Abstract
Very-long-chain fatty acids (VLCFAs) are synthesized as acyl-CoAs by the endoplasmic reticulum-localized elongase multiprotein complex. Two Arabidopsis genes are putative homologues of the recently identified yeast 3-hydroxy-acyl-CoA dehydratase (PHS1), the third enzyme of the elongase complex. We showed that Arabidopsis PASTICCINO2 (PAS2) was able to restore phs1 cytokinesis defects and sphingolipid long chain base overaccumulation. Conversely, the expression of PHS1 was able to complement the developmental defects and the accumulation of long chain bases of the pas2–1 mutant. The pas2–1 mutant was characterized by a general reduction of VLCFA pools in seed storage triacylglycerols, cuticular waxes, and complex sphingolipids. Most strikingly, the defective elongation cycle resulted in the accumulation of 3-hydroxy-acyl-CoA intermediates, indicating premature termination of fatty acid elongation and confirming the role of PAS2 in this process. We demonstrated by in vivo bimolecular fluorescence complementation that PAS2 was specifically associated in the endoplasmic reticulum with the enoyl-CoA reductase CER10, the fourth enzyme of the elongase complex. Finally, complete loss of PAS2 function is embryo lethal, and the ectopic expression of PHS1 led to enhanced levels of VLCFAs associated with severe developmental defects. Altogether these results demonstrate that the plant 3-hydroxy-acyl-CoA dehydratase PASTICCINO2 is an essential and limiting enzyme in VLCFA synthesis but also that PAS2-derived VLCFA homeostasis is required for specific developmental processes.
Keywords: cuticular wax, elongase, sphingolipid, triacylglycerol, leaf development
Very-long-chain fatty acids (VLCFAs) are components of eukaryotic cells and are composed of 20 or more carbons (i.e., >C18). VLCFAs are involved in many different physiological functions in different organisms. They are abundant constituents of some tissues like the brain (myelin) or plant seeds (storage triacylglycerols). VLCFAs are components of the lipid barrier of the skin and the plant cuticular waxes (1). VLCFAs are also involved in the secretory pathway for protein trafficking and for the synthesis of GPI lipid anchor (2). Finally, VLCFAs are components of sphingolipids that are both membrane constituents and signaling molecules (3).
In yeast, VLCFA synthesis is catalyzed in the endoplasmic reticulum (ER) by a membrane-bound multienzyme protein complex referred as the elongase (4). The elongase complex catalyzes the cyclic addition of a C2-moiety obtained from malonyl-CoA to an acyl-CoA. VLCFAs (C20, C22, C24, or higher) are produced from shorter fatty acids (usually C16 or C18) made by the cytosolic fatty acid synthase complex. The two-carbon addition during the elongation cycle requires four independent but sequential enzymatic steps. The first step involves the condensation of the malonyl-CoA with an acyln-CoA precursor, resulting in a 3-ketoacyl-CoA intermediate, which is reduced to form a 3-hydroxy-acyl-CoA. The third enzymatic step is the dehydration of the 3-hydroxy-acyl-CoA to an enoyl-CoA, which is finally reduced to yield an acyln + 2-CoA. The keto and enoyl reductases are encoded respectively by the YBR159w and TSC13/YDL015c genes; the condensing enzymes are coded by a small family of genes, ELO1, 2, and 3, of which ELO2/FEN1/YCR034w and ELO3/SUR4/YLR372w have been shown to be required for the synthesis of VLCFAs (5). Identification of the dehydratase remained elusive until the recent identification of YJL097w/PHS1 as encoding this activity (although a role in sphingolipid biosynthesis had previously been inferred from the biochemical phenotype of phs1 mutant that accumulated the long chain base phytosphingosine [PHS]) (6). The phs1 mutant was also characterized as a cell cycle mutant defective in G2/M phase (7). Ultimate confirmation of the biochemical function of Phs1p as the elongase dehydratase was provided by in vitro activity of recombinant protein and reconstitution of the elongase complex in proteoliposomes (8). Very recently, topology experiments demonstrated that Phs1p has six transmembrane domains with its N- and C-termini in the cytosol and that two conserved amino acids, Y149 and E152, were critical for its activity (9).
In plants, there is a large family of 3-ketoacyl-CoA synthases (KCS) condensing enzymes exemplified by the Arabidopsis gene Fatty Acid Elongase 1 (FAE1), required in seeds for the synthesis of C20+ fatty acids (e.g. erucic acid). The Arabidopsis genome encodes 21 FAE-like KCSs, and although these enzymes are structurally unrelated to the ELO class of condensing enzymes, it has been demonstrated that several Arabidopsis FAE-KCSs can rescue the otherwise lethal yeast elo2Δ/elo3Δ double mutant (10, 11). The Arabidopsis CER10 protein shows significant homology with yeast enoyl-CoA reductase Tsc13p, because it rescues the temperature-sensitive lethality of the tsc13–1 yeast mutant (12) and is involved in VLCFA synthesis (13). The Arabidopsis genome also contains a gene that shares significant homology with the yeast 3-ketoreductase YBR159w, and although it has been demonstrated that this Arabidopsis activity can rescue biochemical phenotypes of the ybr159Δ mutant, the in vivo role of the plant gene remains to be determined (14). The yeast dehydratase PHS1/YJL097w gene also shares significant similarity with the Arabidopsis PASTICCINO2 (PAS2) gene, which was shown to rescue phs1Δ lethality (15). Mutations in the PAS2 gene lead to strong developmental defects associated with ectopic cell division, cell differentiation, and hormonal responses (15–18). Finally, PAS2 was demonstrated to be able to interact with phosphorylated cyclin-dependent kinase (CDK) and subsequently to prevent its dephosphorylation by CDC25-like phosphatase(s), preventing premature entry in mitosis (19). However, the fact that PAS2 shows significant sequence similarity with Phs1p and that it was able to rescue phs1 mutant argues in favor of PAS2 being the dehydratase component of the microsomal elongase complex. Here, we show by reciprocal complementation experiments that PAS2 and PHS1 are functionally exchangeable. The pas2 mutant was also characterized by global reduction of VLCFAs and by the specific accumulation of 3-hydroxyacyl-CoA substrate of the dehydratase. Moreover, PAS2 was found to be associated with ER and to physically interact with the reductase CER10. Collectively our results demonstrate that PAS2 is required for the synthesis of VLCFAs and that these fatty acids are indispensable components of several specific lipid classes.
Results
PASTICCINO2 is the Arabidopsis Orthologue of Yeast PHS1.
PAS2 shares 35% identity with a similar protein in Arabidopsis coded by the gene At5g59770, and both proteins showed a similar degree of similarities with Phs1p (33% and 32% identity, respectively, for PAS2 and At5g59770 coding protein). However, At5g59770 was not able to suppress the lethality of phs1 [supporting information (SI) Fig. S1]. Therefore, PAS2 represents a bona fide functional orthologue of PHS1 in Arabidopsis. However, it was still not clear whether this complementation by PAS2 was associated with the rescue of both cell division and lipid defects present in phs1. To address these questions, we used a phs1 haploid strain expressing the inducible pGAL1:PAS2 construct (designated D1B), which was able to grow only on galactose- and not on glucose-supplemented medium (Fig. S1) (15). When grown on galactose, D1B cells showed wild-type phenotype. Conversely, cells grown on glucose presented the characteristic phs1 multibudded or large-budded phenotypes (Fig. 1A). Likewise, galactose-grown D1B cells displayed a restoration of the normal low levels of free (i.e., nonacylated) long chain bases (LCBs) that accumulated in phs1 mutant (Fig. 1B and Fig. S2). The expression of the Arabidopsis PAS2 gene is thus able to rescue both cell division and lipid defects of the yeast phs1 mutant.
Fig. 1.
Arabidopsis PAS2 and yeast PHS1 are functionally exchangeable. (A) Induction of PAS2 expression in haploid cells carrying mutated phs1 allele and pGAL:PAS2 construct (D1B in presence of galactose [Gal]) complemented phs1 phenotype (D1B in presence of glucose [Glc]). Wild-type PHS1 (D1A) cells were grown on Gal- or Glc-supplemented medium. (B) Expression of PAS2 (D1B Gal) prevents free t18:0 and d18:0 accumulation observed in phs1 (D1B glc). (C) Expression of 35S:PHS1 complemented pas2–1 developmental defects. (Scale bar, 1 mm.) (D) Mutation in PAS2 gene led to LCB and phosphorylated LCB (LCB-P) accumulation that was prevented by the expression of PHS1. LCB values are the average of three samples ± SD.
One scenario we considered was that the plant PAS2 function might still have evolved, compared with its yeast orthologue, by the acquisition of new biochemical functions such as the regulation of cell cycle proteins (15, 19). We thus investigated whether the yeast PHS1 gene could complement the developmental phenotype of the plant pas2 mutant. PHS1 was therefore expressed in the heterozygous pas2/+ plant under the control of the constitutive 35S promoter (Fig. S3). Segregation of pas2/+ plants carrying 35S:PHS1 construct showed that the phenotype of pas2 mutants expressing PHS1 reverted from small dwarf seedlings with fused leaves to plants almost indistinguishable from wild type (Fig. 1C). PHS1 complementation of pas2 phenotype was also observed at the levels of free LCBs that accumulated in pas2 and was almost completely restored to wild-type levels by expression of PHS1 (Fig. 1D and Fig. S2). In conclusion, yeast PHS1 is able to complement pas2 developmental defects and LCB accumulation, demonstrating that both proteins are exchangeable and that PAS2 represents the true PHS1 orthologue. More importantly, PHS1 complementation indicates that pas2 developmental defects are most likely caused by a defective dehydratase activity in the VLCFA microsomal elongase complex.
pas2–1 Mutant Is Characterized by Low VLCFAs.
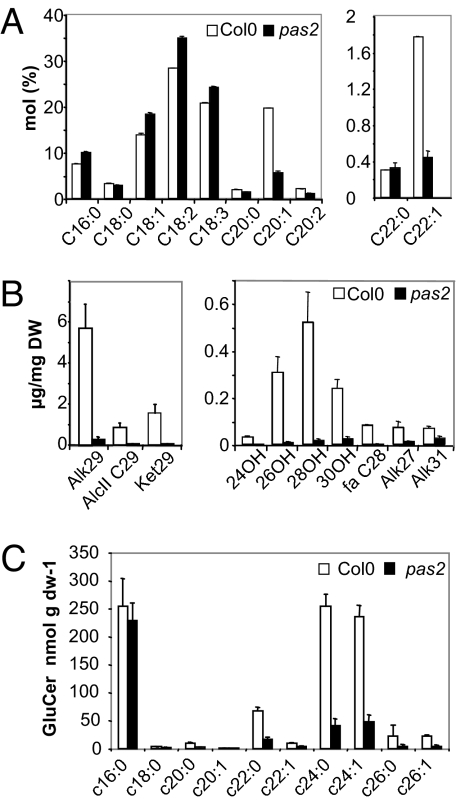
In plants, VLCFAs are involved in several classes of complex lipids, such as seed storage triacylglycerols, cuticular waxes, and sphingolipids, but could also be found in the phospholipid fraction (20). Analysis of VLCFAs in seeds showed that pas2 seeds accumulated lower levels of 22:1 and 20:1 and higher levels of short chains 16:0, 18:1, 18:2, and 18:3 compared with wild type (Fig. 2A). Wax deposition was modified in pas2 mutant stems with a lower density of wax crystals, which were mainly of globule shapes instead of flakes (Fig. S4). This result was consistent with the observation in pas2 of postgenital organ fusion characteristic of cuticule mutants (15). Accordingly, all of the aliphatic components of cuticular waxes were dramatically reduced in the pas2 mutant compared with wild type in both stems and leaves (Fig. 2B and Fig. S5). Finally, VLCFA content from complex sphingolipids was lower in pas2 mutant compared with wild type (Fig. 2C and Fig. S6). The glucosylceramide pool showed an almost complete absence of VLCFAs, whereas in contrast, the glycosyl-inositol-phosphoceramide (GIPC) pool showed only small VLCFA reduction. Almost no difference could be observed in the simple sphingolipid fractions except for 16:0 ceramide and hydroxy-ceramide levels, which were increased in pas2 mutant (Fig. S6). Thus, in agreement with the PAS2 role in fatty acid elongation, three lipid fractions of Arabidopsis that contain VLCFAs were perturbed in the pas2 mutant.
Fig. 2.
pas2–1 mutation leads to a general reduction of VLCFA levels. (A) Total fatty acids from pas2–1 and wild-type seeds were transmethylated and quantified by gas chromatography using an internal standard. (B) Cuticular wax loads on stems of wild type (Col0, white bars) and pas2–1 (black bars). Wax load was analyzed according to fatty acid chain length and side group, that is, primary alcohols (24OH, 26OH, 28OH, 30OH), fatty acid (fa C28), alkanes (Alk27, Alk29, Alk31), secondary alcohol (AlcII C29), and ketone (Ket29). (C) Fatty acyl chain length composition of the glucosylceramide (GluCer) fraction from wild type (Col0, white bars) and pas2–1 (black bars). Fatty acid values are the average of three samples ± SD.
PAS2 Is Involved in 3-hydroxy acyl-CoA Dehydration During VLCFA Elongation.
In yeast, the microsomal elongase complex is localized in the ER, as demonstrated by biochemical and subcellular data (4). In plants, the elongase synthases and the two reductases were also found to be ER-associated proteins (21). Subcellular distribution of PAS2 was thus monitored by expressing the construct pPAS2:PAS2-GFP in both Nicotiana benthamiana and Arabidopsis epidermal cells (22). PAS2-GFP protein fusion was mainly associated with the ER and colocalized with CER10-mRFP1 (Fig. S7 A and B).
Next, we investigated whether PAS2 was directly associated with enzymes of the VLCFA elongase complex by analyzing the interaction between PAS2 and ECR/CER10. Protein-protein interactions were tested in vivo in Arabidopsis by bimolecular fluorescence complementation (BiFC). Transient expression of YFPN-PAS2 and CER10-YFPC under the 35S promoter led to a YFP signal (Fig. S7C). Because the interaction takes place in a closed endomembrane compartment and involved membrane associated proteins, we evaluated the specificity of BiFC assay by using the ER-localized LCB lyase AtDPL1 (23). No YFP signal could be detected between YFPN-PAS2 and AtDPL1-YFPC or between YFPN-AtDPL1 and CER10-YFPC, demonstrating that PAS2 and ECR/CER10 were involved in a genuine interaction.
Finally, if PAS2 is involved like Phs1p in the dehydratase step of the microsomal elongase complex, it should catalyze the conversion of 3-hydroxy-acyl-CoA into an enoyl-CoA form. Consequently, mutations in PAS2 would be predicted to lead to an accumulation of 3-hydroxy-acyl-CoA as intermediates of the elongation cycle. Although no significant quantitative differences among nonhydroxylated acyl-CoAs from 14:0 to 22:1 could be detected between pas2 mutant and wild type (Fig. 3 and Table S1), three additional peaks present in the pas2 profile were identified by MS analysis as C18:0, C20:0, and C22:0 3-hydroxy-acyl-CoAs (Fig. 3 and Fig. S8). No additional peaks could be observed in the complemented pas2–1, 35S:PHS1 line, confirming that the presence 3-hydroxy-acyl-CoAs was caused by a defective dehydratase activity (Fig. S9). These data are not only in agreement with the proposed role of PAS2 as the microsomal elongase dehydratase but also report the presence of intermediates of the elongation cycle in the acyl-CoA pool. Interestingly, the presence of 3-hydroxy-C18:0-CoA in dehydratase-deficient mutants strongly suggests that acyl-CoA with an acyl chain as short as C16 could be a substrate of the Arabidopsis elongase complex.
Fig. 3.
The pas2 mutant accumulates 3-hydroxy Acyl-CoA intermediates. The composition the acyl-CoA pool was determined by extraction, derivatization, and HPLC analysis of acyl-etheno-CoAs. MS confirmed the presence of 3- hydroxy acyl-CoA intermediates in the pas2 mutant (open diamond, open triangle, filled star). The internal standard (Istd) is 17:0 acyl-CoA.
Acyl-CoA Dehydratase Is Essential for Plant Development and Is Limiting for VLCFA Synthesis.
The pas2–1 and pas2–2pep mutations were characterized as leaky alleles because both mutants were still able to accumulate low levels of wild-type transcript (15, 17). We therefore checked the effect of the Salk T-DNA insertion line N617051, which contains an insertion in the 5′ UTR, 174 bases before the initiating methionine of PAS2. Heterozygous plants for this T-DNA insertion mutant were genotyped (Fig. S10), and no homozygous plants could be identified in the progeny. Siliques of heterozygous plants contained about 25% of aborted seeds, suggesting that homozygous pas2–3 mutants were embryo lethal and could not be recovered. (Fig. 4A).
Fig. 4.
Acyl-CoA dehydratase is an essential and limiting activity. (A) Segregation of undeveloped seeds in pas2–3/+ siliques (Bottom) compared with wild type (Top). (B) Ectopic expression of yeast PHS1 (Right) impaired plant growth (Top) and modified leaf size and shapes (Bottom). (Scale bar, 3 cm at Top, 3 mm at Bottom.) (C) SEM of leaf epidermal cells of wild-type and PHS1-expressing plants (Top) and surface detail showing wax deposition in PHS1 plants (Bottom). (Scale bar, 100 at Top, 10 μm at Bottom.) (D) Total fatty acid levels in the roots of pas2–1- and PHS1-expressing plants compared with wild type. (E) Total fatty acid levels in pas2–1- and PHS1-expressing plants compared with wild type. Dry weight and fatty acid values are the average of three samples ± SD.
Because the Arabidopsis dehydratase corresponds to a single gene and its inactivation is lethal, we investigated whether PAS2 might represent a limiting step for VLCFA synthesis. To limit possible transgene-derived silencing associated with the overexpression of PAS2, we used the orthologous yeast PHS1 to monitor the effect of increasing dehydratase activity on VLCFA levels and on plant development. Several independent lines expressing PHS1 under the 35S promoter showed clear growth retardation associated with abnormal leaf development. Leaves from transgenic lines were smaller and crinkled, with pronounced serration and often an asymmetric development compared with that of control plants (Fig. 4B). Epidermal cells from PHS1-expressing transgenic leaves showed a large heterogeneity in cell sizes and shapes (Fig. 4C). Moreover, the surface of PHS1-expressing leaf epidermal cells was decorated with wax crystals, suggesting an increase in cuticular waxes in contrast to wild type (Fig. 4C). Flower development was also modified by PHS1 expression with, for instance, misshapen and unfused carpels (Fig. S11). Detailed analysis of cell surface of unfused carpel showed high accumulation of cuticular waxes (Fig. S11).
To confirm that ectopic expression of PHS1 could modify VLCFA content, we analyzed fatty acid content of roots of young seedlings. The 35S:PHS1 seedlings showed significant changes in the relative distribution of VLCFAs, with higher levels of 22:0 compared with wild type (Fig. 4D). Because VLCFAs are also normally found in mature seeds, we investigated the effect of PHS1 expression on seed size and total fatty acid levels. Expression of PHS1 led to slightly larger seeds, whereas pas2 mutant showed smaller seeds compared with wild type (Fig. S11). Similar to the observation with seedlings, PHS1-expressing seeds showed an increase in VLCFAs, mostly 22:1 (Fig. 4E).
In conclusion, VLCFA dehydratase is not only an essential enzyme for plant growth and development, but it is also a limiting step for VLCFA synthesis because an increased dehydratase expression resulted in enhanced levels of VLCFAs in both vegetative and seed tissues.
Discussion
Collectively, our data demonstrated that the fatty acyl-CoA dehydratase is encoded by the PASTICCINO2 gene, and that it is an essential activity for plants. Similarly, the ketoacyl-CoA reductase activity in the double gl8agl8b maize mutant was also found to be essential (24). However, the loss of function of the enoyl-CoA reductase CER10, which is involved in the ultimate step in the acyl-CoA elongation cycle, is not lethal (unlike the yeast orthologue TSC13), suggesting that there is at least another partially redundant CER10 homologue in Arabidopsis (13). The weak pas2–1 allele is still able to produce some very-long-chain acyl-CoAs, resulting in strong reduction but not complete absence of VLCFAs. However, the channeling of these rare VLCFAs into different lipid classes is not unselective in pas2–1, indicating a previously unobserved level of regulation for the homeostasis of different lipid types. The most straightforward explanation is that a leaky mutation such as pas2–1 would result in significant perturbations to the lipid pools with the highest turnovers. Another hypothesis is that small changes in some lipid pools (like GIPCs) might lead to severe physiologic effects and that its homeostasis is maintained at the expense of less sensitive pools (like glucosylceramides). Alternatively, VLCFAs synthesis is compartmentalized differently and channeled independently for the different lipid pools. It was suggested, for instance, that VLCFA could be channeled into sphingolipids via an association of ceramide synthases with the elongase complex. This hypothesis was raised to explain the accumulation of medium-chain ceramide observed in yeast elongase mutants as in pas2 (25). It was previously thought that the limiting step in the elongase complex involved only the condensing enzymes (10). We demonstrated here that the dehydratase activity is also limiting for VLCFA synthesis. Interestingly, the overexpression of the condensing enzyme FAE1, as with PHS1, led to similar developmental alteration, such as asymmetric leaf shape. However, PHS1 enhanced wax deposition whereas FAE1 suppressed it, suggesting that both enzymes, despite being part of the same complex, could modify VLCFA homeostasis in different ways (20).
Contrary to the cer10 mutant, pas2–1 showed very severe developmental defects. In particular, pas2 was characterized by abnormal cell division that was enhanced in the presence of cytokinins, leading to callus-like structure (15, 16). It has to be noted that a similar phenotype was observed with weak mutations in the PAS3/GURKE gene, which codes the cytosolic acetylCoA carboxylase required for providing the malonyl-CoA to the elongase complex (26). The link between VLCFA and cell division was also reported in yeast (7, 27). In plants, the overexpression of PAS2 delayed cell cycle progression, in particular during mitosis (19). PAS2 was described as interacting directly with phosphorylated cell cycle regulator CDKA (19). The presence of a protein tyrosine phosphatase (PTP) motif that is conserved in eukaryotes led originally to the definition of the PAS2/Phs1p family as PTP-like proteins. However, recent structure/function analysis of Phs1p identified the catalytic residues involved in the dehydratase activity, and they do not belong to the PTP motif (9). We cannot exclude the possibility that the dehydratase function evolved recently from a PTP ancestor and that the PTP motif remained conserved across the PAS2/Phs1p family. Nonetheless, several indications suggest that the PAS2/Phs1p proteins might still be involved in phosphorylation-related processes. First, protein alignment of 31 members of PAS2/Phs1p showed amino acid conservation of the PTP motif (9). Then, the mutation of PAS2/Phs1p mammalian homolog PTPLA led to centronuclear myopathy in dogs, a disease related to mutations in the phosphoinositide phosphatase myotubularin MTM1 (28). PHS1 had the strongest epistatic interaction in yeast with the LCB phosphatase LCB3 (6). Moreover, the stability of the LCB kinase LCB4p is tightly regulated by the CDK PHO85p (29). The involvement of CDK-dependent phosphorylation in the regulation of LCB or VLCFA metabolic enzymes remains to be investigated in plants, but it would provide an attractive model reunifying the apparent divergent PAS2 functions.
The nature of the PAS2-mediated VLCFA pathway that regulates cell division and cell differentiation is still unclear. Mutations downstream in the LCB and sphingolipid pathway will help in understanding the functional role of these different lipids in plant development. The fact that PAS2 fulfills a nonredundant essential activity also opens up the possibility of using tissue-specific RNAi inactivation to probe and better define the multiple roles of VLCFAs in plant form and function.
Methods
Plant Material and Growth Conditions.
The pas2–1 mutants are ethyl methane sulfonate alleles in Col0 background that were maintained as heterozygous stocks. Plants were grown in vitro and in a greenhouse in soil as described previously (30). The pPAS2:PAS2-GFP construct corresponds to the PAS2 genomic sequence with 1014 bp of promoter cloned into pMDC107. The pas2–3 T-DNA insertion line N617051 from the SALK collection was genotyped by PCR with the PAS2-specific primers F20 (5′-AAAAAAGCAGGCTCGAGCTCGTCTAGCTACACC-3′) and R549 (5′-ACC CGGAAAATTCCAAAATC-3′) or T-DNA-specific primer Lba1 (5′-TGGTTCACGTAGTGGGCCATCG-3′). Yeast strains and growth conditions were carried out as described previously (15).
Cytologic Analyses.
Observations were carried out using an inverted TCS SP2-AOBS spectral confocal laser microscope (Leica Microsystems) using either a PL APO 20 × 0.70 NA or 63 × 1.20 NA water-immersion objective. GFP and mCherry fluorescence were respectively recorded after an excitation at 488 and 594 nm (laser Ar/Kry) and a selective emission band of 495–550 nm and 600–643 nm. YFP fluorescence was recorded after an excitation at 514 nm (laser Ar/Kry) and a selective emission band of 520–564 nm. GUS (β-glucuronidase) staining and scanning electron microscopy were carried out as described previously (18). Colocalization and BiFC studies were performed as described previously (22).
Lipid Analyses.
For mass quantification of lipid species, GIPCs, glucosylceramides, and ceramides from A. thaliana wild type and mutant seedlings were extracted, isolated, and quantified as detailed in (31). Analysis of free long chain bases of sphingolipids was adapted from Lester and Dickson (32). Cuticular lipids were extracted and analyzed as described previously (33). Total seed and leaf fatty acid were analyzed as reported by Li et al. (34). AcylCoA profiling and MS/MS analysis were carried out as described previously (8, 35). Detailed description of lipid analysis can be found in the SI Text.
Supplementary Material
Acknowledgments.
We thank Patrick Moreau (Centre National de la Recherche Scientifique-Bordeaux 2), Olivier Grandjean, and Bruno Letarnec for their help and for discussion. J.M. and L.B. were funded, respectively, by 6th European Integrated Project AGRON-OMICS (grant no. LSHG-CT-2006–037704) and Cancéropôle Ile de France. Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council (UK).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805089105/DCSupplemental.
References
- 1.Westerberg R, et al. Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J Biol Chem. 2004;279:5621–5629. doi: 10.1074/jbc.M310529200. [DOI] [PubMed] [Google Scholar]
- 2.Gaigg B, Toulmay A, Schneiter R. Very long-chain fatty acid-containing lipids rather than sphingolipids per se are required for raft association and stable surface transport of newly synthesized plasma membrane ATPase in yeast. J Biol Chem. 2006;281:34135–34145. doi: 10.1074/jbc.M603791200. [DOI] [PubMed] [Google Scholar]
- 3.Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Tehlivets O, Scheuringer K, Kohlwein SD. Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta. 2007;1771:255–270. doi: 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Kohlwein SD, et al. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:109–125. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 7.Yu L, Castillo LP, Mnaimneh S, Hughes TR, Brown GW. A survey of essential gene function in the yeast cell division cycle. Mol Biol Cell. 2006;17:4736–4747. doi: 10.1091/mbc.E06-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denic V, Weissman JS. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 2007;130:663–677. doi: 10.1016/j.cell.2007.06.031. [DOI] [PubMed] [Google Scholar]
- 9.Kihara A, Sakuraba H, Ikeda M, Denpoh A, Igarashi Y. Membrane topology and essential amino acid residues of Phs1, a 3-hydroxyacyl-CoA dehydratase involved in very long-chain fatty acid elongation. J Biol Chem. 2008;283:11199–11209. doi: 10.1074/jbc.M708993200. [DOI] [PubMed] [Google Scholar]
- 10.Millar AA, Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997;12:121–131. doi: 10.1046/j.1365-313x.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- 11.Paul S, et al. Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. J Biol Chem. 2006;281:9018–9029. doi: 10.1074/jbc.M507723200. [DOI] [PubMed] [Google Scholar]
- 12.Gable K, Garton S, Napier JA, Dunn TM. Functional characterization of the Arabidopsis thaliana orthologue of Tsc13p, the enoyl reductase of the yeast microsomal fatty acid elongating system. J Exp Bot. 2004;55:543–545. doi: 10.1093/jxb/erh061. [DOI] [PubMed] [Google Scholar]
- 13.Zheng H, Rowland O, Kunst L. Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell. 2005;17:1467–1481. doi: 10.1105/tpc.104.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn TM, Lynch DV, Michaelson LV, Napier JA. A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann Bot (Lond) 2004;93:483–497. doi: 10.1093/aob/mch071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellec Y, et al. Pasticcino2 is a protein tyrosine phosphatase-like involved in cell proliferation and differentiation in Arabidopsis. Plant J. 2002;32:713–722. doi: 10.1046/j.1365-313x.2002.01456.x. [DOI] [PubMed] [Google Scholar]
- 16.Faure JD, et al. The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development. 1998;125:909–918. doi: 10.1242/dev.125.5.909. [DOI] [PubMed] [Google Scholar]
- 17.Haberer G, Erschadi S, Torres-Ruiz RA. The Arabidopsis gene PEPINO/PASTICCINO2 is required for proliferation control of meristematic and non-meristematic cells and encodes a putative anti-phosphatase. Dev Genes Evol. 2002;212:542–550. doi: 10.1007/s00427-002-0273-9. [DOI] [PubMed] [Google Scholar]
- 18.Harrar Y, Bellec Y, Bellini C, Faure JD. Hormonal control of cell proliferation requires PASTICCINO genes. Plant Physiol. 2003;132:1217–1227. doi: 10.1104/pp.102.019026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Da Costa M, et al. Arabidopsis PASTICCINO2 is an antiphosphatase involved in regulation of cyclin-dependent kinase A. Plant Cell. 2006;18:1426–1437. doi: 10.1105/tpc.105.040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millar AA, Wrischer M, Kunst L. Accumulation of very-long-chain fatty acids in membrane glycerolipids is associated with dramatic alterations in plant morphology. Plant Cell. 1998;10:1889–1902. doi: 10.1105/tpc.10.11.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joubes J, et al. The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol. 2008;67:547–566. doi: 10.1007/s11103-008-9339-z. [DOI] [PubMed] [Google Scholar]
- 22.Marion J, et al. Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. doi: 10.1111/j.1365-313X.2008.03596.x. in press. [DOI] [PubMed] [Google Scholar]
- 23.Tsegaye Y, et al. Arabidopsis mutants lacking long chain base phosphate lyase are fumonisin-sensitive and accumulate trihydroxy-18:1 long chain base phosphate. J Biol Chem. 2007;282:28195–28206. doi: 10.1074/jbc.M705074200. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich CR, et al. Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J. 2005;42:844–861. doi: 10.1111/j.1365-313X.2005.02418.x. [DOI] [PubMed] [Google Scholar]
- 25.Han G, et al. The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase. J Biol Chem. 2002;277:35440–35449. doi: 10.1074/jbc.M205620200. [DOI] [PubMed] [Google Scholar]
- 26.Baud S, et al. gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Rep. 2004;5:515–520. doi: 10.1038/sj.embor.7400124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Feel W, DeMar JC, Wakil SJ. A Saccharomyces cerevisiae mutant strain defective in acetyl-CoA carboxylase arrests at the G2/M phase of the cell cycle. Proc Natl Acad Sci USA. 2003;100:3095–3100. doi: 10.1073/pnas.0538069100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pele M, Tiret L, Kessler JL, Blot S, Panthier JJ. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet. 2005;14:1417–1427. doi: 10.1093/hmg/ddi151. [DOI] [PubMed] [Google Scholar]
- 29.Iwaki S, Kihara A, Sano T, Igarashi Y. Phosphorylation by Pho85 cyclin-dependent kinase acts as a signal for the down-regulation of the yeast sphingoid long-chain base kinase Lcb4 during the stationary phase. J Biol Chem. 2005;280:6520–6527. doi: 10.1074/jbc.M410908200. [DOI] [PubMed] [Google Scholar]
- 30.Smyczynski C, et al. The C terminus of the immunophilin PASTICCINO1 is required for plant development and for interaction with a NAC-like transcription factor. J Biol Chem. 2006;281:25475–25484. doi: 10.1074/jbc.M601815200. [DOI] [PubMed] [Google Scholar]
- 31.Markham JE, Jaworski JG. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:1304–1314. doi: 10.1002/rcm.2962. [DOI] [PubMed] [Google Scholar]
- 32.Lester RL, Dickson RC. High-performance liquid chromatography analysis of molecular species of sphingolipid-related long chain bases and long chain base phosphates in Saccharomyces cerevisiae after derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate. Anal Biochem. 2001;298:283–292. doi: 10.1006/abio.2001.5368. [DOI] [PubMed] [Google Scholar]
- 33.Raffaele S, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008;20:752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Beisson F, Pollard M, Ohlrogge J. Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry. 2006;67:904–915. doi: 10.1016/j.phytochem.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Sayanova O, et al. A bifunctional Δ12, Δ15-desaturase from Acanthamoeba castellanii directs the synthesis of highly unusual n-1 series unsaturated fatty acids. J Biol Chem. 2006;281:36533–36541. doi: 10.1074/jbc.M605158200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.