCL3. Surprisingly, we found that DCL1 represses antiviral RNA silencing through negatively regulating the expression of DCL4 and DCL3. We also implicate DRB4 in antiviral RNA silencing. Finally, we show that both AGO1 and AGO7 function to ensure efficient clearance of viral RNAs and establish that AGO1 is capable of targeting viral RNAs with more compact structures, whereas AGO7 and RDR6 favor less structured RNA targets. Our results resolve several key steps in the antiviral RNA silencing pathway and provide a basis for further in-depth analysis.
Keywords: interpathway regulation, plant antiviral defense
RNA silencing is a cellular mechanism that uses small RNA molecules (21–30 nt in length) as sequence-specific mediators to regulate the expression of a diverse array of genes at the transcriptional, posttranscriptional, or translational levels (1). In plants, these very small RNA species are termed small interfering RNAs (siRNAs) or micro RNAs (miRNAs) depending on the source of their precursors. They are generated by a family of double-stranded RNA (dsRNA)-specific RNases called Dicer-like ribonucleases (DCLs) (2). Once produced, the siRNAs and miRNAs are recruited by Argonaute proteins (AGOs) into RNA-induced silencing complexes (RISCs) to direct the cleavage or translational repression of homologous mRNAs or to remodel the homologous chromosomal DNA to achieve transcriptional silencing (3). Another family of dsRNA-binding proteins (DRBs) has been found to modulate the function of DCLs (4). Plants also encode RNA-dependent RNA polymerases (RDRs) to produce some of the dsRNA precursors that serve as templates for DCLs (2). In Arabidopsis, 4 DCLs, 10 AGOs, 5 DRBs, and up to 6 RDRs have been identified. They participate in at least four different endogenous RNA silencing pathways to achieve spatial and temporal regulation of gene expression throughout the plant life cycle and to condition the plant response to biotic and abiotic stresses (5).
Although the plant RNA silencing mechanism was first revealed through studies aimed to unravel the complexity of plant antiviral defense strategies, the details of plant antiviral RNA silencing pathway(s) are far from resolved. Recent studies have established a primary role for DCL4 and DCL2 in processing dsRNA of virus origin into siRNAs (6–10), although it is not known how activities of these DCLs are regulated. There are also conflicting reports as to which AGOs are necessary for antiviral silencing (11, 12). It also remains to be determined whether any of the DRBs play a role in antiviral silencing. One factor that hinders the dissection of the antiviral RNA silencing machinery is that almost all plant viruses encode suppressors of RNA silencing that, depending on the growth conditions of the plants, could partially or completely disable virus-targeted RNA silencing by the plant host (6, 13–15).
In this report, we implicate several more components of the plant RNA silencing machinery in antiviral defense by using mutant viruses devoid of their silencing suppressors. Specifically, we removed the silencing suppressor from our model virus, turnip crinkle virus (TCV) and used the resulting mutant viral RNAs to infect an array of Arabidopsis plants containing mutations in key silencing pathway genes. A similar approach was first used by Deleris et al. (6) to discover the critical antiviral role of DCL4 and DCL2. Our studies build on their discoveries and clearly demonstrate that DCL1 counteracts the antiviral role of DCL4. We further report that DRB4 contributes significantly to antiviral silencing. Finally, we demonstrate that two AGOs, AGO1 and AGO7, participate in the silencing of viral RNAs. Our results point to a regulatory relationship between components of the silencing pathways.
Results and Discussion
Removal of the TCV Silencing Suppressor Leads to Faster Clearance of Viral RNA in dcl1 Plants.
We initially attempted to determine the antiviral function of silencing pathway genes of Arabidopsis by infecting the corresponding mutant plants with a wild-type (WT) TCV transcript (Fig. 1A), and analyzing both inoculated and systemic leaves (IL and SL) of infected mutants for viral RNA accumulation. To minimize experimental variations, all leaf samples examined throughout this report consisted of pools of six leaves collected from six inoculated plants; all experiments were repeated at least three times with consistent results.
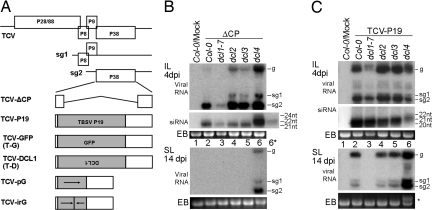
Fig. 1.
Roles of different DCLs in RNA silencing-mediated antiviral defense. (A) Schematic representation of the TCV-derived constructs used in this work. (B and C) RNA blot hybridizations showing accumulation levels of ΔCP (B) and TCV-P19 (C) in different dcl mutant plants. Lane 6* of the siRNA blot in B contained a reduced quantity of RNA (one-fifth of that in lane 6) to better resolve siRNA sizes. Ethidium bromide (EB)-stained rRNA served as the loading control. Also note TCV-P19 RNA accumulation in dcl4 SL is apparent even in EB-stained gel (* in C).
When infected with wtTCV, none of the Arabidopsis mutants tested showed appreciable difference in susceptibility as measured by viral RNA accumulation levels [see supporting information (SI) Fig. S1 for the result with the four dcl mutants]. Similar results have been reported previously for plant as well as animal virus-infected tissues, suggesting that strong suppressors encoded by WT viruses effectively mask the antiviral role of silencing pathway genes of their hosts (6, 16, 17).
It has been shown previously that wtTCV-specific siRNAs accumulated in SL are of a single 22-nt size class (6, 7). Here we show that three distinct size classes of 22, 21, and 20 nt accumulated in the IL (Fig. S1, siRNA blot). We attribute the biogenesis of 22- and 21-nt siRNAs to the function of DCL2 and DCL4, respectively, judging from the fact that the former are absent in dcl2 plants and the latter absent in dcl4 plants (lanes 4 and 6). The 20-nt siRNAs likely resulted from partial degradation of the 21-nt siRNAs, as they are also undetectable in dcl4 plants. These data are generally consistent with the findings of Deleris et al. (6). Therefore, even though TCV viral RNA levels were similar in different dcl mutants, the viral siRNA pattern unique to IL confirmed the antiviral role of DCL4 and DCL2. The critical importance of analyzing IL has also been documented by Diaz-Pendon et al. (10).
We next examined the same set of dcl mutants for their response to TCV-ΔCP (ΔCP), a TCV mutant with its silencing suppressor removed (Fig. 1A) (18). We were then able to detect substantially increased accumulation of genomic (g) as well as subgenomic (sg) RNAs of ΔCP in IL of infected dcl2 and dcl4 plants (Fig. 1B, compare lanes 2, 4, and 6). dcl4 plants showed the highest viral RNA levels, leading to systemic spread of the mutant virus. These results further corroborate previous reports implicating both DCL4 and DCL2 in the antiviral defense (6–8, 10, 17). dcl3 plants also showed a less substantial but consistently reproducible increase in viral RNA levels (lane 5), suggesting that DCL3, in addition to DCL4 and DCL2, functions in some capacity to defend plants against viral infections.
The most unexpected results were observed in dcl1–7 plants, in which ΔCP RNA accumulated to even lower levels than in Col-0 plants (lane 3). Similar results were also obtained with a different dcl1 mutant (dcl1–9) in a different parental background (Ler-0) (Fig. 2, compare lanes 2 with 4, 5 with 7, and 8 with 10). The reduced accumulation of viral RNA in dcl1–7 plants was even more evident when TCV CP was replaced by P19, a silencing suppressor encoded by tomato bushy stunt virus (TBSV). The resulting viral mutant, TCV-P19, accumulated to similar levels in IL of Col-0, dcl2, dcl3, and dcl4 (Fig. 1C, lanes 2 and 4–6). The presence of P19 also enabled viral systemic movement, which was enhanced in dcl4 plants (SL blot, lanes 2 and 4–6). However, the accumulation of TCV-P19 in dcl1–7 IL was much lower, and it failed to move systemically (Fig. 1C, lane 3). No such difference was detectable with wtTCV (Fig. S1, compare lanes 2 and 3), suggesting that DCL1 does not influence TCV replication (19). It has been established that P19 suppresses silencing by binding to siRNAs and inhibiting the formation of RISC complex (20, 21). TCV CP, however, functions to prevent DCL4-mediated processing of viral dsRNA, a process thought to not only feed RISC with siRNAs, but also destroy the viral RNA itself (6, 22). The fact that TCV-P19, but not wtTCV, is associated with a decrease in viral RNA levels in infected dcl1–7 plants strongly suggests that it is because of enhanced processing of viral RNA by other DCLs, thus implying that disrupting DCL1 function up-regulates the antiviral activity of other DCLs.
Fig. 2.
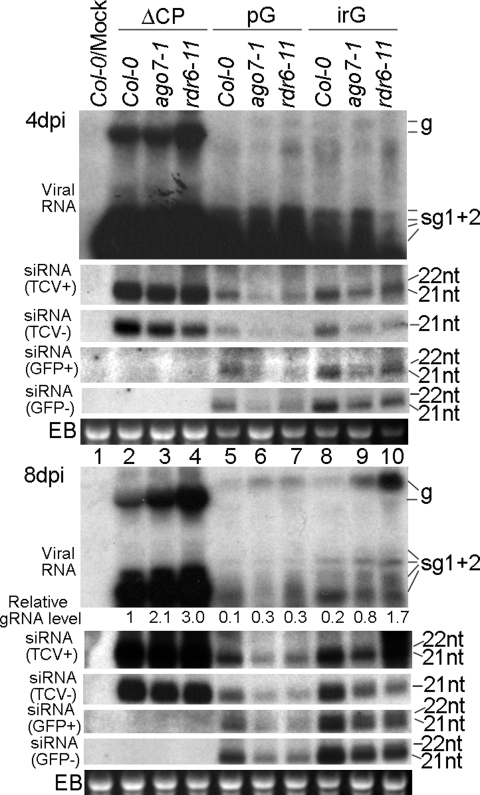
RNA blot hybridizations showing accumulation levels of viral RNAs and siRNAs in ago1–11 and dcl1–9 plants infected with ΔCP, TCV-pG (pG), and TCV-irG (irG). The relative gRNA levels were calculated based on the intensity of respective hybridization signals (see SI Materials and Methods for detailed methods). The same set of siRNA membranes were stripped and hybridized with different probes sequentially to detect siRNAs of different origins and polarities.
The profile of ΔCP-specific siRNAs is generally consistent with previous reports (6, 9, 17). Their size was predominantly 21 nt in all but the dcl4 plants, where the DCL2-generated 22-nt siRNA predominates (Fig. 1B, siRNA blot). Note that the relative concentration of siRNAs in these plants is roughly in proportion to the level of viral RNA, accounting for why dcl1–7-infected plants contained the least siRNA and dcl4 contained the most. This result indicates that in single dcl mutants, the siRNA levels most likely reflect the ongoing siRNA production from viral RNAs rather than the difference in functionality of various DCLs. Together, the above experiments strongly suggest that DCL1 negatively regulates the antiviral activity of other DCLs.
DCL1 Negatively Regulates the Expression of DCL4 and DCL3.
Next we wanted to determine which DCL is primarily responsible for enhanced antiviral silencing in dcl1–7 plants. To accomplish this goal, we first addressed the possibility that the mutated DCL1 itself caused the accelerated degradation of viral RNA, because it has been reported that the DCL1 mRNA level in dcl1–7 plants is actually higher than in Col-0 plants (23). The reason DCL1 mRNA is higher is because the DCL1 mRNA itself is the target of a miRNA (miR162), and the dcl1–7 mutation, an amino acid substitution within the helicase domain of DCL1, drastically reduces the cellular level of miRNAs (23). Because the complete loss-of-function mutant of DCL1 is embryo lethal, we decided to use a virus-induced gene silencing (VIGS) approach to down-regulate the expression level of DCL1. We reasoned that because DCL4 rather than DCL1 is the primary dicer of TCV RNA, a TCV construct containing a portion of DCL1 sequence would be processed efficiently to produce DCL1-targeting siRNAs, which would then result in lower DCL1 expression. The effect of the VIGS-mediated DCL1 down-regulation could then be evaluated by directly examining the RNA accumulation level of the VIGS construct.
To test this, we made the construct TCV-DCL1, in which a 555-bp fragment of DCL1 cDNA (nucleotides 867–1,422) was used to replace the 5′ half of the CP coding region (Fig. 1A). TCV-DCL1, together with a control construct containing the GFP cDNA in the same region (TCV-GFP), was used to infect Col-0 as well as dcl2, dcl3, and dcl4 plants. As shown in Fig. 3A, TCV-DCL1 accumulated to a lower level than TCV-GFP in Col-0 plants (4 dpi blot, lanes 1 and 2), suggesting that DCL1 down-regulation by VIGS released its repression on other DCL(s), which in turn accelerated the degradation of TCV-DCL1. We hence conclude that the lower level of viral RNA accumulation in dcl1–7 plants is caused by the release of DCL1-mediated repression rather than higher DCL1 activity.
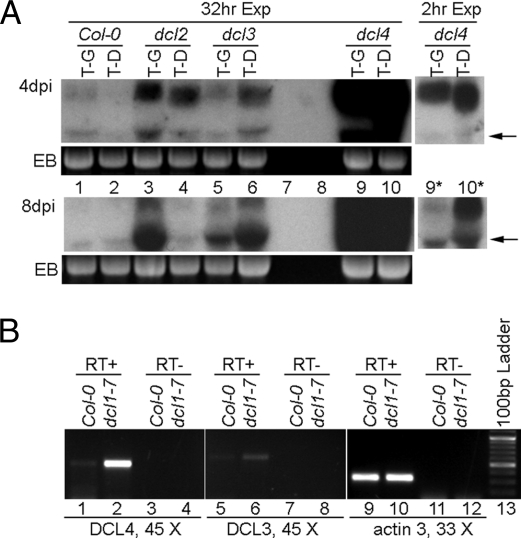
Fig. 3.
DCL1 down-regulates the expression of DCL4 and DCL3. (A) RNA blot hybridization showing accumulation levels of TCV-GFP (T-G) and TCV-DCL1 (T-D) in Col-0, dcl2, dcl3, and dcl4 mutants. Lanes 7 and 8 were purposefully not loaded. Blots covering sample nos. 1–10 were first exposed for 32 h to reveal viral RNA signals in lanes 1 and 2. They were then exposed for 2 h (lanes 9* and 10*) to reduce the over-exposure in lanes 9 and 10. The arrows denote the position of deletion products. (B) sqRT-PCR illustrating the relative levels of DCL4 and DCL3 mRNA in Col-0 and dcl1–7 plants. RT-PCR of actin 3 mRNA was used as the control. Cycle numbers are shown beneath the respective lanes.
The results from infections of dcl mutants with both TCV-GFP and TCV-DCL1 provided strong evidence for enhanced DCL4 activity upon DCL1 down-regulation. In dcl2 plants, both TCV-GFP and TCV-DCL1 RNA levels were appreciably higher than in Col-0 plants (Fig. 3A, lanes 3 and 4 versus lanes 1 and 2). However, the TCV-GFP level remained higher than TCV-DCL1. This result suggests that DCL2 did not contribute significantly to the accelerated viral RNA degradation caused by DCL1 down-regulation. In contrast, both dcl3 and dcl4 plants enabled higher accumulation of TCV-DCL1 relative to TCV-GFP (Fig. 3A, lanes 5, 6, and 9–12), which is opposite to the results observed with both Col-0 and dcl2 plants, suggesting that both DCL4 and DCL3 contribute to enhanced antiviral silencing in response to DCL1 disruption. However, the contribution of DCL4 is predominant because the accumulation levels of both TCV-GFP and TCV-DCL1 in dcl4 plants were at least 10-fold higher than in dcl3 plants (note that the exposure time for lanes 9* and 10* was only 1/16 as long to produce comparable signals with lanes 5 and 6). Finally, the higher levels of TCV-DCL1 than TCV-GFP in dcl3 and dcl4 plants suggests that the former is likely more replication competent, which strengthens the argument that its lower accumulation in WT plants is caused by DCL1 down-regulation.
To further investigate the mechanism of DCL1-mediated suppression of DCL4 and DCL3, we compared their mRNA levels in Col-0 and dcl1–7 plants by using a semiquantitative (sq) RT-PCR procedure. RNA samples extracted from mock-inoculated Col-0 and dcl1–7 leaves (4 dpi) were used as templates. As shown in Fig. 3B, the control actin 3 RT-PCR product (318 bp) was at the same intensity for both Col-0 and dcl1–7 samples (Fig. 3B, lanes 9 and 10). However, the DCL4-specific fragment (550 bp) was readily detectable in dcl1–7 plants under conditions (45 PCR cycles) insufficient for its detection in Col-0 plants (lanes 1 and 2). This result demonstrates that there is a significantly higher level of DCL4 mRNA in dcl1–7 plants. Similarly, the DCL3 fragment (613 bp) was also more intense in dcl1–7 plants, although the difference was much less evident (lanes 5 and 6). All of the RT− (without reverse transcriptase) controls were negative, indicating that the PCR products were mRNA-dependent. We conclude from these results that disruption of DCL1 function leads to higher expression of DCL4 and DCL3 in Arabidopsis leaves. These findings could also account for the reduced susceptibility of dcl1–9 plants to red clover necrotic mosaic virus reported by Takeda et al. (19).
Similarly, VIGS-mediated down-regulation of DCL1 also led to elevated levels of DCL4 and DCL3 mRNA. We were unable to detect any reduction of DCL1 mRNA levels in TCV-DCL1-infected leaves, likely because of limited spread of TCV-DCL1. We thus created TCV-P19-GFP and TCV-P19-DCL1, in which TBSV P19 is included to enhance the spread of the VIGS constructs (Fig. S2A). In addition, we chose a different region of DCL1 cDNA (nucleotides 1,912–2,210) as the insert for TCV-P19-DCL1, aiming to independently verify the results with TCV-DCL1. As expected, TCV-P19-DCL1 accumulated to modestly lower levels than TCV-P19-GFP (Fig. S2B), although both accumulated ≈100-fold more than the comparable constructs lacking the silencing suppressor (data not shown). At 6 dpi, when a reduction of the DCL1 mRNA level was detectable in TCV-P19-DCL1-inoculated leaves (Fig. S2C, compare lanes 5 and 6; DCL1 product highlighted by *), we witnessed a concurrent elevation of DCL4 mRNA level (lanes 9 and 10). The DCL3 mRNA level is also slightly elevated (lanes 13 and 14).
In summary, our results with two different dcl1 mutants and two different VIGS constructs established that disruption of DCL1 function led to enhanced antiviral defense, which we attribute to increased expression of DCL4 and, to a lesser extent, DCL3. Exactly how DCL1 down-regulates the expression of DCL4 and DCL3 awaits further investigation. A simple explanation would be that both DCLs are targeted by miRNAs or other DCL1-dependent endogenous siRNAs (24). In support of this, we found that the DCL4 mRNA level is elevated in ago1–11 mutants as well (Fig. S3). However, no miRNA targeting either DCL4 or DCL3 has been identified. A single DCL4-related small RNA (no.194424) was identified by searching the Arabidopsis Small RNA Project database (http://asrp.cgrb.oregonstate.edu/db/). It is originated from the 18th intron of the DCL4 gene, of messenger sense orientation, and thus not expected to target DCL4 mRNA for degradation. No DCL3-related small RNAs have been identified. Another possibility is that DCL4 (and potentially other silencing pathway genes) could be regulated by one of many transcription factors that are targeted by miRNAs.
DRB4 Is Required for Efficient Antiviral RNA Silencing.
DRB4 has been shown to assist DCL4 in the biogenesis of at least one transacting siRNA and more recently has been found to be targeted by the silencing suppressor encoded by cauliflower mosaic virus (CaMV), a DNA virus (25, 26). In light of these reports, we were interested in learning whether DRB4 also contributes to antiviral defense against RNA viruses. We therefore subjected drb4–1 mutant plants to infections with ΔCP. The dcl4 mutant was included in this experiment for comparison. As shown in Fig. S4, a significant increase of ΔCP RNA level was consistently observed in drb4 plants at both 4 and 8 dpi (compare lanes 2 and 4 in both blots). However, this increase was visibly less than in the dcl4 plants (compare lanes 3 and 4). Compared with Col-0 plants, the drb4 plants showed a slight decrease in the level of 21 siRNAs, despite the significant increase in viral RNA levels (Fig. S4, compare lanes 2 and 4 of the siRNA blot). This result suggests that DRB4 may not be directly involved in siRNA production. Rather, it could stabilize the 21-nt viral siRNAs and deliver them to the RISC complex, a role assigned to its homologs in Drosophila and C. elegans (15, 27).
Both AGO1 and AGO7 Contribute Viral RNA Clearance.
Previous studies concerning the role of AGO proteins in antiviral silencing have been inconclusive (11, 12, 28). To determine which AGO is responsible for the slicing of viral RNAs, we first infected ago1–11 mutant plants with ΔCP transcripts. The ago1–11 plants contain a point mutation within AGO1 gene that leads to partial loss of AGO1 function (29). ΔCP RNA accumulated to much higher levels in ago1–11 plants than in their WT counterparts (Ler-0) (Fig. S5A, lanes 4–6). This enhanced susceptibility in IL also frequently resulted in systemic movement of ΔCP (data not shown). These results unequivocally demonstrated that AGO1 plays a major role in controlling the RNA levels of ΔCP and possibly other viruses (12). However, ago1–11 plants were not more susceptible to wtTCV, again highlighting the critical contribution of viral silencing suppressors to the survival of plant viruses (data not shown). In contrast to dcl mutants, increased viral RNA accumulation in ago1–11 plants was accompanied by significantly decreased siRNA levels (Fig. S5A, siRNA blot, compare lanes 5 and 6), strongly suggesting that viral siRNAs are stabilized by their incorporation into the AGO1-based RISC complex.
Surprisingly, the elevated viral RNA levels, although easily detectable in ago1–11 plants infected with ΔCP, were not seen in the same mutants infected with TCV-GFP (Fig. S5A, lanes 11 and 12). TCV-GFP RNA was as quickly degraded in ago1–11 as in Ler-0 plants. A direct comparison with dcl4 mutants showed that, although in dcl4 plants TCV-GFP also accumulated to lower levels than ΔCP (Fig. S5A, compare lanes 9 with 3), the difference between mutant and WT plants was still very significant (compare lanes 9 with 8). ago1–11 plants, however, permitted higher viral RNA titer only when ΔCP was the inoculum. These results suggest that factors other than AGO1 contribute to the preferential clearance of TCV-GFP viral RNA in ago1–11 plants.
To identify these additional host factors, we searched for alternative AGO(s) capable of slicing TCV-GFP in ago1–11 plants. We hypothesize that AGO7 might contribute to this process because AGO7 is known to function together with DRB4 (25), and we have shown that DRB4 participates in defense against viruses (see Fig. S4). As an additional control, we included an rdr6 mutant (rdr6–11) (30) in this analysis because previous studies have shown that RDR6 plays a critical role in antiviral defense against several viruses including TCV (15, 31, 32). As shown in Fig. S5B, both ΔCP and TCV-GFP RNAs accumulated to modestly higher levels in rdr6–11 and ago7 plants, although the overall level of TCV-GFP RNA was much lower than ΔCP in all infected plants. These results suggest that, although AGO1 is more efficient at slicing RNAs of viral origin, AGO7 appears to act on both viral and nonviral RNA with similar efficiency, likely serving as a surrogate slicer in the absence of AGO1.
Viral RNAs with Nonviral Inserts are Preferentially Targeted in ago1–11 Plants.
To understand how two different AGOs differentiate the two kinds of target RNAs, we entertained the idea that, because viral RNAs tend to have more extensive secondary structures than nonviral RNA, AGO1 might be more capable of tolerating these secondary structures than AGO7. To test this hypothesis, we made two additional TCV-based constructs containing nonviral inserts of the same length (289 nt) but with varying degrees of secondary structure. The first one, TCV-pG, contained a portion of the cycle 3 GFP cDNA (33), whereas the second one, TCV-irG, contained the first 157 nucleotides of the pG insert and a 132-nt homologous fragment derived from the synthetic GFP cDNA (34) fused in the opposite orientation. The two inserted fragments in TCV-irG are 71% complementary so that the whole insert would form an incomplete hairpin with a 25-nt end loop. Both TCV-pG and TCV-irG, together with ΔCP, were then used to infect ago1–11 and ago7 plants.
Several interesting outcomes emerged from analyzing the infected ago1–11 plants (Fig. 2). First, at the early stage of infection, TCV-pG RNA accumulated to approximately one-third the level of TCV-irG, in both Ler-0 and ago1–11 plants (4 dpi viral RNA blot, compare lanes 5 and 6 with lanes 8 and 9; note the relative gRNA levels). This result clearly demonstrates that the more structured nonviral insert was more resistant to degradation. Secondly, whereas ΔCP gRNA levels were increased 1.6-fold in the absence of functional AGO1 (compare lanes 3 with 2), the gRNAs of both TCV-pG and TCV-irG were at similar levels in ago1–11 and Ler-0 plants (compare lanes 6 with 5 and 9 with 8). Furthermore, sgRNAs of TCV-pG and TCV-irG were decreased in ago1–11 plants. This trend was more evident in 8 dpi samples, where even the gRNAs of these two constructs were three- to fourfold less abundant in ago1–11 plants than in Ler-0 plants (Fig. 2, 8 dpi viral RNA blot, compare lanes 6 with 5 and 9 with 8). In addition, although sgRNAs of ΔCP generally accumulated to higher levels than its gRNA, the sgRNAs of both TCV-pG and TCV-irG were far lower than their gRNAs. Collectively, these data suggest that the nonviral inserts in the sgRNAs of TCV-pG and TCV-irG are the primary targets of an RNA clearance pathway to which viral RNAs are more resistant. More importantly, this particular pathway is not only independent of AGO1 function, but is more robust when AGO1 is debilitated. However, as shown in the siRNA blots of Fig. 2, siRNA levels of viral as well as insert origins in all infected ago1–11 plants were decreased, suggesting that additional AGO(s) did not fully compensate for the loss of AGO1. Therefore, other non-AGO components of the silencing pathway may also have contributed to the enhanced degradation of TCV-pG and TCV-irG in ago1–11 plants. In agreement with this assertion, we found that the DCL4 as well as DCL3 mRNA levels are elevated in ago1–11 plants, which could enhance the dicing of TCV-pG and TCV-irG (Fig. S3). These results may also partially explain the inconsistencies in previous reports concerning the antiviral role of AGO1 (11, 28).
AGO7 and RDR6 Contribute Favorably to the Silencing of TCV Mutants Containing Nonviral Inserts.
To further investigate the role of AGO7 as well as RDR6 in silencing TCV mutants, with or without nonviral inserts, we next infected ago7–1 and rdr6–11 plants with ΔCP, TCV-pG, and TCV-irG. Unexpectedly, we witnessed far more significant reduction of both TCV-pG and TCV-irG RNAs in the Col-0 than in the Ler-0 plants (compare lanes 2, 5, and 8 in Fig. 4 with their counterparts in Fig. 2). Exactly why the two ecotypes of Arabidopsis plants responded to insert-containing TCV mutants so differently was not investigated further.
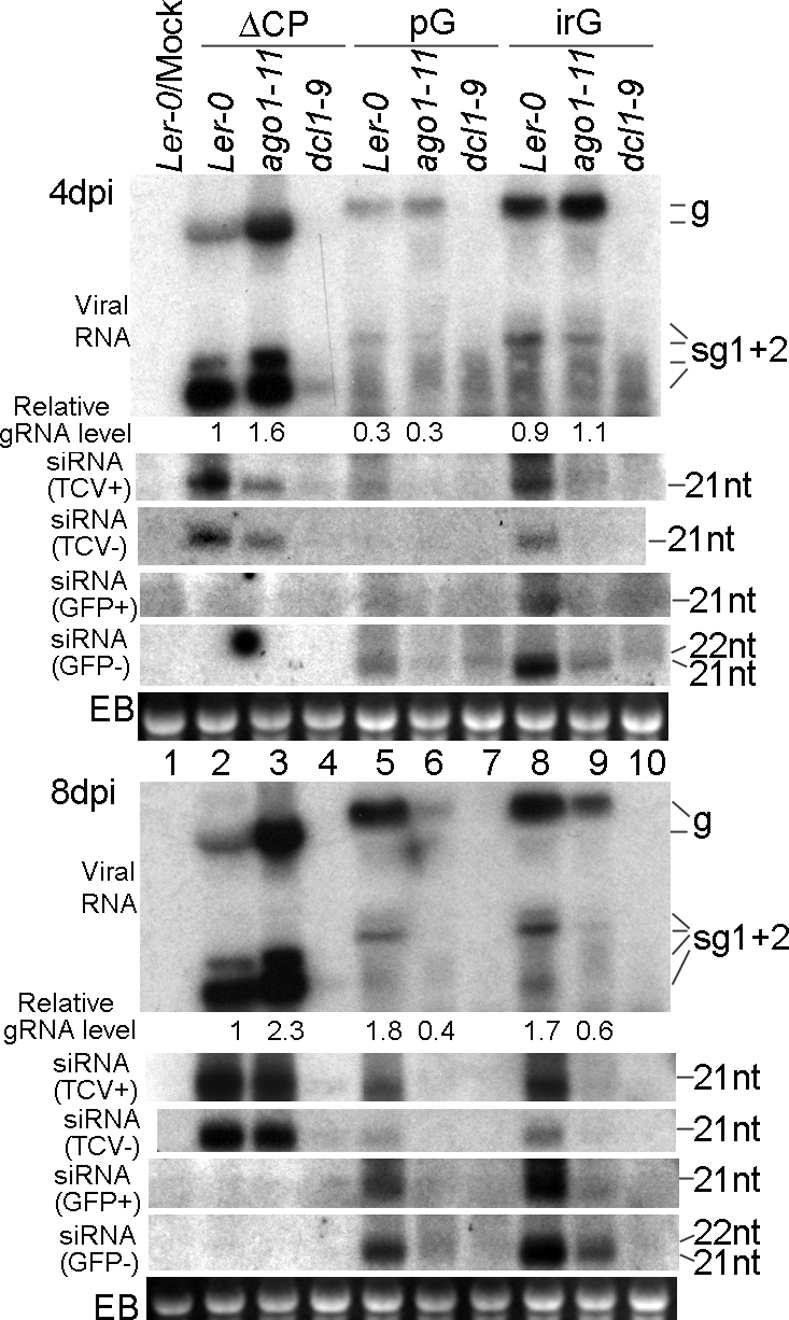
Fig. 4.
RNA blot hybridizations showing accumulation levels of viral RNAs and siRNAs in ago7–1 and rdr6–11 plants infected with ΔCP, TCV-pG (pG), and TCV-irG (irG).
Nevertheless, we were still able to discern a role for both AGO7 and RDR6 in the silencing of all three TCV mutants, each of which accumulated to higher levels in ago7 (two-, three-, and fourfold increases, respectively) and rdr6–11 plants (three-, three-, and eightfold increases, respectively) (Fig. 4, 8 dpi viral RNA blot). Contrary to our expectation, ΔCP and TCV-irG, which are expected to be more structured, were actually more responsive to RDR6 disruption (the rdr6–11 plants) than TCV-pG, which is predicted to be less structured (Fig. 4, 8 dpi viral RNA blot, compare lanes 4 with 2, 10 with 8, and 7 with 5). Although we currently do not have an explanation for this observation, we speculate that other RDRs and/or AGOs could access the pG insert more readily and thus compensate for the loss of RDR6.
Examination of viral siRNAs further implicated RDR6 in the silencing of all three TCV mutants. First, for ΔCP and TCV-irG infections, the TCV-specific, genome sense (TCV+) siRNA levels were almost the same in rdr6–11 and Col-0 plants, whereas the (TCV−) siRNAs were visibly lower (Fig. 4, siRNA blots, compare lanes 4 with 2 and 10 with 8). These results suggest that whereas the production of (TCV−) siRNAs relies more on RDR6, the (TCV+) siRNAs are largely independent of RDR6. In contrast, the TCV-pG infections of rdr6–11 plants were characterized by lower levels of both (TCV+) and (TCV−) siRNAs (Fig. 4, compare lanes 7 with 5 of the siRNA blots). This result suggests to us that the presence of the pG insert in the construct rendered the viral genome more accessible to RDR6, which we interpret to mean that RDR6 is able to proceed along the TCV genome once anchored on the pG portion of the RNA, whereas the hairpin insert in TCV-irG may cause more frequent pauses, partially explaining why TCV-irG is more stable than TCV-pG.
Unlike TCV-specific siRNAs, GFP-specific siRNAs of both polarities showed visible decline in rdr6–11 plants infected with both insert-containing constructs [Fig. 4, siRNA (GFP+) and (GFP−) blots, compare lanes 7 with 5 and 10 with 8]. These results suggest that, whereas TCV gRNAs are diced by DCLs with little help from RDR6, neither pG nor irG inserts are recognized as efficiently by DCLs despite the long, incomplete hairpin of irG. Instead, at least a portion of these inserts have to be converted to dsRNA form by RDR6 before being diced. We conclude from these analyses that the nonviral inserts are more readily targeted by RDR6 than viral RNAs. Taken together, our siRNA data further confirm that viral RNAs, the less structured regions in particular, are subjected to RDR6-mediated antiviral silencing.
Finally, all of the infected ago7 samples accumulated generally lower levels of siRNAs, regardless of the origins and polarities. However, lower siRNA levels were always more pronounced in TCV-pG- and TCV-irG-infected samples than in ΔCP-infected samples, suggesting that AGO7 is more preferentially associated with siRNAs derived from nonviral inserts (Fig. 4, siRNA blots, compare ago7–1 lanes with Col-0 lanes). On the other hand, the majority of TCV-ΔCP-derived siRNAs likely associate with AGO1 because they are still mostly stable in the absence of AGO7 (Fig. 4, siRNA blots, compare lanes 3 and 2).
To summarize, we found that both RDR6 and AGO7 participate in silencing-based defense targeting RNA viruses. Furthermore, both are more preferably associated with nonviral inserts, with a slight bias toward less structured inserts. These observations, together with the finding that ΔCP RNA levels increased drastically in response to AGO1 disruption, led us to hypothesize that AGO1 and AGO7 may differ in their ability to recruit ds siRNAs with varying degrees of mismatch. We argue that, because AGO1 is associated with miRNAs, which are known to have mismatches, it may better tolerate the viral siRNAs derived from highly structured (+) sense viral RNAs, whereas AGO7 likely recruits only fully paired siRNAs (25). We want to stress that this hypothesis does not imply that AGO1 is incapable of recruiting completely paired ds siRNAs as AGO1 is known to be involved in sense trangene-mediated silencing, which is expected to generate fully paired ds siRNAs (28). Rather, AGO1 may tolerate more mismatches than AGO7. Preferential siRNA recruitment has been demonstrated biochemically for two Drosophila AGO proteins; hence, we think it is likely that similar sorting mechanisms exist in Arabidopsis (35). In addition, the functional preference of AGOs could also occur at the slicing step as AGO1 could tolerate more mismatches between siRNAs and their targets, as in the case of miRNA-mediated cleavage. Finally, the siRNAs derived from nonviral inserts could possess different 5′ terminal nucleotides than viral siRNAs and thus be selectively recruited by different AGOs, as revealed by two recent reports (36, 37).
This model predicts that varying genetic requirements for silencing different viruses may in part be caused by the tendency of different viral RNAs to form secondary structures. Viruses with long, relatively unstructured areas in their RNA genomes would be more susceptible to RDR6-AGO7-mediated RNA silencing. On the other hand, viruses with extensive secondary structures would be more frequently targeted by AGO1. This model further predicts that the ds replicative form of RNA viruses likely does not contribute significantly to siRNA production (38).
Conclusion
We have examined the genetic requirements of RNA silencing-based antiviral defense by using TCV mutants devoid of the silencing suppressor to infect Arabidopsis plants with defects in silencing pathway genes. We focused our investigation on the four DCLs, two AGOs, DRB4, and RDR6 that were shown previously to have defined roles in one or more endogenous RNA silencing pathways (5). We found that all four DCLs participate in the antiviral silencing. Whereas DCL2, 3, 4 contribute positively to viral RNA clearance, DCL1 negatively regulates antiviral silencing through down-regulation of DCL4 and DCL3 expression. This observation is in contrast to a report by Moissiard and Voinnet (39), in which DCL1 was found to have a facilitative role in processing the 35S RNA leader of CaMV. The difference may well be because of the fact that CaMV is a DNA virus replicating in the nucleus. In addition, we also established an important role for DRB4 in antiviral silencing. Finally, we revealed that two AGOs, AGO1 and AGO7, function coordinately to ensure efficient clearance of viral RNAs with different degrees of secondary structure. These findings are expected to be conducive to further in-depth investigations.
Materials and Methods
A complete description of the materials and methods used in this study is provided in SI Materials and Methods. Briefly, the Arabidopisis mutant plants containing mutations in several DCL, AGO, DRB, or RDR genes were acquired from various sources and reared in growth chambers under standard conditions. They were infected with in vitro transcripts of TCV cDNA or its derivatives and subjected to RNA blot analyses to determine the accumulation of viral RNA and siRNAs. The mRNA levels of key host genes were evaluated by using sqRT-PCR.
Supplementary Material
Acknowledgments.
We thank Drs. J. C. Carrington, R. A. Martienssen, and O. Voinnet for providing us with seeds of a number of mutant lines, Dr. R. S. Poethig for answering our many questions concerning mutant rdr6–11, and anonymous reviewers of the first version of this manuscript for the invaluable comments and suggestions they made. This work is supported in part by U.S. Department of Energy Grant DE-FG03-98ER20315 and National Institutes of Health Grant P20 RR16469.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805760105/DCSupplemental.
References
- 1.Hammond SM. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 2.Vaucheret H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 3.Peters L, Meister G. Argonaute Proteins: Mediators of RNA Silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Hiraguri A, et al. Specific interactions between Dicer-like proteins and HYL1/DRB- family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 5.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22:268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akbergenov R, et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34:462–471. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz-Pendon JA, Li F, Li W-X, Ding S-W. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2006;19:2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumberger N, Baulcombe DC. Arabidopsis Argonaute1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, et al. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szittya G, et al. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 2003;22:633–640. doi: 10.1093/emboj/cdg74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chellappan P, Vanitharani R, Ogbe F, Fauquet CM. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 2005;138:1828–1841. doi: 10.1104/pp.105.066563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu F, et al. The Nicotiana benthamiana homolog of SDE1/SGS2/RDR6 functions in a temperature-dependent manner to defend both differentiated tissues and shoot apices against viral invasion. J Virol. 2005;79:15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu R, et al. Animal virus replication and RNAi-mediated silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blevins T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu F, Morris TJ. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J Virol. 1997;71:1428–1435. doi: 10.1128/jvi.71.2.1428-1435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeda, et al. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 2005;24:3147–3157. doi: 10.1038/sj.emboj.7600776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panteleo V, Szittya G, Burgyan J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J Virol. 2007;81:3797–3806. doi: 10.1128/JVI.02383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omarov RT, Ciomperlik JJ, Scholthof HB. RNAi-associated ssRNA-specific ribonucleases in Tombusvirus P19 mutant-infected plants and evidence for a discrete siRNA-containing effector complex. Proc Natl Acad Sci USA. 2007;104:1714–1719. doi: 10.1073/pnas.0608117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu F, Ren T, Morris TJ. The coat protein of Turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J Virol. 2003;77:511–522. doi: 10.1128/JVI.77.1.511-522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 24.Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- 25.Adenot X, et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 26.Haas G, et al. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J. 2008 doi: 10.1038/emboj.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, et al. A bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 28.Morel JB, et al. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- 30.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 32.Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by Potato Virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 34.Chiu W-L, et al. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 35.Forstemann K, Horwich MD, Wee LM, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mi S, et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montgomery TA, et al. Specificity of Argonaute7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 38.Molnar A, et al. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moissiard G, Voinnet O. RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci USA. 2006;103:19593–19598. doi: 10.1073/pnas.0604627103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.