Abstract
The nicotinic acetylcholine receptor (nAChR) is a cation-selective channel central to both neuronal and muscular processes and is considered the prototype for ligand-gated ion channels, motivating a structural determination effort that spanned several decades [Unwin N (2005) Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol 346:967–989]. Purified nAChR must be reconstituted in a mixture containing cholesterol to function. Proposed modes of interaction between cholesterol and the protein range from specific binding to indirect membrane-mediated mechanisms. However, the underlying cause of nAChR sensitivity to cholesterol remains controversial, in part because the vast majority of functional studies were conducted before a medium resolution structure was reported. We show that the nAChR contains internal sites capable of containing cholesterol, whose occupation stabilizes the protein structure. We detect sites at the protein–lipid interface as conventionally predicted from functional data, as well as deeply buried sites that are not usually considered. Molecular dynamics simulations reveal that occupation of both superficial and deeply buried sites most effectively preserves the experimental structure; the structure collapses in the absence of bound cholesterol. In particular, we find that bound cholesterol directly supports contacts between the agonist-binding domain and the pore that are thought to be essential for activation of the receptor. These results likely apply to those other ion channels within the Cys-loop superfamily that depend on cholesterol, such as the GABA receptor.
Keywords: protein–lipid interaction, ligand-gated ion channel, cys loop receptor
Critical for memory and cognition, the nAChR is implicated in neurological disorders (1), addiction (2, 3), and the mechanism of anesthetics (4). Early efforts to reconstitute the nAChR noted that cholesterol as well as charged lipids (such as phosphatidic acid) are required for proper function (5). Proposed mechanisms for the role of cholesterol in nAChR function fall into two general types: indirect and direct. Indirect mechanisms are those in which a cholesterol-induced change in fluid or elastic properties of the lipid bilayer modulates the protein behavior. Although early studies (6, 7) indicated that current flow through the channel is insensitive to bilayer fluid properties, more recent studies (8, 9) have proposed that this result depends on the choice of order parameters. The direct mechanism requires cholesterol binding to the nAChR and effecting structural and/or dynamic changes. Proponents of this theory have determined that (i) cholesterol molecules in proximity to the protein exchange very slowly with the bulk lipid bilayer (10) (electron spin resonance difference spectroscopy); (ii) cholesterol accesses sites on the protein that cannot be occupied by phospholipids (11) (fluorescence quenching); and (iii) some cholesterol bound to the nAChR interacts with the phospholipids in the surrounding bilayer (12) (substitution of cholesterol with phospholipid–cholesterol hybrids). This final observation was interpreted as indicating that cholesterol does not occupy deeply buried sites within the nAChR, and such sites were generally considered to be ruled out (8, 12, 13). Such an interpretation was consistent with earlier predictions that cholesterol-binding sites were superficial, at the lipid–protein interface (Fig. 1A). Unfortunately, experiments that do not consider the possibility of deeply buried sites generally preclude their observation; for instance, a recent photolabeling study of cholesterol binding to the nAChR (14) used incubation times that would allow for occupation by labeled cholesterol of external sites but not internal ones, as was already indicated by the cholesterol extraction experiments of Leibel et al. (15).
Fig. 1.
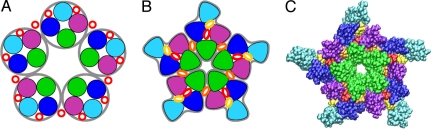
Proposed cholesterol binding sites. (A) The hypothetical binding sites for cholesterol proposed in 1993, adapted from ref. 42, with cross-section parallel to the membrane. M1 is purple, M2 is green, M3 is blue, and M4 is cyan. Cholesterol is represented by red circles. This cartoon reflects the very low resolution of nAChR structural data available at the time, as well as the hypothesis that cholesterol-binding sites are superficial (lipid/protein interface) rather than deeply buried in the protein. (B) Basic shape of the nAChR transmembrane domain as revealed by EM structure 2BG9 (19), with potential binding sites identified by our calculations based on that structure: sites A (yellow), sites B (orange), sites C (red). Sites such as B and C have generally been considered nonexistent since the publication of ref. 12. (C) Transmembrane portion of the nAChR in atomic detail, following the model of 2BG9, with cholesterol docked to proposed binding sites.
Focusing on the abundant nAChR from the electric organ of Torpedo, Unwin and coworkers (16–19) have gradually provided electron microscopy (EM) data of increasing resolution. They determined that each of the subunits (labeled αβ, β, δ, αδ, and γ) in the heteropentamer consists of an extracellular agonist-binding domain composed mostly of β-sheets, a transmembrane (TM) domain composed of a four-helix bundle, and an intracellular vestibule domain. The helices in the TM domain are labeled M1–M4, with the M2 helices lining the ion pore, the M4 helices forming the outer shell, and M1 and M3 joining adjacent subunits. The most recent structure at a medium resolution (4 Å) shows that the TM helices are not tightly packed, and there are numerous gaps in the density not filled by amino acids. These gaps, which are comparable in size with the central pore, were originally assumed (18) to be filled with water. The EM density map (Fig. 2), however, shows that the contents of these gaps are significantly more dense and/or ordered than the contents of the pore, which is known to contain water at room temperature. Furthermore, the density in the gaps is most similar to that of the surrounding lipid, which is consistent with the hydrophobic nature of the residues that line the gaps.
Fig. 2.
EM density map used by Miyazawa, Fujiyoshi, and Unwin (18) to derive a structure of the nAChR TM domain (PDB ID code 1OED, a precursor to the full structure 2BG9). The view is a slice through the TM domain, in the membrane plane, showing regions of high (blue), intermediate (white), and low density (red). Dark-blue circles include protein backbone and most side-chain density; the composition of material filling gaps between α-helices is unspecified but presumed to be water. However, the density in these gaps (white or pale blue) is higher than in the pentameric pore (red) and similar to that of the lipid surrounding the protein. Data retrieved from the Electron Microscopy Database, entry EMD-1044.
Bridging these functional and structural perspectives, we hypothesize that these gaps in protein density are occupied by cholesterol not resolved in the EM structure, rather than by water. Docking of cholesterol to the nAChR structure 2BG9, both manually and using AutoDock (20), revealed three binding sites per subunit (Fig. 1 B and C). We label them in order of increasing depth within the nAChR: site A (in the groove behind M4 and in direct contact with the phospholipid); site B (at the interface between subunits, bordered by M1 and M2 of one subunit and M2 and M3 of the adjacent subunit); and site C (in the subunit center, bordered by M1, M2, M3, and site A). The proposed binding sites are comparable in size and hydrophobicity with the cholesterol-binding site of the protein cryptogein (21), one of the few protein structures featuring resolved cholesterol. They are consistent with experimental observation in that they should be slow to exchange, could not be occupied by phospholipid, and yet (as shown below) would still allow cholesterol–phospholipid interactions.
Recently, an x-ray structure of a related prokaryotic pentameric ligand-gated channel, ELIC (PDB ID code 2VL0), has been reported (22). ELIC exhibits neither the large interhelical gaps nor the tapered shape of the nAChR TM domain (Fig. 3A). These differences are compatible with our hypothesis: Prokaryotic membranes (as well as the environment used to crystallize ELIC) are cholesterol-free and therefore should not reveal these signatures of embedded cholesterol.
Fig. 3.
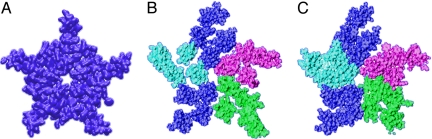
Density gaps in pentameric ligand-gated ion channels. (A) Molecular isodensity surface of the extracellular half of the prokaryotic ELIC channel TM domain (homopentameric), derived from 2VL0, showing few gaps in protein density. (B) Same representation for nAChR, derived from 2BG9, and colored by subunit type: α: blue, β: purple, δ: green, γ: cyan. (C) nAchR isodensity surface after 25 ns of MD simulation in a hydrated lipid bilayer. Protein collapses to close most gaps, representing a significant departure from the starting configuration.
Previous molecular dynamics (MD) simulations (23, 24) of the nAChR embedded in a phospholipid bilayer without cholesterol have revealed an exceptionally dynamic oligomeric complex that deviates significantly from the starting structure over a few ns, calling the stability of the experimental structure into question. MD simulations of the complete nAChR structure (2BG9) with various amounts of bound cholesterol (0, 5, 10, and 15 cholesterol molecules), embedded in a phosphatidylcholine membrane, were performed to investigate the possible improved stability of the proposed nAChR–cholesterol complex. nAChR with cholesterol occupying all identified binding sites behaves most consistently with the experimental structure, according to several measures of stability.
Results
Docking.
Both automated and manual docking found many available sites for cholesterol within the nAChR. Binding sites predicted by both methods are depicted in supporting information (SI) Fig. S1. Results of the two methods differ with respect to pentameric symmetry, overlap of sites, and cholesterol orientation. High-scoring sites as found by Autodock only partially reflect the fivefold pseudosymmetry. Although there is no evidence for cholesterol binding to be perfectly symmetric, gaps in the transmembrane domains are similar among the five subunits, suggesting a comparable cholesterol occupancy. Several overlapping clusters tend to be detected at the most accessible sites (A), or in one case, one bound cholesterol is predicted to overlap with both an A site and the adjacent C site. The manual search complies with the additional restraint of defining nonoverlapping sites that are likely to be occupied simultaneously. Finally, in several cases, manually inserted and Autodock-predicted bound cholesterol reside in the same general position (dictated by the bulky sterol scaffold) but point in opposite directions, with Autodock predicting the hydroxyl group to point toward the cytoplasmic side of the receptor. Although it cannot be definitively ruled out, such a binding mode would result in the hydroxyl group residing in a hydrophobic region of the protein, offering few hydrogen-bonding partners, whereas the alkyl chain would interact with the mostly polar environment of the interface region between the TM and binding domains. The Autodock scoring function, an empirical estimate of the free energy of binding, was evaluated on manually docked, energy-minimized nAChR–cholesterol complexes: The scores were between −14.1 and −9.8 kcal/mol, with only the more superficial A sites less favorable than −11 kcal/mol. As a comparison, the best site found by Autodock had a score of −14.2 kcal/mol, indicating that manually and automatically docked sites have similar physical plausibility.
Although the low structural resolution in the 2BG9 model of nAChR (19) would be a significant obstacle in more typical docking of a small ligand to a tight binding site, the cholesterol sites are sufficiently large that they are well defined at this resolution. Introducing side-chain flexibility at this stage would further reduce limitations due to low resolution, at the cost of increasing the number of degrees of freedom to be sampled. The manual approach focuses on overall consistency of the binding mode of multiple cholesterol molecules with the transmembrane domain. Local interactions with the side chains are optimized in the subsequent MD simulations, where the full flexibility of both the protein and cholesterol, as well as interactions with water molecules, are described and sampled explicitly. Based on statistics from the simulations, a list of residues most involved in protein–cholesterol interactions is provided in Fig. S2. Each cholesterol consistently interacts with at least 10 (mostly hydrophobic) residues. A PDB file containing coordinates for the fully occupied nAChR-cholesterol complex after energy minimization is available as Dataset S1.
Deviation from Experimental Structure.
Purely on the basis of the medium resolution of the EM structure (19), which allows reasonable confidence in backbone coordinates but not necessarily in the orientation of side chains, nonnegligible drift from the starting configuration over the course of the simulation can be expected. Previous simulations (23, 24) of the nAChR have exhibited such distortions in the backbone coordinates, however, that one may question whether the structure represented in 2BG9 is anywhere near a local free-energy minimum. In the case of a protein as large as the nAChR, ascertaining that such a free-energy minimum has been reached may well require more than the 25 ns explored by most of the simulations presented here. To this end, a much longer simulation (90 ns) of the control system was run, with very gradual lifting of restraints in several stages. Despite the substantial differences in equilibration protocol and simulation time, the short and long control simulations behaved very similarly, both in terms of overall deviation (see Fig. S3) and specific events (breaking of interfacial contacts, collapse of the TM bundle). Therefore, although the absolute stability or instability of the structure is difficult to assess, the similar behavior resulting from two such different simulation protocols suggests a deep trend rooted in major features of the 2BG9 structure, beyond inaccuracies in its fine, atomic-scale detail.
Here, we propose that the consistent drift of the protein from the starting structure is due in part to the large gaps in protein density in the TM domain. In our hypothesis, occupancy of gaps by cholesterol should reduce this drift. The actual effect of cholesterol on the RMSD of the TM domain, broken into subunits, is subtle (Fig. S4). Statistically, the only firm conclusion that can be drawn is that full occupancy by cholesterol does not increase or accelerate drift from the initial conformation (Fig. S5). This is a significant result, however, which becomes clear if one considers that in a protein with a more typical packing efficiency, introducing 15 cholesterol molecules into the hydrophobic core—if possible at all—would undoubtedly result in major distortion of the structure when allowed to relax. In contrast, the organization of the nAChR TM bundle appears as well preserved (if not better) in the simulation if it contains 15 cholesterol molecules, as is apparent from a simplified visualization of the TM helices (Fig. S6).
In the EM structure, the M4 helix tilts away from the core of the TM domain, resulting in a large gap open to the hydrophobic core of the surrounding membrane. This tilt is not preserved upon simulation unless spacer molecules enforce the separation between M4 and the TM core. The evolution of M4 tilt was determined by first aligning the M4 helices to the initial structure to minimize overall RMSD. A vector was then fit to each helix at initial time t0 and final time tf; the angle θ between these vectors was calculated and averaged over the five subunits. M4 helices in the control simulation deviate an average of 9 ± 2° from their starting configuration in 2BG9. Adding five cholesterol molecules to the sites A adjacent to M4 reduces the deviation to 6 ± 2°, and filling both the A sites and the C sites adjacent to the A sites (as well as the interfacial B sites for a total of 15 cholesterol molecules) reduces the deviation further to 3 ± 1°. As with other measures, the result for only 10 cholesterol molecules filling just the A and B sites is an outlier (11 ± 2°); close examination of the trajectory reveals that the force exerted on M1 and M3 by site B cholesterol distorts the protein unless it is balanced by a counteracting force from site C cholesterol. The reduced deviation in systems with cholesterol demonstrates that the tilted orientation of M4 (which gives the molecule its tapered shape) is most consistent with spacer molecules such as cholesterol embedded in the extracellular half of the protein.
Evolution of Contacts Critical for Gating.
Experimentally, contact between the β1–β2 loop of the agonist-binding domain and the M2–M3 loop of the TM domain has been implicated in transmission of the agonist-binding signal to the pore (18). In 2BG9, however, the M2–M3 loop of each subunit is largely unsupported and surrounded by water, except for its contact with the β1–β2 and Cys loops. The contact between the M2–M3 and β1–β2 loops, therefore, appears somewhat tenuous because of the absence of more structured regions on both sides. This could explain why previous simulations of the α4β2 channel have found the M2–M3 loop to be particularly mobile (23), and our MD simulations illustrate the fragility of this contact in the absence of cholesterol. Within the first 25 ns of the control simulation, the K45–P280 contact of the γ-subunit breaks as the TM loop collapses to fill the empty space that comprises the A and C binding sites (Fig. 4). Without this contact, the secondary structure of the β-sheets in the agonist-binding domain begins to unravel, and mechanical coupling between the agonist-binding domain and the TM domain appears to be significantly weakened. Occupation of the cholesterol A binding site is sufficient to prevent this decoupling, because it is not observed in any subunits of any of the systems with cholesterol bound. However, cholesterol initially docked into A sites has a tendency to tilt into adjacent unoccupied C sites, so partial occupation of the C site, which lies directly below the broken contact, may be necessary for its preservation.
Fig. 4.
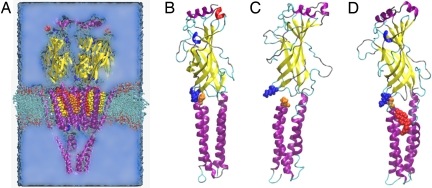
Mechanical decoupling of TM and agonist-binding domains in system without cholesterol. (A) All simulations contained the nAChR structure as described in 2BG9 (cartoon, colored by structure: α-helix is purple, β-sheet is yellow, turn is cyan, coil is silver, 310-helix is blue, π-helix is red), water (blue medium), ions (not shown), phospholipid (licorice), and 0–15 cholesterol molecules (space-filling, colored as in Fig. 1). (B–D) Agonist-binding and TM domain of γ subunit are shown at start of all simulations (B), at the end of control simulation without cholesterol (C), and at the end of simulation with cholesterol bound to sites A, B, and C (D). Shown in space-filling representation are K45 (blue), P280 (orange), and cholesterol in binding site C (red). The K45–P280 contact anchors the agonist-binding domain to the TM domain. When it is broken, as at the end of the control simulation shown in C, the agonist-binding domain tilts away from its starting orientation and the TM domain, so that the only communication between the two domains is through the flexible peptide chain linking M1 to the agonist-binding domain. Furthermore, β-sheets in the agonist-binding domain begin to unravel, reducing mechanical cohesion within the agonist-binding domain. Breaking of the K45–P280 contact is caused by the collapse of the loop containing P280 into the cholesterol-binding sites; occupation of these sites by cholesterol (D) precludes such a collapse.
The salt link between E45 (also on the β1–β2 loop) and R209 (on the β1–M1 linker) in the α-subunits has been implicated in gating dynamics (25). This salt link is preserved in all systems with cholesterol sites occupied but breaks in the αβ-subunit of the control simulation after 17 ns (7 ns of unrestrained simulation), assuming an N–O distance of 15 Å, and does not reform for the remainder of the simulation. In our longer (90 ns) simulation of the control system (with a different equilibration protocol), the salt link broke in both α-subunits upon lifting of the restraints, and in a previous 15-ns unrestrained simulation of the α4β2 receptor, one of the two α-salt links broke. (R.L., unpublished work). Summarizing previous and present results, the salt link broke in four of six cholesterol-free α-subunits, and none of six α-subunits treated with cholesterol. The breaking of this salt link in the control is accommodated by adjustments of helices and loops in the TM domain within the empty A and C binding sites.
Expulsion of Water from Gaps Not Occupied by Cholesterol.
After 25 ns in the control simulation, the protein has significantly redistributed its density. Most gaps that were reported in 2BG9 close as the individual subunits collapse, and some interfacial gaps between the subunits widen to a distance that would be expected to damp mechanical communication (Fig. 3 A and B). Such behavior supports our hypothesis that occupation by water does not justify the presence of these gaps in the structure. Collapse of the subunits is reduced with the placement of cholesterol in the proposed binding sites. This effect can be quantified by measuring the evolution of the volume occupied by the gaps, which was estimated by assigning all atoms in the extracellular half of the TM domain to cubic bins of side length 1.5 Å, according to their position. Bins devoid of protein density were counted to determine the empty volume at a given time. The empty volume at the starting time was subtracted from subsequent measurements to determine the change in gap volume. The bin size was chosen so that empty bins represented only significant gaps in the protein structure. In the course of the control simulation, the gap volume shrinks by 40 nm3, indicating that the protein has collapsed to fill the large gaps. In comparison, when five A sites are filled by cholesterol, the gap volume decreases by only 28 nm3, and with 15 cholesterol molecules in the A, B, and C sites, the gap volume increases by 18 nm3. This quantity deviates most in the system with 10 cholesterol molecules in the A and B sites (increasing by 68 nm3), reflecting the asymmetric forces exerted on M1 and M3 by site B cholesterol without the counteracting force from site C. According to these results, gaps in protein density observed in 2BG9 are more consistent with 15 bound cholesterol molecules than any of the other treatments.
Phospholipid Accessibility from Buried Sites.
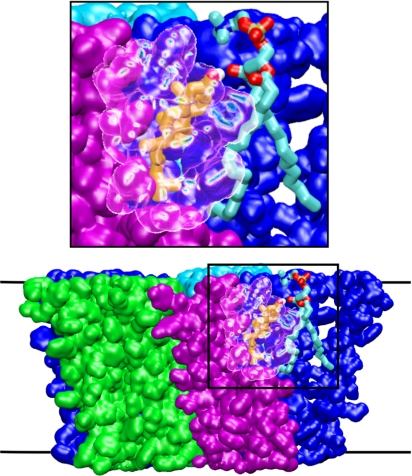
The possibility of deeply buried cholesterol-binding sites has been largely discounted because of the findings of an experiment (12) in which it was shown that nAChR functions when reconstituted in a mixture lacking cholesterol but containing hybrid phospholipids with one fatty acid replaced by cholesterol hemisuccinate. It has not been shown, however, that cholesterol molecules occupying particularly buried sites within each subunit (such as those we call C) were washed away before reconstitution; the negative effect of purification on receptor function simply indicates that those cholesterol molecules that were removed were essential. Furthermore, both sites A and B could feasibly be occupied by phospholipid-tethered cholesterol in the experiment. This is clear from instances in our simulations (Fig. 5) where cholesterol in a B site, although it is fairly deeply buried, forms a hydrogen bond with a phospholipid in the membrane through the intersubunit gap, yielding a molecular complex comparable with the hybrid lipid used in the experiment of ref. 12. This result highlights the ambiguity inherent in referring to “buried sites,” given the porous nature of the nAChR apparent from the latest structures (18, 19): The cholesterol steroid group may reach deep within the protein, whereas the hydroxyl group is exposed to water or phospholipid.
Fig. 5.
Hydrogen bonding between cholesterol with tail deeply buried in the nAChR and an external phosphatidylcholine molecule. The cholesterol is in site B and is colored orange, except for the OH group, colored by atom (oxygen is red, hydrogen is white). Phospholipid is colored by atom type (carbon is cyan, phosphorus is brown, nitrogen is blue). Part of the protein β-subunit is shown as a transparent surface.
Discussion
The primary finding of this study is that there are sites capable of containing cholesterol in the nAChR, including deep within the transmembrane bundle. We show that the experimental structure for the nAChR can easily accommodate up to 15 cholesterol molecules through docking calculations. Through MD simulation, we show that the published experimental structure is consistent with numerous cholesterol molecules buried within the receptor. Furthermore, these cholesterol molecules bolster the receptor structure, preserving contacts previously deemed critical for receptor function.
If cholesterol occupies deeply buried sites within Cys-loop ion channel proteins, as we propose, then experiments should be highly sensitive to the method of cholesterol extraction/depletion or supplementation. Depletion experiments, which primarily remove cholesterol from the membrane rather than from the presumably higher-affinity protein sites, may show a different dependence on cholesterol than reconstitution experiments, which are expected to more thoroughly remove bound cholesterol. Indeed this has been observed (26, 13). Likewise, the results of a reconstitution experiment should be sensitive to details of the reconstitution process such as the detergent used, insofar as they affect the completeness of cholesterol extraction or repletion. Consequently, experimental results that could have been deemed inconsistent can be reconciled by considering embedded cholesterol.
The overall effect of embedded cholesterol in the nAChR is to stabilize the structure determined by cryo-EM and preserve motifs that are known or presumed to be essential for gating. The present results are consistent with multiple experimental observations regarding cholesterol bound to the nAChR. These findings are likely applicable to other members of the Cys-loop ligand-gated ion channel superfamily [the GABA receptor, for example, also requires cholesterol for gating (27)].
Materials and Methods
Docking of Cholesterol.
To determine possible cholesterol sites within the transmembrane domain of nAChR, two approaches were used independently: Automated docking provided a systematic search, whereas a manual search allowed for more criteria to be taken into account.
For the automated docking, cholesterol coordinates were built by using Chem3D Pro with the Chemdraw plugin (CambridgeSoft), and the molecule was minimized in vacuo using MM2 within Chem3D. Gasteiger charges were calculated and added for the atoms in the ligand using Autodock Tools (20). The protein model was prepared for docking by merging nonpolar hydrogens into groups for the Kollman united atom force field (28) and adding Kollman charges and Stouten desolvation terms for each atom. Docking was performed by using Autodock 3.02 (29). A 220 × 220 × 250-point grid, with 0.375-Å spacing was used to search the whole receptor protein model. All torsions were allowed for the ligands as part of a flexible Lamarckian genetic algorithm (LGA)-based docking routine. Three hundred LGA runs were carried out. Modes of binding were separated with a 0.5-Å RMSD cut-off. The lowest-energy structure from each cluster was selected, and each mode was visually compared using VMD (30).
Manual docking was performed by using VMD (30). Unlike the automated docking, this process included symmetry considerations and aimed at describing simultaneous occupation of all favorable sites rather than docking one molecule at a time. The main criteria were avoiding steric clashes and searching for sites related through the pentameric pseudosymmetry. Individual cholesterol molecules were translated and rotated manually as rigid bodies, whereas clashes were monitored by displaying protein atoms within a cut-off of the cholesterol molecule. The cut-off was increased in steps until the locally optimal position and orientation were determined precisely. When necessary, contacts involving the flexible alkyl tail of cholesterol were preferred over those involving the rigid steroid ring, thus accounting for the possibility of relaxing such contacts in the ensuing simulations. Docked cholesterol molecules were kept in a general orientation comparable with that of lipids in the outer leaflet of the membrane, i.e., the hydroxyl group pointing toward the extracellular domain.
System Setup.
Atomic coordinates for the nAChR are taken from the 2BG9 structure. The β8–β9 loop in the agonist-binding domain of the β-, δ-, and γ-subunits are not resolved in Unwin's cryo-EM data, and hence are missing from the 2BG9 entry. These unstructured loops were modeled by using MODELLER (31, 32); they sample a wide range of conformations during the course of the simulations, indicating that the outcome of the simulations is not very sensitive to the modeled initial coordinates. The M3–MA loop is also missing from the structure; rather than attempting to model this loop of 100 residues, we placed restraints on both ends of the MA helix in the vestibule domain (see Simulation Details below).
Atomic coordinates for cholesterol were taken from the results of the manual docking.
A 12 × 12-nm2 1-palmitoyl-2-oleoyl-sn-glycerol-phosphatidyl-choline (POPC) bilayer was briefly relaxed as in ref. 23. For each simulation, containing 0 cholesterol molecules, 5 cholesterol molecules in sites A, 10 in sites A and B, or 15 in sites A, B, and C, the nAChR/cholesterol complex was inserted into the bilayer so that aromatic girdle residues on the nAChR aligned with the lipid–water interface. Water and lipid molecules that overlapped the nAChR complex with 15 cholesterol molecules were removed from all systems, so that each simulation had the same number of phospholipids and water molecules. Each system was then fully solvated to a box size of 12 × 12 × 18 nm3 using Solvate (33); unoccupied cholesterol-binding sites were filled with water. Sodium and chloride counterions were added to neutralize the systems and provide a 0.15 mole/liter solution. Each solvated system contained ≈230,000 atoms.
Simulation Details.
The simulations used the CHARMM22-CMAP force field with torsional cross-terms (34, 35) for the protein, CHARMM27 (36) for phospholipids, the Cournia et al. (37) model for cholesterol, ion parameters from Beglov and Roux (38), and the modified TIP3P water potential (39, 40). Minimization and dynamics were conducted with the NAMD2 package (41). Periodic boundary conditions were applied, with particle-mesh Ewald long-range electrostatics and a cut-off of 1.2 nm for Lennard–Jones potentials, with a smooth switching function starting at 1.0 nm. Simulations were conducted at a constant temperature of 300 K and pressure of 1 bar. Bonds involving hydrogen atoms were constrained to their equilibrium length using the SHAKE/RATTLE algorithm. Multiple-time-step integration was carried out using r-RESPA, with a base time step of 2 fs and a secondary time step of 4 fs for long-range interactions.
Each system was initially energy minimized for 1,000 steps, and then all atoms except Cα were released for 10 ns of dynamics. During this period, a harmonic potential with a force constant of 1 kcal/mol/Å2 restrained Cα atoms around their initial positions in the 2BG9 structure. Furthermore, the area of the simulation box in the xy-plane (parallel to the membrane) fluctuated according to the constant isotropic pressure ensemble with box reshaping allowed. Subsequently 15 ns of dynamics simulation were conducted with no restraints on the TM or agonist-binding domain of the protein. The vestibule domain was subjected to restraints that maintained its shape. Two Cα atoms were chosen per subunit, one at the intracellular membrane–water interface and one at the vestibule tip. The distances between any 2 of the 10 chosen atoms were subjected to harmonic restraints with a force constant of 1 kcal/mol/Å2. During this part of the simulation, the box area was restrained, and only the height of the box was allowed to fluctuate, to maintain a pressure of 1 bar. A much slower relaxation protocol, in which restraints were lifted over 90 ns, was tested on the control system and did not improve stability. All MD runs were conducted on 768 cores of the 64-bit Intel cluster Abe at the National Center for Supercomputing Applications, University of Illinois at Urbana–Champaign, Urbana, IL.
Supplementary Material
Acknowledgments.
Prof. William DeGrado is gratefully acknowledged for his critical reading of the manuscript. The work was supported by grants from the National Institutes of Health and by the National Science Foundation through TeraGrid resources provided by the National Center for Supercomputing Applications.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803029105/DCSupplemental.
References
- 1.Gotti C, Fornasari D, Clementi F. Human neuronal nicotinic receptors. Prog Neurobiol. 1997;53:199–237. doi: 10.1016/s0301-0082(97)00034-8. [DOI] [PubMed] [Google Scholar]
- 2.Larsson A, Engel JA. Neurochemical and behavioral studies on ethanol and nicotine interactions. Neurosci Biobehav Rev. 2004;27:713–720. doi: 10.1016/j.neubiorev.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Francis MM, Cheng EY, Weiland GA, Oswald RE. Specific activation of the α7 nicotinic acetylcholine receptor by a quaternary analog of cocaine. Mol Pharmacol. 2001;60:71–79. doi: 10.1124/mol.60.1.71. [DOI] [PubMed] [Google Scholar]
- 4.Yamakura T, Bertaccini E, Trudell JR, Harris RA. Anesthetics and ion channels: Molecular models and sites of action. Annu Rev Pharmacol Toxicol. 2001;41:23–51. doi: 10.1146/annurev.pharmtox.41.1.23. [DOI] [PubMed] [Google Scholar]
- 5.Dalziel AW, Rollins ES, McNamee MG. The effect of cholesterol on agonist-induced flux in reconstituted acetylcholine receptor vesicles. FEBS Lett. 1980;122:193–196. doi: 10.1016/0014-5793(80)80435-2. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Ballester G, et al. A role for cholesterol as a structural effector of the nicotinic acetylcholine receptor. Biochemistry. 1994;33:4065–4071. doi: 10.1021/bi00179a035. [DOI] [PubMed] [Google Scholar]
- 7.Sunshine C, McNamee MG. Lipid modulation of nicotinic acetylcholine receptor function: The role of membrane lipid composition and fluidity. Biochim Biophys Acta. 1994;1191:59–64. doi: 10.1016/0005-2736(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 8.Baenziger JE, Morris ML, Darsaut TE, Ryan SE. Effect of membrane lipid composition on the conformational equilibria of the nicotinic acetylcholine receptor. J Biol Chem. 2000;275:777–784. doi: 10.1074/jbc.275.2.777. [DOI] [PubMed] [Google Scholar]
- 9.daCosta CJB, Ogrel AA, McCardy EA, Blanton MP, Baenziger JE. Lipid–protein interactions at the nicotinic acetylcholine receptor. A functional coupling between nicotinic receptors and phosphatidic acid-containing lipid bilayers. J Biol Chem. 2002;277:201–208. doi: 10.1074/jbc.M108341200. [DOI] [PubMed] [Google Scholar]
- 10.Marsh D, Barrantes FJ. Immobilized lipid in acetylcholine receptor-rich membranes from torpedo marmorata. Proc Natl Acad Sci USA. 1978;75:4329–4333. doi: 10.1073/pnas.75.9.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones OT, McNamee MG. Annular and nonannular binding sites for cholesterol associated with the nicotinic acetylcholine receptor. Biochemistry. 1988;27:2364–2374. doi: 10.1021/bi00407a018. [DOI] [PubMed] [Google Scholar]
- 12.Addona GH, Sandermann H, Kloczewiak MA, Husain SS, Miller KW. Where does cholesterol act during activation of the nicotinic acetylcholine receptor? Biochim Biophys Acta. 1998;1370:299–309. doi: 10.1016/s0005-2736(97)00280-0. [DOI] [PubMed] [Google Scholar]
- 13.Burger K, Gimpl G, Fahrenholz F. Regulation of receptor function by cholesterol. Cell Mol Life Sci. 2000;57:1577–1592. doi: 10.1007/PL00000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamouda AK, Chiara DC, Sauls D, Cohen JB, Blanton MP. Cholesterol interacts with transmembrane alpha-helices M1, M3, and M4 of the Torpedo nicotinic acetylcholine receptor: Photolabeling studies using [3H]azicholesterol. Biochemistry. 2006;45:976–986. doi: 10.1021/bi051978h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leibel WS, Firestone LL, Legler DC, Braswell LM, Miller KW. Two pools of cholesterol in acetylcholine receptor-rich membranes from torpedo. Biochim Biophys Acta. 1987;897:249–260. doi: 10.1016/0005-2736(87)90421-4. [DOI] [PubMed] [Google Scholar]
- 16.Toyoshima C, Unwin N. Ion channel of acetylcholine receptor reconstructed from images of postsynaptic membranes. Nature. 1988;336:247–250. doi: 10.1038/336247a0. [DOI] [PubMed] [Google Scholar]
- 17.Unwin N. Acetylcholine receptor channel imaged in the open state. Nature. 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- 18.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 19.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 20.Morris GM, Goodsell D, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of autodock 2.4. J Comput Aid Mol Des. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 21.Lascombe MB, et al . The 1.45 Å resolution structure of the cryptogein-cholesterol complex: A close-up view of a sterol carrier protein (scp) active site. Acta Crystallogr D. 2002;58:1442–1447. doi: 10.1107/S0907444902011745. [DOI] [PubMed] [Google Scholar]
- 22.Hilf RJC, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 23.Law RJ, Henchman RH, McCammon JA. A gating mechanism proposed from a simulation of a human α7 nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 2005;102:6813–6818. doi: 10.1073/pnas.0407739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corry B. An energy-efficient gating mechanism in the acetylcholine receptor channel suggested by molecular and Brownian dynamics. Biophys J. 2006;90:799–810. doi: 10.1529/biophysj.105.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lummis SCR, et al. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 26.Barrantes FJ. Cholesterol effects on nicotinic acetylcholine receptor. J Neurochem. 2007;103(Suppl 1):72–80. doi: 10.1111/j.1471-4159.2007.04719.x. [DOI] [PubMed] [Google Scholar]
- 27.Sooksawate T, Simmonds MA. Effects of membrane cholesterol on the sensitivity of the GABA(A) receptor to GABA in acutely dissociated rat hippocampal neurones. Neuropharmacology. 2001;40:178–184. doi: 10.1016/s0028-3908(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, et al. New-generation amber united-atom force field. J Phys Chem B. 2006;110:13166–13176. doi: 10.1021/jp060163v. [DOI] [PubMed] [Google Scholar]
- 29.Morris GM, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 30.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 31.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiser A, Sali A. Modloop: Automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 33.Grubmüller H, Heymann B, Tavan P. Ligand binding: Molecular mechanics calculation of the streptavidin–biotin rupture force. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 34.MacKerell A, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 35.MacKerell AD, Feig M, Brooks CL. Improved treatment of the protein backbone in empirical force fields. J Am Chem Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 36.Feller SE, MacKerell AD., Jr An improved empirical potential energy function for molecular simulations of phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- 37.Cournia Z, Smith JC, Ullmann GM. A molecular mechanics force field for biologically important sterols. J Comput Chem. 2005;26:1383–1399. doi: 10.1002/jcc.20277. [DOI] [PubMed] [Google Scholar]
- 38.Beglov D, Roux B. Finite representation of an infinite bulk system: Solvent boundary potential for computer simulation. J Chem Phys. 1994;100:9050–9063. [Google Scholar]
- 39.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 40.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 41.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narayanaswami V, McNamee MG. Protein–lipid interactions and Torpedo californica nicotinic acetylcholine receptor function. 2. Membrane fluidity and ligand-mediated alteration in the accessibility of gamma subunit cysteine residues to cholesterol. Biochemistry. 1993;32:12420–12427. doi: 10.1021/bi00097a021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.