Abstract
The human fungal pathogen Histoplasma capsulatum grows in a sporulating filamentous form in the soil and, after inhalation of infectious spores, converts to a pathogenic yeast form inside host macrophages in response to temperature. Here we report the identification of two genes (RYP2 and RYP3) required for yeast-phase growth. Ryp2 and Ryp3 are homologous to each other and to the Velvet A family of regulatory proteins in Aspergillus species and other filamentous fungi. Wild-type H. capsulatum grows as filaments at room temperature and as yeast cells at 37°C, but ryp2 and ryp3 mutants constitutively grow as filaments independent of temperature. RYP2 and RYP3 transcripts accumulate to higher levels at 37°C than at room temperature. This differential expression is similar to the previously identified RYP1 transcript, which encodes a transcriptional regulator required for the yeast-phase expression program. Ryp1 associates with the upstream region of RYP2, and each of the three RYP genes is required for the differential expression of the others at 37°C. In addition to responding to the elevated temperature of the mammalian host, RYP2 and RYP3 are essential for viable spore production and regulation of sporulation at room temperature. This regulatory function is strikingly similar to the role of the Aspergillus Velvet A protein family in spore development in response to light, with the notable distinction that the H. capsulatum circuit responds to temperature.
Keywords: development, fungal pathogenesis, gene regulation, microbial pathogenesis, signal transduction
Developmental programs are triggered in response to environmental stimuli in a vast array of organisms. The systemic dimorphic fungal pathogens, which grow as filaments in the soil, initiate a dramatically different developmental program during colonization of mammalian hosts (1). An outstanding question in fungal evolution is how these pathogens, which include Histoplasma capsulatum, modified ancestral pathways to develop morphology and pathogenesis programs that foster disease. Here we describe the identification and characterization of two temperature-responsive regulators of morphology in H. capsulatum. Of note, these factors are homologous to a regulator in the filamentous fungus Aspergillus nidulans that controls cellular development in response to light (2).
H. capsulatum is a primary fungal pathogen endemic to the Mississippi and Ohio River Valleys. It grows in the soil as filamentous cells that produce vegetative spores. When the spores or filaments are aerosolized, the particles can be inhaled into the lungs of a mammalian host, where the fungal cells convert into a budding-yeast form that is able to survive and replicate within host macrophages. The ability of cells to adopt the yeast growth program is thought to be critical for H. capsulatum virulence (3, 4). Interestingly, the morphologic switch from the filamentous form to the yeast form can be recapitulated in culture by changing the temperature (room temperature growth triggers the soil form of the organism, whereas growth at 37°C triggers the host form of the organism). We recently identified Ryp1, a key transcriptional regulator required for yeast-phase growth at 37°C (5). Ryp1 is required for the vast majority of the yeast expression profile, but little else is known about the regulatory network that governs how temperature activates growth in the yeast phase.
In this study, we use a genetic screen to identify key regulators of morphological development in H. capsulatum. We identified two genes (RYP2 and RYP3) that are required for yeast-phase growth at 37°C. We show that both RYP2 and RYP3 transcripts are more highly expressed in yeast cells at 37°C than in filaments grown at room temperature. Furthermore, we show that regulation of RYP1, RYP2, and RYP3 is interdependent, suggesting that each is required for the expression and function of the others.
Notably, Ryp2 and Ryp3 are homologous to the Velvet A (VeA) family of regulators (which includes VeA, VelB, and VosA) from the filamentous fungus A. nidulans. Although Aspergillus species do not change their morphology in response to temperature, these fungi do exhibit VeA-dependent regulation of developmental programs, such as the production of spores, in response to environmental conditions such as light (6, 7). In addition, the VeA orthologs in the filamentous fungi Fusarium verticillioides, Acremonium chrysogenum and Neurospora crassa regulate hyphal morphology and spore formation (8–10). Taken together with this study, these data indicate that the Velvet proteins are conserved fungal factors that regulate distinct developmental pathways in response to diverse environmental stimuli.
Results
Identification of RYP2 and RYP3 as Regulators of Cell Fate.
A genetic screen was used to identify genes required for yeast-form growth at 37°C. Insertional mutagenesis was performed on wild-type H. capsulatum G217B by using A. tumefaciens-mediated transformation (11). Whereas wild-type cells growing in the yeast form give rise to shiny, smooth colonies, morphology mutants that are trapped in the filamentous form grow as dull, fuzzy colonies. Of the 40,000 insertion mutants assessed at 37°C, 15 constitutively filamentous mutants were identified and further analyzed. Inverse PCR and Southern analysis were used to identify two mutants [M9 and M15, supporting information (SI) Fig. S1] with independent insertions in the same ORF, which we named RYP2 (required for yeast-phase growth), and one mutant (F14, Fig. S1) with a 20-kb deleted region. The chromosomal deletion in F14 eliminated three ORFs, one of which was homologous to the RYP2 ORF, one of which had no significant BLAST homology, and one of which represented the remnants of a retrotransposon. We hypothesized that the RYP2 homolog might be responsible for the F14 mutant phenotype, and named the corresponding ORF RYP3.
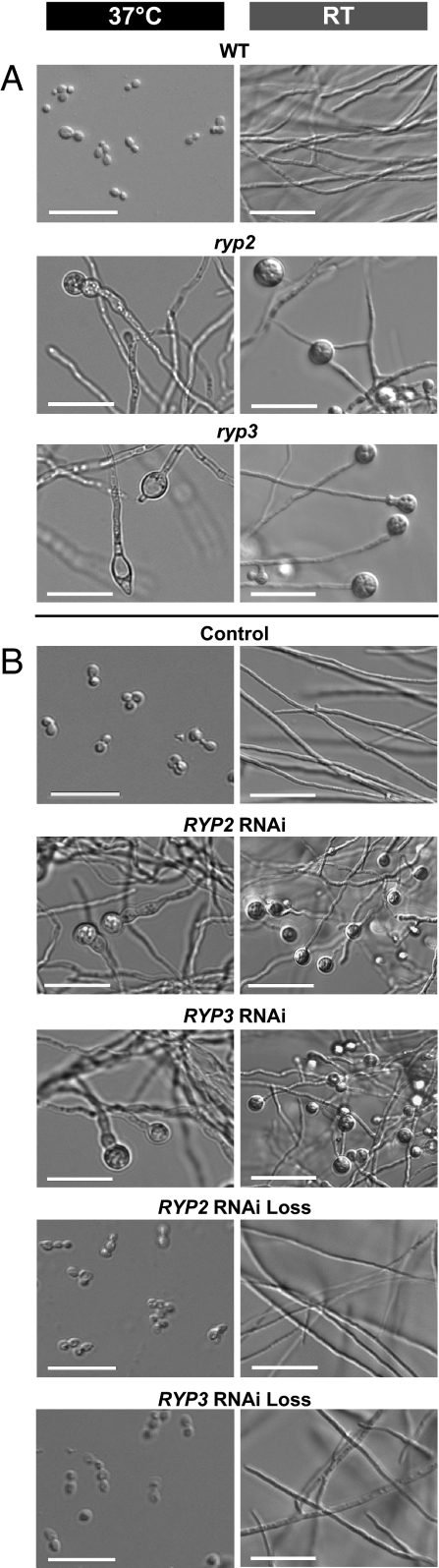
Microscopic analyses were performed to compare the morphology of wild-type and mutant cells at 37°C (Fig. 1A). Whereas wild-type cells grew in the yeast form, the ryp2 and ryp3 mutants failed to produce yeasts and instead grew as filaments, indicating that the mutants do not sense or respond appropriately to temperature. The mutant filaments displayed terminal bulbous structures of unknown significance. In addition to the prominent 37°C morphology defects, the ryp2 and ryp3 mutants also displayed a phenotype at room temperature. Whereas wild-type strains do not produce many spores under submerged, shaking culture conditions at room temperature, both the ryp2 and ryp3 mutants showed robust, inappropriate sporulation under these conditions (Fig. 1A). These phenotypes were reminiscent of the ryp1 mutant (5), and suggest that RYP2 and RYP3 regulate developmental processes at both room temperature and 37°C.
Fig. 1.
RYP2 and RYP3 are required for yeast phase growth at 37°C and regulating sporulation at room temperature. (A) Microscopic analysis of wild-type (G217B), a representative ryp2 insertion mutant (M9), and the ryp3 insertion mutant (F14) identified by the screen. The cells were grown in aerated liquid cultures at either 37°C or room temperature (RT). (B) Microscopic analysis of the control, RYP2, or RYP3 RNA interference (RNAi) strains, and RNAi loss strains that no longer contain the RNAi constructs, grown under the same conditions as in (A). Strains shown are representative of at least four independent isolates.
Because the original ryp3 insertion mutant contained a large chromosomal deletion (Fig. S1), it was critical to test whether the mutant phenotype was linked to absence of the RYP3 gene. No markers were available for complementation of the mutant phenotype, and we were unable to obtain gene disruptions of RYP2 or RYP3 due to the inefficient nature of this technology in H. capsulatum. We generated episomal plasmids that expressed RNA interference (RNAi) constructs for each gene (Fig. S2) and transformed these plasmids into H. capsulatum yeast cells. Targeted knockdown of RYP2 or RYP3 by RNAi recapitulated the mutant phenotypes (Fig. 1B). Upon loss of the episomal RNAi plasmids, the wild-type phenotype was restored (Fig. 1B).
Ryp2 and Ryp3 Are Homologous to VeA.
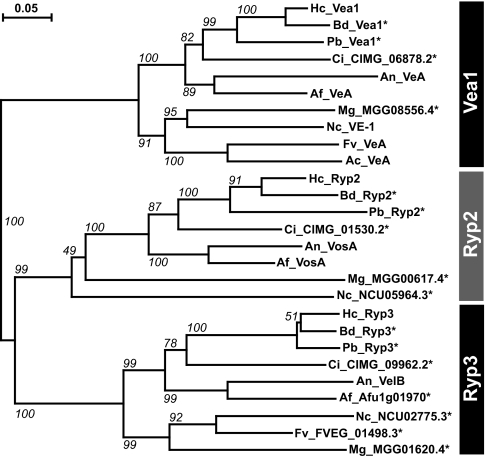
Ryp2 and Ryp3 are both homologous to Velvet A (VeA), a regulator of asexual and sexual development in A. nidulans (2). Specifically, Ryp2 is the ortholog of the VeA family member VosA [which regulates the production and viability of asexual spores (7)], and Ryp3 is orthologous to VelB [which is required for sexual spore formation (12)]. In addition to Ryp2 and Ryp3, H. capsulatum has a third homolog of unknown function, which we named Vea1 because it is most closely related to VeA. The three-member VeA family is highly conserved among other dimorphic and constitutively filamentous fungi (Fig. 2), but it is not present in strictly yeast-phase species such as Saccharomyces cerevisiae (2).
Fig. 2.
Ryp2 and Ryp3 are homologous to VeA and are part of a family of conserved proteins in both dimorphic and filamentous fungi. The phylogenetic tree shows the relationship of the three VeA homologs (Vea1, Ryp2, and Ryp3) in an array of fungal species [H. capsulatum (Hc), B. dermatititis (Bd), P. brasiliensis (Pb), C. immitis (Ci), A. nidulans (An), A. fumigatus (Af), M. grisea (Mg), N. crassa (Nc), F. verticillioides (Fv), and A. chrysogenum (Ac)]. Bootstrapping values for 1,000 iterations are shown. An * indicates predicted protein.
Because H. capsulatum genes tend to contain multiple small introns, it was necessary to sequence the Vea1, Ryp2, and Ryp3 cDNAs to determine the spliced coding sequences and predicted translation products (GenBank accession numbers EU543494–EU543496, Fig. S2). Protein alignments revealed three highly conserved blocks of amino acids along with regions of charged residues and proline-rich stretches (Fig. S3). Sequence motifs that would imply a particular biochemical function were not observed in Vea1, Ryp2, or Ryp3.
RYP2 and RYP3 Are Differentially Expressed in Response to Temperature.
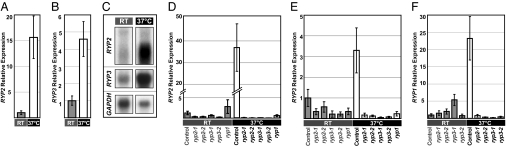
To determine whether the expression levels of RYP2 or RYP3 were regulated in response to temperature and/or cell morphology, we used quantitative reverse transcriptase PCR (qRTPCR) and Northern blot analyses to evaluate RYP2 and RYP3 transcripts in wild-type cells grown at either 37°C (yeast cells) or room temperature (filaments). Expression of RYP2 was ≈15-fold higher in wild-type yeast cells compared to filaments, whereas expression of RYP3 was ≈5-fold higher in yeast vs. filaments (Fig. 3 A and B). Nonetheless, both genes showed expression at room temperature, consistent with a role at this temperature in addition to 37°C (Fig. 3C).
Fig. 3.
RYP2 and RYP3 expression is regulated in response to temperature and by other RYP genes. All strains were grown in aerated conditions at either 37°C or RT as in Fig. 1. (A) and (B) Quantification of the relative expression of RYP2 and RYP3 in wild-type cells as measured by qRTPCR signal normalized to levels of TEF1. (C) Northern blot analysis of RYP2, RYP3, and GAPDH transcripts in wild-type cells. (D and E) Relative expression levels of RYP2 and RYP3 in the control strain, two independent RYP2 or RYP3 RNAi isolates, and the ryp1 mutant measured by qRTPCR signal normalized to levels of TEF1. (F) Relative RYP1 expression in the control and RYP2 or RYP3 RNAi isolates measured by qRTPCR signal normalized to levels of TEF1.
Expression of RYP Genes Is Interdependent.
We were interested in understanding the basis of the enriched expression of RYP2 and RYP3 at 37°C in yeast cells. RYP1 is a transcriptional regulator required for yeast-phase growth at 37°C in H. capsulatum (5). Because the ryp1 mutant strain displayed phenotypes similar to those of ryp2 and ryp3, we investigated whether RYP1 was required for differential expression of RYP2 and 3. In parallel, we analyzed the expression of each RYP gene in the absence of the others. The level of each RYP transcript was analyzed in the control strain, stably integrated RYP2 or RYP3 RNAi strains, and the ryp1 mutant strain (Fig. 3 D–F). Surprisingly, each RYP gene was required for the high level of expression of the other RYP genes at 37°C. This decreased RYP gene expression in each of the mutants could be a secondary consequence of their filamentous growth (Fig. 3 A–C, ref. 5). However, in that case, the expression level of the RYP genes in the mutants at 37°C would be equivalent to that of wild-type filaments. Instead, in each of the ryp mutants, the expression level of all of the RYP genes at 37°C was significantly lower than that of wild-type filaments (Table S1). For example, in the ryp2 mutant growing filamentously at 37°C RYP3 transcript levels were, on average, 7-fold lower than in wild-type filaments. Similarly, in the ryp3 mutant, levels of RYP2 transcript were ≈24-fold lower than in wild-type filaments. A similar, but less dramatic, trend was observed for the RYP1 transcript, which was 2–3-fold lower in the ryp2 and ryp3 mutants compared to wild-type filaments. These data imply that loss of RYP gene expression is not simply a secondary consequence of filamentous growth, and that regulation of each of the RYP genes is interdependent.
Ryp1 Associates with the RYP2 Promoter.
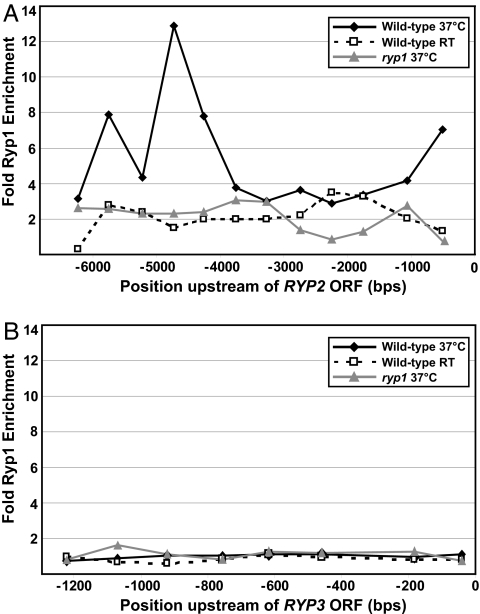
Because Ryp1 is a transcriptional regulator that associates with DNA (5), we tested whether it associates with the upstream regions of either RYP2 or RYP3. Using chromatin immunoprecipitation (ChIP) and quantitative PCR (qPCR) analysis, we compared the occupancy of Ryp1 at discrete locations upstream of the RYP2 or RYP3 ORF to the occupancy of Ryp1 at a reference control ORF. The entire intergenic region between either RYP2 or RYP3 and the most proximal upstream ORF was examined (Table S2). Interestingly, Ryp1 was associated with DNA upstream of the RYP2 ORF, but not upstream of the RYP3 ORF, at 37°C (Fig. 4). Enhanced association was observed at ≈500, 4,500, and 5,500 bp upstream of the RYP2 ATG; the long-range association events at −4500 and −5500 were reminiscent of the binding characteristics of Wor1, the Candida albicans Ryp1 ortholog (13, 14). Similar association events were obtained by using ChIP-on-chip studies aimed at determining all Ryp1 target loci (M. Gutierrez, M. Voorhies, and A. Sil, unpublished work). These results indicate that Ryp1 may directly regulate RYP2, but not RYP3, expression.
Fig. 4.
The transcriptional regulator Ryp1 associates with the region upstream of RYP2, but not RYP3. ChIP was performed with α-Ryp1 antibodies in wild-type cells grown at 37°C or at RT and in ryp1 cells grown at 37°C. Ryp1 ChIP enrichment was measured by qPCR at 500-bp intervals across the 6-kb intergenic region immediately upstream of the RYP2 ORF (A) or at 150-bp intervals across the 1.2-kb intergenic region immediately upstream of the RYP3 ORF (B). The enrichment values shown are for each position upstream of the ORFs relative to the reference gene TEF1.
Production of Viable Spores Requires RYP2 and RYP3.
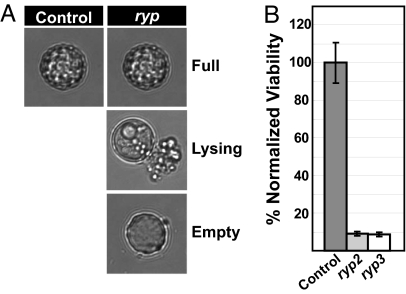
The observation that deregulated spore production occurs in ryp2 and ryp3 mutants at room temperature (Fig. 1) suggested that Ryp2 and Ryp3 may control sporulation. In addition to inhibiting spore production under submerged, shaking conditions, we wanted to determine whether Ryp2 and Ryp3 were required for the formation of normal spores under conditions that promote sporulation. Control and integrated RYP2 and RYP3 RNAi strains were allowed to produce spores under standard conditions. Spores from each strain were isolated and microscopic analysis revealed that the control strain produced both small smooth and larger tuberculate spores, whereas the ryp2 and ryp3 mutant strains predominately produced large tuberculate spores (data not shown). Although most of the control spores appeared full of cellular material, the majority of the mutant spores were in varying stages of lysis (Fig. 5A). The viability of the spores was tested at regular intervals for 7 days after harvesting. Both the ryp2 and ryp3 mutant spores displayed severe viability defects (at least a 10-fold decrease) when compared to control spores (Fig. 5B). These data indicate that, like A. nidulans VosA, Ryp2 and Ryp3 are required for the production of normal viable spores (7).
Fig. 5.
RYP2 and RYP3 are required for the production of viable spores. Cells carrying control, RYP2, or RYP3 RNAi constructs were grown on sporulation plates for at least 4 weeks at room temperature (RT). Spores were harvested in PBS and quantified. (A) Representative microscopic images of the spore populations observed from the control and ryp (ryp2 and ryp3) mutant strains. (B) The spores were stored at 4°C for 7 days and plated to calculate viable CFUs at Days 0, 1, 3, 5, and 7. The viability data were similar for the entire time course, and only the Day 0 time point is shown. The percent viable spores were calculated as described in Materials and Methods. The plot shown is representative of two independent experiments with two RYP2 or RYP3 RNAi isolates.
The Switch to Yeast-Form Growth in the Presence of Host Cells Requires RYP2 and RYP3.
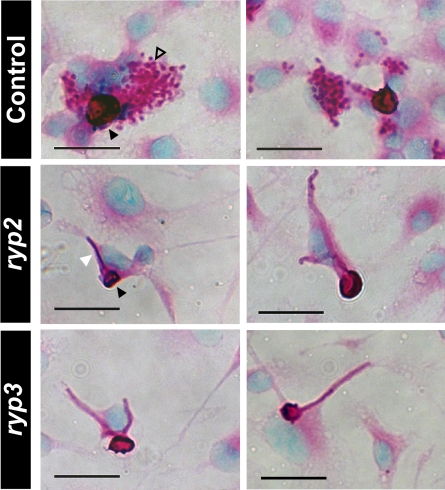
Temperature is a key signal that stimulates H. capsulatum yeast-form growth in the host, but the existence of other uncharacterized host-dependent signals has been suggested (15, 16). To determine whether host factors produced by macrophages could induce a morphologic change in the ryp mutants, we infected murine bone marrow derived macrophages with spores from control or the integrated RYP2 or RYP3 RNAi strains and observed them over a period of 72 h. Although the majority of ryp2 and ryp3 spores were inviable and did not germinate, it was possible to assess and quantify the cellular morphology of 150 germinating spores associated with macrophages for each strain. As expected, the control spores predominantly germinated to give rise to yeast cells (71 ± 3%) when associated with macrophages (Fig. 6), whereas the remainder gave rise to filaments. In the ryp2- or ryp3-infected macrophage populations, spores germinated to give rise only to filaments, and yeast cells were never observed (Fig. 6). These data suggest that the ability of macrophages to promote yeast-phase growth is dependent on RYP2 and RYP3.
Fig. 6.
Induction of yeast-phase growth in macrophages depends on RYP2 and RYP3. Bone marrow-derived macrophages were infected with spores from the control strain, RYP2, or RYP3 RNAi isolates at an MOI of 0.1 for 72 h and stained at the 48-h timepoint with PAS and methyl green. Two representative images are shown for each infection. The infections were performed in triplicate with 2 independent RYP2 or RYP3 RNAi isolates. Black arrowheads point to representative spores, the open arrowhead points to a cluster of yeast cells, and the white arrowhead points to a representative filament.
Discussion
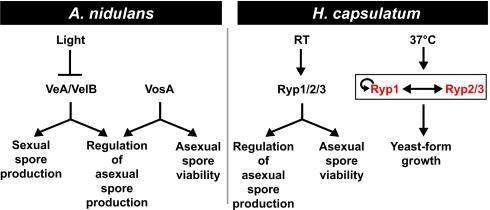
Functional genomic comparisons and molecular studies have revealed that transcriptional networks can undergo dramatic rewiring during evolution (17–22). An outstanding question in fungal biology is how the systemic dimorphic fungal pathogens, which initiate a host-colonization program in response to temperature, have evolved temperature-dependent regulatory circuits from the ancestral state (23). Here we describe the identification of two conserved fungal proteins, Ryp2 and Ryp3, which are required for H. capsulatum to grow in the parasitic yeast form in response to host temperature. In the ascomycete lineage, Ryp2 and Ryp3 are highly conserved in systemic dimorphic and filamentous fungi (Fig. 2), but clear homologs are not present in fungi that grow exclusively as yeast, such as the Saccharomycetales (2). In filamentous fungi, homologs of Ryp2 and Ryp3 control spore formation and viability (2). Interestingly, we also show that in addition to their role in yeast-form growth, Ryp2 and Ryp3 are required for appropriate spore production and viability during H. capsulatum filamentous growth. These data indicate that Ryp2 and Ryp3 have distinct functions under different environmental conditions. Furthermore, it is likely that the systemic dimorphic fungi have adapted a conserved module that controls sporulation in filamentous fungi for use in regulation of yeast-phase growth at 37°C (Fig. 7).
Fig. 7.
Regulatory diagram comparing homologous developmental regulators in A. nidulans and H. capsulatum. Although the inputs (light and temperature) into these regulatory pathways are different, the outputs are remarkably similar (regulation of cellular development). Light inhibits A. nidulans VeA accumulation in the nucleus, resulting in the inhibition of sexual spore development and the production of asexual spores. VeA has been shown to bind VelB (the Ryp3 ortholog), and both are required for sexual spore formation in the absence of light. VosA, the Ryp2 ortholog, is required to produce viable asexual spores and to inhibit inappropriate asexual sporulation. In H. capsulatum, Ryp2, Ryp3, and the transcriptional regulator Ryp1 function at room temperature (RT) to produce viable asexual spores and to inhibit inappropriate asexual sporulation. At 37°C, each of these Ryp transcripts accumulates to higher levels than at RT, as indicated by the red color. This transcript accumulation is interdependent, as described in the text and indicated by the bidirectional arrow. In addition, Ryp1 associates with the Ryp2 promoter, implying that Ryp1 directly regulates Ryp2 accumulation. We showed previously that Ryp1 associates with its own promoter at 37°C, suggesting the presence of a positive feedback loop as indicated by the circular arrow (5).
Molecular studies in filamentous fungi including Aspergillus, Fusarium, and Neurospora have defined the roles of Ryp2 and Ryp3 homologs in the regulation of cellular development (2, 6–10, 12). These proteins are members of the Velvet A family, which includes VeA, VelB, and VosA. The role of VeA and VelB in spore and hyphal development has been best characterized in A. nidulans, in which they are required to promote sexual spore development as well as to inhibit the formation of asexual spore structures (Fig. 7) (2, 12). A. nidulans VosA has been shown to be required for appropriate asexual sporulation and spore viability (Fig. 7) (7). The first breakthrough regarding the biochemical functions of these proteins comes from a recent study showing that VeA binds VelB as well as the putative methyl transferase LaeA, which is thought to regulate gene expression (12). Our data are consistent with an analogous role for Ryp2 and Ryp3 in the regulation of yeast-phase and spore transcriptional programs.
We observed that Ryp2 and Ryp3 are required for the differential expression of the transcriptional regulator Ryp1 at 37°C. Ryp1 is required for the majority of the yeast-phase specific gene expression program (5), including the higher levels of expression of Ryp2 and Ryp3 at 37°C. Indeed, we determined that the enhanced expression of all three RYP genes at 37°C is interdependent: When accumulation of any single RYP transcript is disrupted, the other two RYP transcripts are no longer highly expressed. These data may help explain the apparent lack of functional redundancy between RYP2 and RYP3, as disruption of either results in failure to transcribe both genes. Interestingly, Ryp1 associates with the upstream regulatory region of RYP2 but not RYP3, suggesting that Ryp1 may directly regulate RYP2, but not RYP3. Taken together, these data suggest that Ryp1, 2, and 3 could function in a complex to regulate the transcription of yeast-specific genes, analogous to the complex formed by VeA, VelB, and LaeA. In addition, ryp2 and ryp3 mutants undergo inappropriate sporulation, much like that observed with ryp1 mutants (5), suggesting that the three Ryp proteins may share a role in regulation of sporulation at room temperature. Of note, although Ryp1 is conserved across all fungal orders, Ryp2 and Ryp3 are found only in the filamentous and systemic dimorphic fungi, indicating that transcriptional regulation by Ryp1 orthologs in other fungi, such as the C. albicans Wor1, does not involve proteins homologous to Ryp2 or Ryp3.
Because RYP1 is required for expression of virulence genes, and because RYP2 and RYP3 are required for Ryp1 accumulation at 37°C, it is likely that RYP2 and RYP3 are also required for the expression of virulence genes. It would be interesting to directly assess the role of RYP2 and RYP3 in virulence. However, conventional mouse models of Histoplasma pathogenesis require infection with discrete fungal particles, such as spores or yeast cells, but not filaments. Mutants lacking ryp2 or ryp3 cannot grow in the yeast form, and the severe viability defect of ryp2 and ryp3 mutant spores made it impossible to use these cells in a mouse infection. Future development of conditional alleles of RYP2 and RYP3 will help determine the role of these genes in pathogenesis.
Temperature is the best characterized signal known to stimulate yeast-form growth in H. capsulatum. The existence of host-specific signals other than high temperature is implied by the observation that conversion of H. capsulatum cells to the yeast form occurs faster within host cells than in culture (24). Because phagocytosis of ryp2 or ryp3 mutant spores by macrophages fails to trigger yeast-form growth, either ryp2 and ryp3 mutants are refractory to temperature-independent host signals, or these signals are not sufficient to shift morphology in the absence of the temperature- and RYP-dependent program. In sum, these data indicate that Ryp2 and Ryp3 are essential regulators of yeast-form growth. In addition, the requirement of these regulators to produce viable asexual spores, which are thought to be the most common infectious particle during the natural course of infection, highlights the important role of these regulators in H. capsulatum biology and pathogenesis.
Materials and Methods
A complete description of all materials and methods can be found in the SI Text. H. capsulatum G217B (ATCC 26032) and G217Bura5Δ (WU15), both generously provided by William Goldman (Washington University, St. Louis, MO), were grown in the yeast or mycelial form by changing the temperature of the culture environment.
Yeast-Form Cultures.
G217B and G217Bura were grown in Histoplasma macrophage medium (HMM) liquid or on HMM agarose plates (25). G217Bura5Δ cultures were supplemented with 0.2 mg/ml uracil (Sigma–Aldrich). Broth cultures were grown at 37°C with 5% CO2 on an orbital shaker at 120–150 rpm. Plates were grown in a humidified incubator at 37°C with 5% CO2. Cells were thawed fresh from frozen stock and passaged on HMM agarose plates every 1–2 weeks for up to 2 months.
Mycelial-Form Cultures.
H. capsulatum yeast cultures were converted into filamentous mycelial cultures by inoculating Sabouraud Dextrose (Difco) agar plates with yeast cells and growing at room temperature (22–26°C) for at least 2 weeks. The mycelial cells were then inoculated into liquid HMM broth and grown for 2–4 weeks at room temperature on an orbital shaker at 120 rpm to establish robust mycelial cultures. Cultures were passaged every 1–2 weeks at 1:20 dilution. ryp2, ryp3, and ryp1 mutant strains were grown on either HMM or 3M (26) agar plates in a humidified incubator at 37°C. Liquid cultures of the filamentous mutants were grown in either HMM broth or HMM supplemented with 200 μg/ml hygromycin B (Roche) at 37°C with 5% CO2 on an orbital shaker at 120 rpm. Cultures were passaged every 1–2 weeks at 1:20 dilution.
A. tumefaciens-mediated transformation was used to generate insertion mutants as previously described (11). RNAi (27), RNA isolation, Northern analysis, qRTPCR and chromatin immunoprecipitation were performed as previously described (5). Standard sporulation conditions were used to produce spores from control and mutant RNAi strains. Spores were harvested in PBS and inoculated onto BHI plus 10% sheep's blood agar plates at regular intervals over the 7-day time course. Macrophages were infected with control or RNAi mutant spores at a multiplicity of infection (MOI) of 0.1. Cells were fixed and stained at 48 and 72 h post infection.
Supplementary Material
Acknowledgments.
We thank Van Nguyen and Diane Inglis for invaluable experimental assistance and advice; Mark Voorhies for exceptional assistance with sequence analysis; William Goldman (Washington University, St. Louis, MO), Chad Rappleye (Ohio State University, Columbus, OH), Bruce Klein (University of Wisconsin, Madison, WI), Thomas Sullivan (University of Wisconsin, Madison, WI), and Paul Hooykaas (Leiden University, Leiden, The Netherlands) for the generous gifts of strains, plasmids, and/or protocols; and Jeff Cox, Hiten Madhani, Rebecca Zordan, Amy Kistler, Aaron Hernday, and members of the A.S. laboratory for helpful comments on the manuscript. This work was supported by the National Institutes of Health Grants 5T32AI060537-04 (to R.H.W.) and R01AI066224 and PO1AI063302 (to A.S.); the Sandler Program in Basic Sciences; and Howard Hughes Medical Institute Biomedical Research Support Program Grant 5300246 (to the University of California School of Medicine, San Francisco).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.M. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EU543495 (RYP2), EU543496 (RYP3), and EU543494 (VEA1)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0806221105/DCSupplemental.
References
- 1.Holbrook ED, Rappleye CA. Histoplasma capsulatum pathogenesis: Making a lifestyle switch. Curr Opin Microbiol. 2008;11:318–324. doi: 10.1016/j.mib.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Calvo AM. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet and Biol. 2008;45:1053–1061. doi: 10.1016/j.fgb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Medoff G, et al. Irreversible block of the mycelial to yeast phase transition of H capsulatum. Science. 1986;231:476–479. doi: 10.1126/science.3001938. [DOI] [PubMed] [Google Scholar]
- 4.Nemecek JC, Wuthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci USA. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HS, et al. The. veA gene activates sexual development in Aspergillus nidulans. Fungal Genet and Biol. 2002;37:72–80. doi: 10.1016/s1087-1845(02)00029-4. [DOI] [PubMed] [Google Scholar]
- 7.Ni M, Yu JH. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One. 2007;2:e970. doi: 10.1371/journal.pone.0000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayram O, Krappmann S, Seiler S, Vogt N, Braus GH. Neurospora crassa ve-1 affects asexual conidiation. Fungal Genet and Biol. 2008;45:127–138. doi: 10.1016/j.fgb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Dreyer J, Eichhorn H, Friedlin E, Kurnsteiner H, Kuck U. A homologue of the Aspergillus velvet gene regulates both cephalosporin C biosynthesis and hyphal fragmentation in Acremonium chrysogenum. Appl Environ Microbiol. 2007;73:3412–3422. doi: 10.1128/AEM.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, et al. FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides. Mol Microbiol. 2006;62:1418–1432. doi: 10.1111/j.1365-2958.2006.05447.x. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan TD, Rooney PJ, Klein BS. Agrobacterium tumefaciens integrates transfer DNA into single chromosomal sites of dimorphic fungi and yields homokaryotic progeny from multinucleate yeast. Eukaryot Cell. 2002;1:895–905. doi: 10.1128/EC.1.6.895-905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayram O, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 13.Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci USA. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard DH. Observations on tissue cultures of mouse peritoneal exudates inoculated with Histoplasma capsulatum. J Bacteriol. 1959;78:69–78. doi: 10.1128/jb.78.1.69-78.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pine L, Webster RE. Conversion in strains of Histoplasma capsulatum. J Bacteriol. 1962;83:149–157. doi: 10.1128/jb.83.1.149-157.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: A case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 18.Tuch BB, Li H, Johnson AD. Evolution of eukaryotic transcription circuits. Science. 2008;319:1797–1799. doi: 10.1126/science.1152398. [DOI] [PubMed] [Google Scholar]
- 19.Martchenko M, Levitin A, Whiteway M. Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryot Cell. 2007;6:291–301. doi: 10.1128/EC.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rokas A. Evolution: Different paths to the same end. Nature. 2006;443:401–402. doi: 10.1038/443401a. [DOI] [PubMed] [Google Scholar]
- 21.Ihmels J, Bergmann S, Berman J, Barkai Comparative gene expression analysis by differential clustering approach: Application to the Candida albicans transcription program. PLoS Genet. 2005;1:e39. doi: 10.1371/journal.pgen.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raijman D, Shamir R, Tanay Evolution and selection in yeast promoters: Analyzing the combined effect of diverse transcription factor binding sites. PLoS Comput Biol. 2008;4:e7. doi: 10.1371/journal.pcbi.0040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot Cell. 2007;6:2169–2174. doi: 10.1128/EC.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hempel H, Goodman NL. Rapid conversion of Histoplasma capsulatum, Blastomyces dermatitidis, and Sporothrix schenckii in tissue culture. J Clin Microbiol. 1975;1:420–424. doi: 10.1128/jcm.1.5.420-424.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon-Chung KJ. Emmonsiella capsulata: Perfect state of Histoplasma capsulatum. Science. 1972;177:368–369. doi: 10.1126/science.177.4046.368. [DOI] [PubMed] [Google Scholar]
- 26.Worsham PL, Goldman WE. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol. 1988;26:137–143. [PubMed] [Google Scholar]
- 27.Marion CL, Rappleye CA, Engle JT, Goldman WE. An alpha-(1,4)-amylase is essential for alpha-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol Microbiol. 2006;62:970–983. doi: 10.1111/j.1365-2958.2006.05436.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.