Abstract
Recent years have seen much interest in the study of systems characterized by multiple interacting components. A class of statistical models called graphical models, in which graphs are used to represent probabilistic relationships between variables, provides a framework for formal inference regarding such systems. In many settings, the object of inference is the network structure itself. This problem of “network inference” is well known to be a challenging one. However, in scientific settings there is very often existing information regarding network connectivity. A natural idea then is to take account of such information during inference. This article addresses the question of incorporating prior information into network inference. We focus on directed models called Bayesian networks, and use Markov chain Monte Carlo to draw samples from posterior distributions over network structures. We introduce prior distributions on graphs capable of capturing information regarding network features including edges, classes of edges, degree distributions, and sparsity. We illustrate our approach in the context of systems biology, applying our methods to network inference in cancer signaling.
Keywords: Bayesian networks, biological networks, graphical models, protein signaling
A class of models called graphical models (1–3) is widely used for formal statistical inference regarding systems of multiple interacting components. A graphical model consists of a graph, describing probabilistic relationships between variables, and parameters specifying conditional distributions implied by the graph. In many settings, questions of interest concern the graph itself. For example, in molecular biology, we may be interested in saying something about which molecules or combinations of molecules influence one another; in the social sciences we may be interested in relationships between various economic and demographic factors.
Inference on graphical model structure (4–8) is widely recognized to be a challenging problem, partly because of the vast space of possible graphs for even a moderate number of variables. Yet, equally, an understanding of the relevant scientific domain may suggest that not every possible graph is equally plausible, and that certain features should be regarded as a priori more likely than others. Where available, such knowledge is a valuable resource, making the question of how to capture and exploit it an important one. In this article, we address precisely this question. We focus on directed graphical models called Bayesian networks, and use Markov chain Monte Carlo (MCMC) for network inference. We seek to take account of detailed information concerning network features such as individual edges, edges between classes of vertices, and sparsity. In many settings, such beliefs follow naturally from a consideration of the underlying science or semantics of the variables under study. We present priors for beliefs of this kind and show examples of how these ideas can be used in practical settings.
Network Inference
We begin by reviewing basic ideas and notation for Bayesian networks, with an emphasis on inference regarding network features. A Bayesian network (1, 2) consists of (i) a directed acyclic graph G = (V(G), E(G)), whose vertices V represent random variables X1, …, Xp of interest, and whose edge-set E contains edges describing conditional independencies between those variables, and (ii) parameters θ that specify conditional distributions implied by G. In particular, the graph G implies that each variable is conditionally independent of its nondescendants given its immediate parents. Importantly, this means that the joint distribution over X1, …, Xp can be factorized into a product of local terms, such that p(X1, …, Xp | G) = Πi=1pp(Xi | PaG(Xi)), where PaG(Xi) is the set of parents of Xi in graph G.
The goal of network inference is to make inferences regarding the graph G itself. Let X represent a p × n data matrix, where n is the number of multivariate samples available. Using Bayes' theorem, the posterior probability P(G | X) of graph G is given (up to proportionality) by p(X | G)P(G), where p(X | G) is the (marginal) likelihood and P(G) a prior distribution over directed acyclic graphs; we refer to the latter as a “network prior.”
Assume that the form of the conditional distributions p(Xi | Pa(Xi)) is known, and let θ represent a complete set of model parameters. Then, the marginal likelihood p(X | G) can be evaluated by integrating over model parameters θ. This article is concerned with inferences regarding the graph G itself, and the ideas presented here are applicable for any choice of conditional distributions and parameter priors under which the marginal likelihood can be evaluated. In our experiments, we follow previous authors (4) in assuming parameter independence and using multinomial conditionals and Dirichlet priors. This gives the following well known closed-form marginal likelihood:
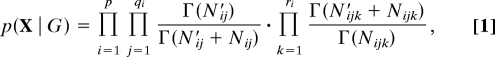 |
where Nijk is the number of observations in which Xi takes the value k, given that PaG(Xi) has configuration j; qi is the number of possible configurations of parents PaG(Xi); and ri is the number of possible values of Xi. N′ijk are Dirichlet hyperparameters. Finally,
and
MCMC on Networks.
The number of possible graphs grows superexponentiallye with the number of variables p, precluding an exhaustive enumeration of the posterior distribution beyond p = 6 or so. Thus, although we can evaluate the posterior probability of a graph up to a multiplicative constant, we cannot consider every possible graph during inference. This motivates the use of computational methods to characterize posterior distributions over graphs; here, we use a Metropolis–Hastings sampler (5, 7, 10) for this purpose.
Let η(G) denote a neighborhood around a directed acyclic graph G, consisting of every directed acyclic graph that can be obtained by adding, deleting or reversing a single edge in G. Define proposal distribution Q as follows:
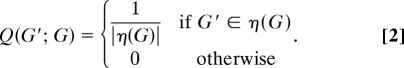 |
The acceptance ratio is then 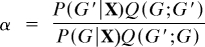 . A proposed graph G′, drawn from Q, is accepted with probability min(1, α) and otherwise rejected. If accepted, G′ is added to the sequence of samples drawn and becomes the current graph. Else, G is added to the sequence of samples and remains the current graph. The proposal distribution Q gives rise to an irreducible Markov chain, since there is positive probability of reaching any part of the state space (5, 7). Standard results (10) then guarantee convergence of the Markov chain to the desired posterior P(G | X).
. A proposed graph G′, drawn from Q, is accepted with probability min(1, α) and otherwise rejected. If accepted, G′ is added to the sequence of samples drawn and becomes the current graph. Else, G is added to the sequence of samples and remains the current graph. The proposal distribution Q gives rise to an irreducible Markov chain, since there is positive probability of reaching any part of the state space (5, 7). Standard results (10) then guarantee convergence of the Markov chain to the desired posterior P(G | X).
Inferences Regarding Network Features.
The sampling procedure gives rise to samples G(1), …, G(T). An important property of these samples is that, provided the Markov chain has converged to its stationary distribution, they provide a means by which to compute the expectation of essentially any network feature. Specifically, if 𝔼[φ(G) | X] is the expectation, under the posterior P(G | X), of a function φ(G), then
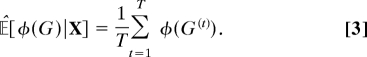 |
is an asymptotically valid estimator of 𝔼[φ(G) | X].
An important special case of Eq. 3, which we shall make use of below, concerns the posterior probability of an individual edge e, or P(e | X). Summing over graphs gives P(e | X) = 𝔼[IE(G)(e) | X] (where IA denotes the indicator function for set A). Applying Eq. 3 then gives the following asymptotically valid estimate of posterior edge probability:
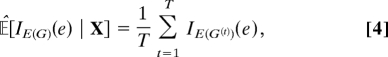 |
where, G(t) = (V(G(t)), E(G(t))).
Informative Priors on Networks
We begin with a motivating example highlighting some of the kinds of prior beliefs that are encountered in practice and that we might like to take account of during inference. We then introduce network priors in a general way, before looking at examples of such priors for specific kinds of prior information. Finally, we look briefly at the use of MCMC proposal distributions based on network priors.
A Motivating Example.
Table 1 shows 14 proteins that are components of a biological network called the epidermal growth factor receptor or EGFR system (11, 12). Here, each protein is a ligand, a receptor, or a cytosolic protein; for our present purposes, these may be regarded as well defined classes of variable. Our goal is to infer structural features of the biological network in which these components participate.
Table 1.
Some components of the epidermal growth factor receptor system
| Protein | Type |
|---|---|
| EGF | Ligand |
| AMPH | Ligand |
| NRG1 | Ligand |
| NRG2 | Ligand |
| EGFR | Receptor |
| ERBB2 | Receptor |
| ERBB3 | Receptor |
| ERBB4 | Receptor |
| GAP | Cytosolic protein |
| SHC | Cytosolic protein |
| RAS | Cytosolic protein |
| Raf | Cytosolic protein |
| MEK | Cytosolic protein |
| ERK | Cytosolic protein |
The biochemistry of the system provides us with some prior knowledge regarding network features, which we would like to take account of during inference. Some illustrative examples of the kind of knowledge that might be available include the following.
S1. Ligands influence cytosolic proteins via ligand–receptor interactions. As a consequence, we do not expect them to directly influence cytosolic proteins. Equally, we do not expect either receptors or cytosolic proteins to directly influence ligands.
S2. Certain ligand–receptor binding events occur with particularly high affinity; these include EGF and AMPH with EGFR, NRG1 with ERBB3, and NRG1 and NRG2 with ERBB4. Equally, the receptors EGFR, ERBB3, and ERBB4 are all capable of influencing the state of ERBB2 (via heterodimer formation and transphosphorylation). In addition, there is much evidence indicating that Raf can influence MEK, which in turn can influence ERK.
S3. Since we observe ligand-mediated activity at the level of cytosolic proteins, we expect to see a path from ligands to receptors, and from receptors to cytosolic proteins.
These beliefs correspond to information regarding network structure: S1 contains information concerning edges between classes of vertices, S2 contains information regarding individual edges, and S3 contains higher-level information regarding paths between classes of vertices.
General Framework.
We now introduce a general form for our network priors. Let f(G) be a real-valued function on graphs that is increasing in the degree to which graph G agrees with prior beliefs (a “concordance function”). Then, for potentially multiple concordance functions {fi(G)}, we suggest a log-linear network prior of the form
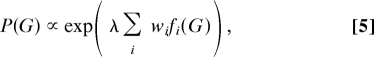 |
where λ is a parameter used to control the strength of the prior. Here, in the spirit of ref. 13, we use weights wi to control the relative strength of individual concordance functions, with w1 set to unity to avoid redundancy. We discuss setting strength parameters below. [Note that the only way in which the prior enters into MCMC-based inference is via the prior odds P(G′)/P(G) in favor of proposal G′; it is therefore sufficient to specify the prior up to proportionality.]
Individual Edges.
Suppose we believe that certain edges are a priori likely to be present or absent in the true data-generating graph. Let E+ denote a set of edges expected to be present (“positive edge-set”) and E− a set of edges expected to be absent (“negative edge-set”). We assume that these two sets are disjoint. Then, we suggest the following network prior:
Here, the concordance function is a counting function on individual edges, with the prior attaining its maximum value if and only if G contains all of the positive edges and no negative edges.
Edges Between Classes of Vertices.
The network prior given by Eq. 6 may also be used to capture beliefs regarding edges between classes of vertices.f Let {Ck} be a set of classes into which vertices v ∈ V can be categorized, with C(v) denoting the class to which vertex v belongs. Suppose we wish to penalize graphs displaying edges between vertices of class i and j. This can be accomplished by using the prior specified by Eq. 6 with a negative edge-set E− containing all such edges:
Positive priors on edges between vertex classes can be defined in a similar fashion.
Network Sparsity.
In many settings, parsimonious models are desirable both for reasons of interpretability and amelioration of overfitting. Since Bayesian networks factorize joint distributions into local terms conditioned on parent configurations, model complexity can grow rapidly with the number of parents. Controlling the in-degree of graphs is therefore a useful means of controlling model complexity.g The in-degree indeg(v) of a vertex v ∈ V is the number of edges in edge-set E leading into v; that is, indeg(v) = |{(vi, vj) ∈ E : vj = v}|. Let Δ(G) = maxv∈V(G)indeg(v) be the maximum in-degree of graph G. Then, the following network prior penalizes graphs having in-degree exceeding λindeg but remains agnostic otherwise:
The priors introduced up to this point are sufficient for the experiments presented below. However, to illustrate the full generality of network priors, we briefly discuss two further types of prior information.
Higher-Level Network Features.
In some cases, we may wish to capture prior knowledge concerning higher-level network features that cannot be described by reference to sets of individual edges. To take but one example, we may believe that there ought to be at least one edge between certain classes of vertices, as in S3. Let EC be a set of ordered pairs of classes such that (Ci, Cj) ∈ EC means that we believe there ought to be at least one edge from class Ci to class Cj. Then, we suggest using the prior in Eq. 5 with concordance function f(G) = Σ(Ci,Cj)∈ECIℤ+[Σ(v1,v2)∈E(G)δ((C(v1), C(v2)), (Ci, Cj))] (where ℤ+ is the set of positive integers and δ the Kronecker delta function).
Degree Distributions.
We may have reason to believe that the degree distribution of the underlying network is likely to be scale-free. The degree deg(v) of a vertex v is the total number of edges in which vertex v participates. The degree distribution of a graph G is a function πG(d) = |{v ∈ V(G) : deg(v) = d}| describing the total number of vertices having degree d. A graph is said to have a scale-free degree distribution if πG follows a power-law with πG(d) ∝ d−γ, γ > 0 such that log(πG(d)) is approximately linear in log(d). Accordingly, the negative correlation coefficient between log(πG(d)) and log(d) is a natural choice for a concordance function for the scale-free property, giving a network prior P(G) ∝ exp(−λ r(log(πG(d)), log(d))) (where r(·, ·) denotes the correlation coefficient of its arguments).
Prior-Based Proposals.
The prior P(G) provides information regarding which graphs are a priori more likely. Yet, the proposal distribution in Eq. 2 is uniform over neighborhood η(G). A natural idea, then, is to exploit prior information in guiding the proposal mechanism; here, we suggest one way of doing so that we have found empirically to be useful in accelerating convergence. We suggest a proposal distribution of the form
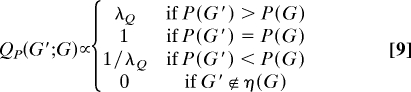 |
where, λQ ≥ 1 is a parameter controlling the degree to which the proposal mechanism prefers a priori likely graphs.
The proposal distribution specified by Eq. 9 ensures that all graphs in η(G) have a nonzero probability of being proposed, thereby preserving irreducibility and convergence to the desired posterior. Now, large values of λQ will result in frequent proposals of a priori likely graphs, but because of the “Hastings factor” Q(G; G′)/Q(G′; G) will also lead to low acceptance rates for such graphs. However, consideration of the form of the acceptance ratio yields a simple heuristic for determining λQ. Let Δf denote the median nonzero value of the absolute difference |f(G′) − f(G)| in the values of the concordance function for G′ and G (this can be determined during diagnostic sampling runs). Then, setting λQ = max(1, π exp(½λΔf)), π < 1, suffices to ensure that (i) a priori likely graphs do not suffer low acceptance ratios, and (ii) if the overall prior is too weak to permit a prior-based proposal, the proposal distribution in Eq. 9 reverts to the uniform distribution of Eq. 2. For example, for the counting function in Eq. 6 and neighborhoods constructed by single edge changes, |f(G′) − f(G)| is typically unity, giving λQ = max(1, π exp(λ/2)). (When using prior-based proposals, by default we set π = 1/2.)
Constructing a Prior.
We now consider two aspects of constructing a network prior: the qualitative question of what information to include, and the quantitative question of how to decide on a value for the strength parameter λ.
In a scientific domain, information to include in the prior must be derived from what is understood regarding the system under study. Although this process of extracting domain information is necessarily a subjective enterprise, we favor a conservative approach in which only information about which there is a broad consensus is included in the prior. We provide an example from cancer signaling below.
We address the question of prior strength in two steps. We first engage in a process of elicitation aimed at setting the strength parameter λ approximately to within an order of magnitude; this is accomplished in consultation with collaborators and by reference to the well known Jeffreys' scale (14), which relates odds ratios to intuitive degrees of belief. We then carry out a sensitivity analysis to check that results obtained are robust to changes in λ around the elicited value; we show examples of sensitivity analysis below. In the present article, we set parameters wi to unity. However, the formulation presented in Eq. 5 allows for the weighting of multiple sources of prior information; this is an important topic in its own right but one that we do not address further here.
A Simulation Study
Data.
We simulated data for the p = 14 variables described previously in Table 1, using the data-generating graph shown in Fig. 1C. Details of our data-generating model are as follows: the random variables are binary {0, 1}; all conditional distributions are Bernoulli, with success parameter p depending upon the configuration of the parents. In particular, root nodes are sampled with p = 0.5, whereas for each child node, p = 0.8 if at least one parent takes on the value 1, and p = 0.2 otherwise. Sample size was n = 200.
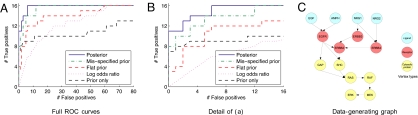
Fig. 1.
Receiver operating characteristic (ROC) curves for synthetic data. True positive rates are plotted against false positive rates across a range of thresholds, providing a comprehensive summary of sensitivity and specificity. Also shown is the true data-generating graph.
Priors.
The graph shown in Fig. 1C is based on the epidermal growth factor receptor system alluded to in the motivating example above. We constructed informative network priors corresponding to the beliefs S1 and S2 described above. We used S1 and S2 to define a negative edge-set E− and positive edge-set E+, respectively; these edge-sets were then used to specify a network prior using Eq. 6. S3 was not used in these experiments. To investigate the effects of priors containing erroneous information, we also constructed a mis-specified prior that included incorrect information regarding individual edges.h This allowed us to consider a realistic scenario in which the prior is largely reasonable but contains a number of entirely false beliefs. In all cases, λ was set to unity. We based all inferences on a single, long run of T = 50,000 iterations for each prior, with 5,000 samples discarded as “burn-in” in each case.i
ROC Analysis.
Our knowledge of the true data-generating graph allowed us to construct receiver operating characteristic or ROC curves from calls on individual edges. Let G* = (V*, E*) denote the true data-generating graph. As before, let P(e | X) denote the posterior probability of an edge e = (vi, vj). Then, the set of edges called at threshold τ ∈ [0, 1] is Eτ = {e : P(e | X) ≥ τ}, the number of true positives is |Eτ ∩ E*| and the number of false positives is |Eτ \ E*|. ROC curves were constructed by plotting, for each sampler, the number of true positives against the number of false positives parameterized by threshold τ; these are shown in Fig. 1 A and B. We also show results obtained by using absolute log odds ratios |ψij| for each pair (i, j) of variables; these are a natural measure of association for binary variables and provide a simple, baseline comparison. Finally, we show results obtained by drawing samples from the prior itself (“prior only”).
These ROC curves are obtained by comparison with the true edge-set E* and in that sense represent “gold-standard” comparative results. The posterior distribution provides substantial gains in sensitivity and specificity over both prior alone and data alone (i.e., the flat prior), suggesting that inference is indeed able to usefully combine data and prior knowledge.
Prior Sensitivity.
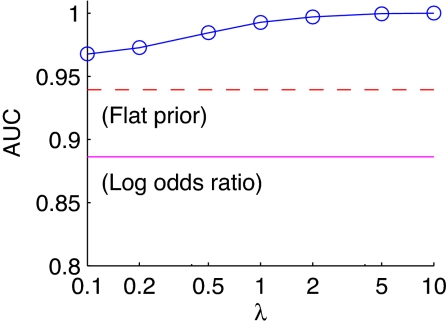
We investigated sensitivity to the strength parameter λ by performing ROC analyses as described above for a range of values of λ from 0.1 to 10. Fig. 2 shows the resulting area under the ROC curve (AUC) plotted against λ, for the correctly specified prior. The good results obtained by using the informative prior hold up across a wide rangej of values of λ.
Fig. 2.
Sensitivity analysis for synthetic data. Area under the ROC curve (AUC) is plotted against the strength parameter, for an informative prior. The area under the ROC curve captures, as a single number, the correctness of calls on edges across a range of thresholds; higher scores indicate lower error rates. For comparison, we show also AUC results for a flat prior and log-odds ratios as horizontal lines.
A Biological Network
Protein signaling networks play a central role in the biology of cancer. There remain many open questions regarding cancer-specific features of signaling networks, especially at the level of protein phospho-forms and isoforms. In this section, we present some results obtained in an analysis of protein signaling in breast cancer, using the methods introduced above.
Data.
Proteomic data were obtained for the 11 protein phospho-forms and isoforms shown in Fig. 3; these included two receptors, PDGF and C-MET (both are receptor tyrosine kinases or RTKs); two phospho-forms of AKT; two isoforms of MKK; two isoforms of ERK; MNK1; and two downstream proteins known to be involved in translational control, ELF4E and EIF2B. The data were obtained from an assay performed by Kinexus on a panel of 18 breast cancer cell lines. Preprocessing comprised (i) setting all zeros to 1/100 of the smallest nonzero value, (ii) taking logs, and (iii) discretizing where possible into active and inactive states or else around the median for each protein. This gave rise to binary data for each of the 11 proteins.
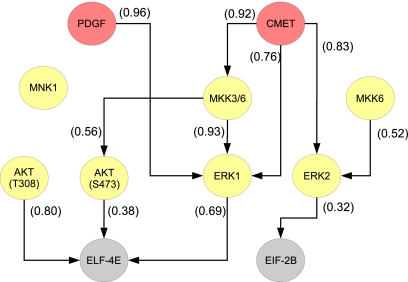
Fig. 3.
Posterior mode for protein data. Edges are annotated with posterior edge probabilities.
Priors.
Our prior beliefs concerning the network can be summarized as follows. The receptors are expected to have edges going only to ERKs and AKTs. This reflects known biology in which RTKs influencek these proteins (15, 16). The AKTs and ERKs are in turn expected to have edges going only to the downstream proteins ELF4E and EIF2B, and in the case of ERK only, additionally to MNK1; MNK1 is expected to have edges going only to ELF4E and EIF2B (17). Our prior beliefs concerning MKKs are few: we expect only that they should not have edges going directly to the receptors. We constructed a network prior corresponding to these beliefs using Eqs. 6 and 7. In addition, because of the small sample size, we used sparsity-promoting prior Eq. 8 with λindeg = 3. Following the prior elicitation strategy discussed above, we set λ = 3. As before, we used short diagnostic runs (T = 10,000) to check for convergence, followed by a single long run of T = 50,000 iterations, with a “burn-in” of 5,000 samples. A prior-based proposal was used, using Eq. 9 with λQ set (automatically) to max(1, ½exp(λ/2)) = 2.24. (The resulting acceptance rate was 0.23.)
Single Best Graph.
Fig. 3 shows the single most probable graph encountered during sampling. Each edge e is annotated with the corresponding posterior probability P(e | X). Note that some edges in the posterior mode have relatively low probability: this highlights the danger of relying on simple mode-finding rather than posterior simulation for inference in problems of this kind.
Network Features.
Eq. 3 provides a means by which to compute probabilities or posterior odds concerning network features. To take but one example in the present context, a biologically important question concerns the influence of MKK on ERK phosphorylation. We computed the posterior odds in favor of MKK → ERK connectivity (i.e., at least one edge from MKKs to ERKs) versus no such connectivity (no edge from MKKs to ERKs). The posterior odds in favor of MKK → ERK connectivity are 42, suggesting that MKK directly or indirectly influences ERK activation in the cell lines under study. Interestingly, the corresponding odds under the flat prior are just under 2. Although no prior information was provided concerning MKK → ERK connectivity specifically, network inferences of this kind are embedded within an overall graph embodying the joint distribution of all variables under study and therefore implicitly take account of specified prior beliefs, even when these concern other parts of the network.
Prior Sensitivity.
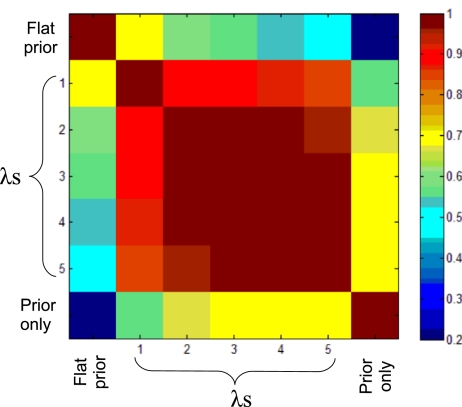
To investigate sensitivity to prior strength, we looked at the agreement between results obtained under different values of strength parameter λ. We considered five values of strength parameter λ, as well as a flat prior and samples drawn from the prior only (with λ = 3). This gave seven different prior settings, each of which led to a set of posterior edge probabilities. Fig. 4 shows Pearson correlations for these posterior edge probabilities, for all pairs of prior settings: values close to unity indicate posteriors that are effectively very close. Inferences using the informative prior with different values of λ are in very close agreement, yet differ from both the flat prior and from the informative prior alone. This gives us confidence that (i) inference integrates both data and prior information, and (ii) results are not too sensitive to the precise value of λ.
Fig. 4.
Prior sensitivity for protein data. Seven different prior settings (informative prior with strength parameter = 1, 2, 3, 4, 5; flat prior; and prior only) each give rise to a set of posterior edge probabilities. The image shows Pearson correlations between these posterior edge probabilities for all pairs of prior settings.
Discussion
In this article, we discussed the use of informative priors for network inference. In our view, informative network priors play two related roles. First, they allow us to capture valuable domain knowledge regarding network features. Second, they facilitate a refining or sharpening of questions of interest, in effect playing a role analogous to formulating an initial set of hypotheses but with much greater flexibility. Indeed, our investigation started out as a simpler, correlational analysis of components in cancer signaling. The complex nature of relationships between such components motivated us to move toward a multivariate approach, while the need to sharpen our questions in light of rich but uncertain biochemical knowledge motivated the work presented here on network priors. Our work forms part of a growing trend in the computational biology literature (including refs. 19–21) toward network inference schemes that take account of prior information of various kinds.
In many settings of interest, from molecular biology to the social sciences, relatively small sample sizes add to the challenge of network inference. This motivated us to focus our simulation experiments on the small-sample setting; we found that informative priors permit effective inference under these conditions, offering substantial gains over flat priors. We note also that the sample size of the protein phosphorylation data analyzed here was orders of magnitude smaller than in a previous application of Bayesian networks to protein signaling (18); this further motivated a need to make use of existing knowledge regarding the system.
We note that the network priors introduced here permit a prior preference for one graph over another even when both graphs imply the same conditional independence statements. For example, if we believe that a variable A is capable of physically influencing B, or that A precedes B in time, we may express a preference for A → B over B → A. In our experience, in the context of practical scientific inquiry, it can be useful to incorporate outside information of this kind.
Friedman and Koller (6) have proposed an interesting approach to network inference, in which samples are drawn from the space of orders, where an order ≺ is defined as a total order relation on vertices such that if Xi ∈ PaG(Xj) then i ≺ j. The appeal of this approach lies in the fact that the space of orders is much smaller than the space of graphs. On the other hand, the use of order space means that network priors must be translated into priors on orders, and inferences on graph features carried out via order space. Furthermore, the authors' own experiments show that sampling in order space offers no real advantage at smaller sample sizes.
There remains much to be done in extending the methods presented in this article to higher-dimensional problems. One approach to rendering such problems tractable would be to place strong priors on some parts of the overall network. This would, in effect, amount to using background knowledge to focus limited inferential power on the least well understood, or scientifically most interesting, parts of the system. Our current applied efforts are directed toward questions in cancer biology where we have found the ability to specify priors directly on networks and make posterior inferences on network features to be valuable in casting biologically interesting questions within a statistical framework.
Acknowledgments.
We thank Rich Neve, Paul Spellman, Laura Heiser, and other members of Joe Gray's laboratory at Lawrence Berkeley National Laboratory for a productive, ongoing collaboration and for providing the proteomic dataset used in this article. S.M. was supported by a Fulbright–AstraZeneca postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
The number Np of possible directed acyclic graphs with p vertices is given by the recurrence formula (see ref. 9 for details): Np=Σ pi=1(−1)i+1(pi)2i(p−i)N(p−i) with N1 = 1. This gives, e.g., N10 ≈ 4.2 × 1018 and N14 ≈ 1.4 × 1036.
Examples of knowledge pertaining to vertex classes are abundant in molecular biology, where the classes may represent distinct types of molecule thought to influence one another in specific ways.
An alternative is to consider the total number of edges in the graph; see ref. 8 for an example of this approach.
Specifically, it includes in its negative edge-set edges from Raf to MEK and from MEK to ERK, and in its positive edge-set an edge from Ras to ERK.
For diagnostic purposes, we first performed several short (T = 10,000) runs with different starting points. In each case, we found that monitored quantities converged within a few thousand iterations, giving us confidence in the results obtained by using the subsequent single, longer run.
This takes place via SH2-domain-containing molecules that are not analyzed here.
Indeed, given the exponential form of the prior, this represents a very wide range of strength regimes.
References
- 1.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. San Francisco: Morgan Kaufmann; 1988. [Google Scholar]
- 2.Lauritzen SL. Graphical Models. New York: Oxford Univ Press; 1996. [Google Scholar]
- 3.Jordan MI. Graphical models. Stat Sci. 2004;19:140–155. [Google Scholar]
- 4.Heckerman D, Geiger D, Chickering DM. Learning Bayesian networks: The combination of knowledge and statistical data. Mach Learn. 1995;20:197–243. [Google Scholar]
- 5.Madigan D, York J, Allard D. Bayesian graphical models for discrete data. Int Stat Rev. 1995;63:215–232. [Google Scholar]
- 6.Friedman N, Koller D. Being Bayesian about network structure. A Bayesian approach to structure discovery in Bayesian networks. Mach Learn. 2003;50:95–125. [Google Scholar]
- 7.Giudici P, Castelo R. Improving Markov chain Monte Carlo model search for data mining. Mach Learn. 2003;50:127–158. [Google Scholar]
- 8.Jones B, et al. Experiments in stochastic computation for high-dimensional graphical models. Stat Sci. 2005;20:388–400. [Google Scholar]
- 9.Robinson RW. In: New Directions in Graph Theory. Harary F, editor. New York: Academic; 1973. pp. 239–273. [Google Scholar]
- 10.Robert CP, Casella G. Monte Carlo Statistical Methods. Berlin: Springer; 2004. [Google Scholar]
- 11.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg RA. The Biology of Cancer. New York: Garland Science; 2007. [Google Scholar]
- 13.Jensen ST, Chen G, Stoeckert CJ., Jr Bayesian variable selection and data integration for biological regulatory networks. Ann Appl Stat. 2007;1:612–633. [Google Scholar]
- 14.Jeffreys H. The Theory of Probability. 3rd Ed. Oxford: Oxford Univ Press; 1961. [Google Scholar]
- 15.Heldin CH, Östman A, Rönnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim Biophys Acta. 1998;1378:F79–F113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- 16.Xiao GH, et al. Anti-apoptotic signaling by hepatocyte growth factor/Met via the phosphatidylinositol 3-kinase/Akt and mitogen-activated protein kinase pathways. Proc Natl Acad Sci USA. 2001;98:247–252. doi: 10.1073/pnas.011532898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonenberg N, Gingras AC. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 18.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 19.Tamada Y, et al. Estimating gene networks from gene expression data by combining Bayesian network model with promoter element detection. Bioinformatics. 2003;19:227–236. doi: 10.1093/bioinformatics/btg1082. [DOI] [PubMed] [Google Scholar]
- 20.Bernard A, Hartemink AJ. Proceedings of the Pacific Symposium on Biocomputing 2005. Singapore: World Scientific; 2005. Informative structure priors: Joint learning of dynamic regulatory networks from multiple types of data; pp. 459–470. [PubMed] [Google Scholar]
- 21.Werhli AV, Husmeier D. Reconstructing gene regulatory networks with Bayesian networks by combining expression data with multiple sources of prior knowledge. Stat Appl Genet Mol Biol. 2007;6:15. doi: 10.2202/1544-6115.1282. [DOI] [PubMed] [Google Scholar]