Abstract
Gene expression levels appear to be under pervasive stabilizing selection. Yet the genetic architecture underlying abundant gene expression diversity within and between populations remains elusive. Here, we investigated the role of dominance in the segregation of cis- and trans-regulation within and between populations. We used chromosome substitution lines of Drosophila melanogaster to show that (i) >70% of the genes that are differentially expressed between two homozygous lines are masked in the heterozygous, suggesting that one of the substituted chromosomes contains a recessive allele; (ii) such large masking is already obtained with heterozygous chromosomes originating from the same population, with the time of divergence between chromosomes in heterozygous lines making only a small but significant contribution to the masking of variation observed in homozygous lines; (iii) variation in gene expression due to trans-regulation is biased toward greater deviations from additivity because of recessive and dominant alleles, whereas variation due to cis-regulation shows higher additivity; and (iv) genetic divergence between second chromosomes is associated with increased cis-regulation, whereas the level of trans-regulation shows little increase over the time scale studied. Our results indicate that cis-acting alleles may be preferentially fixed by positive natural selection because of their higher additivity, and that the disruption of gene expression by recessive variation with pervasive trans-effects may be important for understanding gene expression variation within populations. We suggest that widespread regulatory effects of recessive low-frequency homozygous variation may provide a general mechanism mediating disease phenotypes and the genetic load of natural populations.
Keywords: genetic load, genome architecture, regulatory evolution, recessive, natural selection
The ubiquity of regulatory variation within and between populations is well documented and manifested as abundant gene expression differences among individuals (1). The relevance of stabilizing selection restricting divergence in regulatory variation that would otherwise be expected from the relatively large effect of mutations on gene expression is also unequivocal (2–5). The list of biological attributes that appear to constrain or promote regulatory diversity in natural populations is already long and includes, for example, attributes of the protein–protein interaction and regulatory networks, transcription rates, sex and tissue of expression, motifs in the promoter, and the biological function of the genes (5–7). The rate of gene expression evolution and protein evolution might also be associated (6, 8).
Variation in gene expression levels, including the differential rate, timing, and tissue of expression, contributes to higher-level phenotypic differences (9). Such connections not only highlight the significance of gene expression levels as an underlying phenotype associated with ecologically and evolutionary important variation, but they also underscore the relevance of mRNA abundance as a phenotype in and of itself. For this reason, gene expression levels merit a detailed analysis of their evolution and genomic architecture. The genomic architecture of a trait refers to the myriad of genetic properties underlying a complex phenotype (10); it describes the mapping of underlying variation in genetic parameters onto variation among phenotypes. Such mappings mediate the interaction of genomes with the environment and are of fundamental interest, because they reveal links between natural selection and genome evolution.
No single trait had surfaced as a good candidate for a detailed description of its genetic architecture. One of the reasons is that classical higher-level morphological traits often undergo complex morphogenetic processes that obscure the mapping between genotypes and phenotypes. However, genetic variation affecting gene expression levels can often be ascertained unambiguously (11), and a detailed description of the genetic architecture including the effects of particular genes may be accomplished. Hence, variation in the expression of a focal gene may be attributed to variation in a number of proximal attributes allowing a link between natural selection on phenotypes and correlated effects on genome evolution to be inferred. This attractive prospect has helped gene expression level emerge as a model trait of choice to test models and concepts in the evolution of genomic architecture and of complex phenotypes more generally.
Genetic factors controlling variation in a focal gene may segregate in linkage with the gene (cis-effects) or segregate independently of the focal gene (trans-effects), with evidence indicating that cis-regulation contributes disproportionately to gene expression divergence between species relative to its contribution within species (12). This is in agreement with expectations that cis-regulatory alleles may be preferentially fixed because of their weaker pleiotropic effects (13). However, fundamental differences in the mutation rates and coefficients of dominance of cis- and trans-regulatory variation may also play significant roles underlying different contributions of cis- and trans-variation to gene expression diversity within and among species.
Here, we used genome-wide gene expression variation measured across Drosophila melanogaster chromosome substitution lines for the second and third chromosomes to empirically address these issues. We hypothesize that a greater additivity of alleles with cis-regulatory effects might underlie a preferential fixation of cis-regulatory loci (14), and that the disruption of regulatory networks by recessive variation with pervasive trans-effects might be a relevant mechanism mediating the genetic load of natural populations. This, together with the much higher rate by which trans-regulatory variation is produced (5), may underlie larger levels of trans-regulation within populations. These hypotheses regarding the distribution of coefficients of dominance of segregating loci underlying cis- and trans-variation could explain the distinct contributions of cis- and trans-regulation within and among species. A simple mathematic model for describing our data is also developed.
Results
Chromosome Substitution Lines and Comparison Sets.
We constructed second-chromosome substitution lines of D. melanogaster according to the mating scheme shown on supporting information (SI) Fig. S1. This resulted in homozygous lines that differed in the origin of the second chromosome while being identical with respect to all other chromosomes and cytoplasm. Any genetic variation observed between lines must therefore be attributable to genetic differences residing in the second chromosome. We checked for the homogeneity of the background chromosomes by typing the X and third chromosome of each line with PCR markers with variable length and sequencing of the amplified fragments. Verification was done for both the original strains and the derived second-chromosome substitution lines. These tests confirmed that the second chromosomes were substituted successfully into an identical and homozygous background.
Two homozygous lines (PS1/PS1 and PS2/PS2) were randomly chosen among second chromosomes originating from a single population in Pennsylvania (15). Six heterozygous lines were obtained by mating males of these selected homozygous lines, PS1/PS1 and PS2/PS2, with females of three others homozygous lines (PS3/PS3, CS/CS, and Z53/Z53). The chromosomes PS2, PS1, and PS3 are closely related chromosomes, because they derive from a single natural population, whereas CS and Z53 are “divergent chromosomes,” because they derive from different natural populations in North America (CS, collected in Ohio in the 1940s) or in Africa (Z53, collected in Zimbabwe); Indeed, the Zimbabwe strain is highly divergent from and shows some degree of premating isolation with North American populations (16). We then measured differences in transcript abundance between adult males across these eight lines (Fig. 1). Gene expression data were collected by hybridization on microarrays following the design shown in Fig. S2.
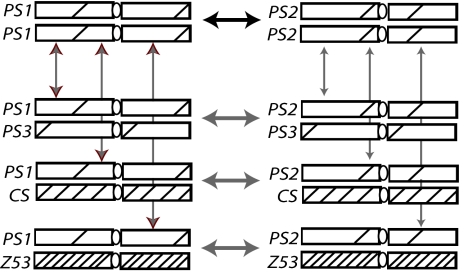
Fig. 1.
Second-chromosome genotypes for which gene expression data were collected. Contrasts of genotypes on each of the columns show the heterozygous effects of chromosomes PS3, CS, and Z53. The number of genes differently expressed in each of these contrasts is expected to increase with decreasing identity of the heterozygous chromosome. Contrasts across genotypes on each of the rows show the differences between chromosomes PS1 and PS2 on various backgrounds. In the absence of dominance, the number of genes differentially expressed in each of these contrasts is expected to remain constant.
We focused on three sets of pairwise comparisons and developed a mathematical model to describe these contrasts in terms of genetic identities in cis- and trans-regulatory loci, dominance, and the average number of genes affected by each cis- and trans-regulatory locus (see Model in SI Text and Figs. S3–S5). The homozygous-homozygous set contains the comparison between the two selected homozygous lines (PS1/PS1-PS2/PS2), with the number of differentially expressed genes expected to be proportional to the level of genetic divergence (1–IPS1,PS2) between the two substituted chromosomes, where IPS1,PS2 is the identity between chromosomes PS1 and PS2. The homozygous-heterozygous set includes six pairwise comparisons, PS1/PS1 vs. PS1/PS3, PS1/PS1 vs. PS1/CS, PS1/PS1 vs. PS1/Z53, PS2/PS2 vs. PS2/PS3, PS2/PS2 vs. PS2/CS, and PS2/PS2 vs. PS2/Z53. In this set, the number of differentially expressed genes is expected to increase with divergence but will also depend on dominance relationships between alleles carried by the two substituted chromosomes. The heterozygous-heterozygous set contains three comparisons (PS1/PS3 vs. PS2/PS3, PS1/CS vs. PS2/CS, and PS1/Z53 vs. PS2/Z53), where in this case the number of differentially expressed genes is expected to depend not only on the divergence between PS1 and PS2 but also on the dominance of PS3 (or CS or Z53) alleles over PS1 and PS2. Note that, in all of these contrasts, there are only two chromosomes varying, such that the number of genes differentially expressed in each contrast is attributable to the differences between these chromosomes (see Model in SI Text).
Number of Genes Differentially Expressed and Estimates of Cis- and Trans-Regulation.
In the set of homozygous-heterozygous contrasts, allelic variation within a single heterozygous second chromosome results in expression differences in an average of 420 genes [P < 0.001; false discovery rate (FDR) <0.05]. Although the second chromosome corresponds to 39% of the analyzed transcripts, it accounts for ≈63% (265 genes) of the changes in genes with changes in expression. Conversely, the X and third chromosomes correspond to 16% and 45% of the analyzed transcripts but account for only ≈10% and 27% of the genes differentially expressed, respectively. The finding of significant enrichment for second-chromosome transcripts and the deficit of purely trans-variation because of the X and third chromosome transcripts (P ≪ 0.0001; Fisher's exact test) extends previous findings for third-chromosome substitution lines (17) and the X-chromosome (18). This bias for the second chromosome is modestly affected by the different backgrounds, PS1 or PS2, because their second chromosome contains, respectively, 61% and 65% of the differentially expressed genes. Similar figures are found in the set of heterozygous-heterozygous contrasts, whereas the homozygous-homozygous contrasts show an increase in trans-regulation (see below).
Gene expression differences can be inferred to be due to effects in trans (i.e., all genes in the third and X chromosomes, plus a fraction of genes on the second chromosome) and in cis (a fraction of genes on the second chromosome) (see Fig. S3). If trans-factors have the same probability to regulate genes on the second, third, fourth, and X chromosomes, we can estimate the number of differentially expressed genes regulated in cis by simply using the total number of genes carried by the different chromosomes (see Model in SI Text). Hence, among the 265 genes that are on average differentially expressed on the second chromosome, we expect that 99 genes are trans-regulated [(420–265) × (39%/(1–39%)] and that 166 genes are cis-regulated. Consequently, gene expression variation observed in the second chromosome is greatly enriched in cis-regulatory effects (65% cis-regulatory vs. 35% trans-regulatory, P < 0.001, binomial test).
Masking of Homozygous Expression Variation in Heterozygous Substitution Lines.
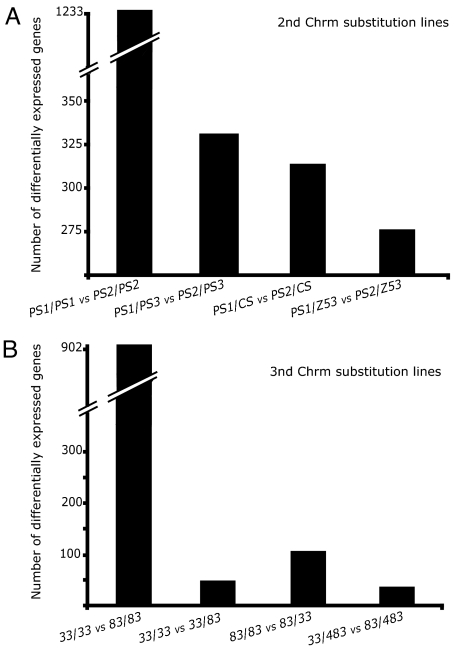
The comparisons between the homozygous-homozygous set (PS1/PS1 vs. PS2/PS2) and the heterozygous-heterozygous set (PS1/PS3 vs. PS2/PS3, PS1/CS vs. PS2/CS, PS1/Z53 vs. PS2/Z53) allow us to tackle the question of dominance in gene expression. Dominance can be defined at the level of loci underlying phenotypic differences or at the level of the phenotype itself. At the level of the phenotype, recessive/dominant is synonymous with masking/lack of masking of the gene expression differences. Hence, gene expression differences that are expressed between homozygous genotypes and across a range of heterozygous genotypes are dominant. However, gene expression differences expressed between homozygous genotypes that are not maintained across heterozygous genotypes are recessive. We found 1,233 gene expression differences between PS1/PS1 and PS2/PS2 (P < 0.001; FDR < 0.05), whereas significantly fewer gene expression differences were observed in the contrasts between PS1/PS3 vs. PS2/PS3 (467 genes), PS1/CS vs. PS2/CS (684), and PS1/Z53 vs. PS2/Z53 (377). In particular, >70% of the genes that show expression differences in the homozygous-homozygous comparison are masked in the heterozygous-heterozygous comparison, with the most divergent chromosome (Z53) showing a stronger masking of the differences between PS1/PS1 and PS2/PS2 than the more closely related (PS3) (Fig. 2A).
Fig. 2.
Masking of homozygous gene expression differences in heterozygous. Number of gene expression differences in contrasts between genotypes from (A) second- and (B) third-chromosome substitution lines. Note the change of scale in the y axis.
To confirm the occurrence of masking of expression differences in an independent dataset, we turned to data available from third-chromosome substitution lines from Hughes et al. (17). In this dataset, gene expression measurements were taken from three homozygous lines (33/33, 83/83, and 483/483) and their three heterozygous lines (33/83, 33/483, and 83/483). In agreement with our expectations, we find that the majority (≈95%) of gene expression differences observed between homozygous third-chromosome genotypes are masked in heterozygous comparisons (Fig. 2B). In both second- and third-chromosome substitution lines, the masking effect appears quite insensitive to the fold differences observed in the homozygous-homozygous contrasts. Specifically, for differentially expressed genes showing fold changes >2, the dampening in the masking effects is reduced only slightly from 95% (when all fold changes are considered) to 90% (when only genes with >2-fold change are considered) in the case of third-chromosome data. Similarly, masking in the second-chromosome data fluctuates ≈70%, regardless of higher or lower fold-change cutoffs.
A key challenge is to infer modes of additive or dominant/recessive gene action from the masking or lack of masking of gene expression phenotypes. In particular, large and numerous gene expression differences can, in some cases, be shown to result from a single polymorphic point mutation segregating in a natural population (19). If we assume that observed differences in gene expression are produced by noninteracting cis- or trans-regulatory loci, we can build straightforward relationships between phenotypes and genotypes (see Model in SI Text). Regulatory loci with alleles with dominant/recessive relationships to other alleles will result in gene expression differences that are masked or not, depending on the frequencies of the recessive and the dominant alleles. If the recessive allele is rare, there is a higher probability of observing a masked phenotype (Table S1). However, if the recessive allele is common, we expect that phenotypic differences will not be masked (Table S1). Hence, masked phenotypes likely result from regulatory loci with recessive alleles segregating at low frequency; Phenotypes that are not masked might result from regulatory loci with additive effects or loci with dominant alleles segregating at low frequency. We observed that, among the 1,233 genes differentially expressed between PS1/PS1 and PS2/PS2, 99 genes are never masked in heterozygous comparisons (phenotypically dominant). Furthermore, in this set of genes, fold changes in heterozygous-heterozygous comparisons are highly correlated to fold changes observed in the homozygous-homozygous comparison (Fig. S6). However, we observed 624 genes that are systematically masked (phenotypically recessive) in all heterozygous-heterozygous comparisons.
Masked Phenotypes and Cis-/Trans-Regulation.
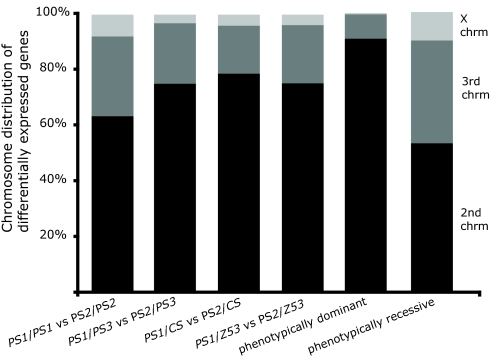
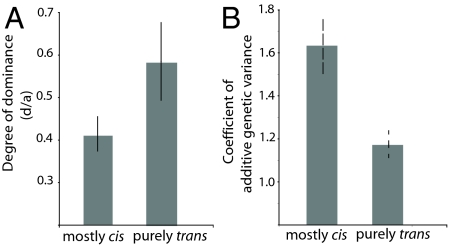
We found a close association between the masking of gene expression phenotypes and cis-/trans-regulation. First, there is a consistent decrease in purely trans-effects (i.e., genes differentially expressed on the X and third chromosomes) from 36% of all expression differences between PS2 and PS1 to an average of 23% among those that are not masked in heterozygous-heterozygous comparisons (Fig. 3; P < 0.0001, Fisher's exact test). Remarkably, this proportion further drops to a meager 10% among the 99 “phenotypically dominant” genes whose expression differences are not masked in any of the heterozygous comparisons (Fig. 3; P < 0.0001). However, the proportion of purely trans-regulation climbs significantly to 47% among the 624 genes with masked expression phenotypes (Fig. 3; P = 0.0007). We find that third-chromosome substitution data show identical patterns regarding the masking of gene expression differences resulting from cis- or trans-regulation. The results presented thus far suggest that cis-regulatory loci might include alleles that are skewed toward additivity, whereas trans-regulatory loci might have rare recessive alleles with impact on gene expression. If within-species allelic variation underlying differences in trans were indeed biased toward being recessive, it would more frequently be masked, such that we can predict it might result in higher degrees of dominance (d/a) and lower levels of heritability and additive genetic variance (20). This is what we found using data on gene expression inheritance (17) and estimates of additive genetic variance (21). First, the degree of dominance (d/a) is significantly lower for differentially expressed genes carried by the substituted chromosome than for the other genes with purely trans-effects (mean absolute d/a = 0.29 and 0.37, respectively; P < 0.001, Wilcoxon test; Fig. 4A). Second, genes herein identified as harboring cis-regulatory alleles in the second chromosome show significantly higher levels of heritability and additive genetic variance than genes subject to trans-regulation (mean coefficient of additive variation = 1.63 and 1.17, respectively; P < 0.05, Wilcoxon test; Fig. 4B).
Fig. 3.
Distribution of gene expression differences among second (black), third (dark gray), and X (light gray) chromosomes in various contrasts. The “phenotypically dominant” column refers to the differences between PS2/PS2 and PS1/PS1 that persist across all heterozygous PS2-PS1 contrasts. Rightmost bar (“phenotypically recessive”) refers to the differences between PS2/PS2 and PS1/PS1 that are present only in the homozygous but in none of the heterozygous PS2-PS1 contrasts.
Fig. 4.
Effects of cis- and trans-regulatory variation on modes of gene expression inheritance. (A) Average degree of dominance of cis and trans gene expression variation (d/a) values from third-chromosome substitution data of Hughes et al. (17). (b) Average level of additive genetic variation in the set of not masked cis-regulated genes and in the set of masked trans-regulated genes. Segregating additive genetic variation estimated by Wayne et al. (21). Black bars represent two times the standard error of the mean.
Finally, we addressed the relationship between the magnitude of the gene expression difference as measured by fold changes in mRNA abundances and patterns of masking (Table S2). First, regardless of their being masked or not, fold changes are significantly higher for genes carried by the substituted chromosome than for the background chromosomes (P < 0.001; Kruskal–Wallis test), although the magnitude of the difference is very small in the case of masked variation. Second, masked differences show lower fold changes than differences that are not masked. Hence, this suggests that, on average, cis-regulation might underlie larger fold changes than trans-regulation.
Divergent Chromosomes and Cis-/Trans-Regulation.
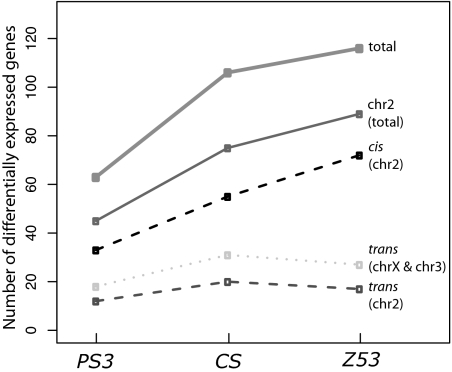
The homozygous-heterozygous set (PS1/PS1 vs. PS1/PS3, PS1/PS1 vs. PS1/CS, PS1/PS1 vs. PS1/Z53, PS2/PS2 vs. PS2/PS3, PS2/PS2 vs. PS2/CS, and PS2/PS2 vs. PS2/Z53) allows us to compare the effects on gene expression of three divergent second chromosomes (PS3, CS, and Z53) in two different genetic backgrounds (PS1 or PS2; plus a common X, third, and fourth chromosomes). Accordingly, a single dosage of a divergent second chromosome produces more gene expression differences than a single dosage of a more closely related chromosome (Fig. 5). For instance, the most divergent (Z53) chromosome relative to the background PS1 and PS2 produced on average 55% more differences in gene expression than the least divergent (PS3) second chromosome tested. Genetic divergence among chromosomes might also affect the relative contributions of cis- and trans-regulation. Accordingly, the fraction of all gene expression differences that are observed strictly in trans (i.e., attributable to the X and third chromosomes) significantly decreased from 33% in the case of the heterozygous effect of chromosome PS3 to some 18% in the case of chromosome Z53 (P < 0.001; Fisher's exact test). This is because the increase in differential expression with time is mostly due to a sharp increase in cis-regulatory variation, whereas the magnitude of trans-regulatory variation remained approximately constant at various levels of divergence (Fig. 5).
Fig. 5.
Differential accumulation of cis-and trans-regulatory variation with increasing genetic divergence. Chromosome distribution of the heterozygous effect of second chromosomes from strains PS3, CS, and Z53 that is common to both PS1 and PS2 second-chromosome backgrounds. Contrasts are done along the vertical axis in Fig. 1. See text for details.
Discussion
Genetic Load and Recessive Regulatory Variation Within Natural Populations.
Our results for both second- and third-chromosome substitution lines of D. melanogaster indicate that the majority of gene expression differences observed between homozygous genotypes are masked when heterozygous genotypes are contrasted. Four relevant points bear on this result. First, chromosomes extracted from the same population already possess enough variation to mask >70% of all gene expression differences between homozygous lines. This suggests that many of the differences observed between homozygous lines are due to recessive mutations segregating at low frequency. Second, gene expression variation arising because of trans-effects is particularly sensitive to masking in heterozygous. This suggests that either there are a large number of recessive alleles with a few effects in trans, or that there are a few recessive loci with numerous trans-effects. Third, the masking effect of chromosome 3 appears higher than that of chromosome 2. This finding could be explained if the second chromosome were enriched for cis-effects relative to the third chromosome (22) or could be due to technical differences between the two datasets compared. Fourth, in agreement with what one might have expected from chromosome-wide heterozygosity, the most divergent chromosome (Z53) is the most efficient at masking the recessive differences between PS1 and PS2.
Chromosome-wide homozygosity has long been known to result in lowered fitness of Drosophila chromosome substitution strains (23, 24), and widespread heterozygous masking of deleterious effects on regulatory networks may provide a molecular mechanism for understanding the genetic load in natural populations. In particular, our results may shed light on two hypotheses regarding the mechanism for inbreeding depression and genetic load, namely homozygosity of overdominant loci vs. homozygosity of low-frequency deleterious recessives (25). In the first model, one might expect that an overall increase in chromosome-wide heterozygosity might result in sharp increases in the masking of differences between chromosomes. Although we do observe a continuous increase, with chromosomes PS3, CS, and Z53 being increasingly more efficient at masking differences between PS1 and PS2, the reduction is of quite small magnitude. Conversely, the second model might predict that a closely related chromosome originating from the same population should already result in a substantial masking of the gene expression differences between two homozygous chromosomes, virtually as much as distantly related chromosomes from other populations. This is what we observed. Hence, although the data might suggest a small effect of genome-wide heterozygosity on the masking of homozygous variation, the hypothesis of low-frequency recessive alleles appears the most forceful. Note that any kind of molecular variation, which should not be restricted to transcription factors (26, 27), can underlie the recessive trans-effects in homozygous genotypes. Accordingly, structural variation that has been recently uncovered as copy number polymorphism both in humans and fruit flies (28) might be a promising source of abundant mutations with recessive trans-effects. This is because structural variation typically undergoes mutation rates orders of magnitude higher than single-nucleotide substitutions (29). Hence, under mutation-selection balance, the equilibrium level of copy-number polymorphism is expected to be substantially higher than that of point mutations. All in all, results indicate that heterozygous genotypes as typically found in Drosophila, and other organisms harbor much greater gene expression diversity than is readily apparent from their gene expression phenotypes and have implications for understanding fitness costs of homozygosity. Indeed, it suggests that the disruption of gene expression levels by recessive homozygous genotypes might be a pervasive mechanism mediating the genetic load of natural populations.
Dominance, Mutational Variance, and Evolutionary Accumulation of Cis- and Trans-Regulation.
There are fundamental properties of cis- and trans-effects that must help shape the evolutionary dynamics of cis and trans gene expression variation in natural populations. First, differences in the mutational variance for these two modes of gene expression regulation impose critical upper and lower boundaries on the relative amount of cis- and trans-variation that is possible across various timescales. Second, differences in the degree of dominance of gene expression differences resulting from cis- vs. trans-regulation impose fundamental differences in the population genetics of cis- and trans-variation.
There are two ways in which a pattern of over- and underrepresentation of trans vs. cis within populations may be produced. There may be too little cis-variation within populations relative to trans, or there may be too much cis-variation between populations relative to trans. We argue that cis- and trans-regulation undergo distinct population genetic dynamics across short and long timescales, which lead to a relative overabundance of trans-regulation within population and a relative overabundance of cis-regulation between populations. This inference follows from two observations. First, the mutation variance for trans-variation is substantially larger than the mutation variance for cis-variation. Indeed, small rates of single-nucleotide substitution (30) indicate that only a small fraction of the gene expression diversity generated in mutation-accumulation experiments may be ascribed to cis-variation. Accordingly, the trans-mutational target size has a much large contribution to gene expression variation among yeast mutation-accumulation lines than the cis-target size (5). Furthermore, the few hundred (2, 4) or a few thousand generations (5) with minimal selection observed in mutation-accumulation studies is already enough for trans-mutational variance to reach a level of variation beyond that typically detected among natural genotypes evolved under stabilizing selection (3). This also suggests that trans-effects might be more evolutionarily reversible.
Second, our findings that differences due to trans-regulation show higher degrees of dominance, whereas cis-variation arises from regulatory loci that are more additive (or with rare dominant alleles) may suggest a simple way by which too much trans within populations and too much cis between populations can be reconciled. Accordingly, despite selection against trans-regulatory variation within species being particularly strong because of a presumably larger pleiotropic effect of these mutations (13), substantial recessive variation with large trans-effects might still be maintained concealed in heterozygous in natural populations under mutation-selection balance. However, although cis-regulatory variation is produced at a slower rate than trans-variation, positive selection may act most efficiently on cis-regulatory variation, because allelic variation underlying cis differences might have greater additivity such that differences because of cis loci are less sensitive to genomic background. Hence, the higher additivity of cis-regulatory variation might underlie its preferential fixation between populations.
Materials and Methods
Fly Stocks, Second-Chromosome Extraction, and Genotyping.
Second chromosomes originating from five strains were substituted into an identical background with respect to the remaining autosomes, sex chromosomes, mitochondrial DNA, and cytoplasm (Fig. S1). Homogeneity of the background was verified by using primer pairs for PCR product-length polymorphisms (31). The strains from which second chromosomes were extracted were: BPL1d (PS1), BPL2a (PS2), and BPL8f (PS3) [containing second chromosomes originally collected from a wild population in Pennsylvania (15)], Canton-S (a commonly used laboratory strain collected in Ohio, in the 1940s), and Z53 (a strain collected in Zimbabwe). Males from strain PS1/PS1 and PS2/PS2 were then crossed with females from strain PS3/PS3, CS/CS, and Z53/Z53 to produce the F1 heterozygous genotypes PS3/PS1, PS1/CS, PS1/Z53, PS2/PS3, PS2/CS, and PS2/Z53. Males with genotypes PS1/PS1 and PS2/PS2 and their heterozygous combination with PS3, CS, and Z53 were profiled by microarrays (Fig. S2). Flies were grown under 24-h-light temperature (25°C) and humidity-controlled incubators. Newly emerged males were collected daily and allowed to age for 2 days, after which they were flash-frozen in liquid nitrogen and stored at −80°C.
Microarray Platform, Hybridizations, Quality Control, and Analyses.
An ≈18,000-feature array spotted primarily with PCR products designed for single exons was used. Detailed description of the PCR products can be found elsewhere (32). Spotting of the complete set on polyL-lysine coated slides (Erie) was carried out according to standard protocols (www.microarray.org). Total RNA was extracted from flash frozen males stored at −80°C using TRIZOL (Gibco-BRL, Life Technologies) and according to the manufacturer's recommendations. Total RNA samples were checked for quality by spectrophotometric analyses with A260/A280 ratios close to 2. The cDNA synthesis and hybridization reactions were carried out using 3DNA protocols and reagents (Genisphere) according to the manufacturer's recommendations. Upon hybridization, slides were scanned by using an Axon 4000B scanner (Axon Instruments) and the GenePix Pro 6.0 software (Axon Instruments). Hybridizations were carried out in a balanced loop design with dye swaps for a total of 32 hybridizations producing eight replicate measurements per expression (Fig. S2). Foreground fluorescence Cy5 and Cy3 intensities were normalized by the Loess method implemented in the library Limma of the statistical software R (33). Raw data were deposited in the National Center for Biotechnology Information GEO database, series reference number GSE12191. Significance of variation in gene expression across strains was assessed by using the Bayesian Analysis of Gene Expression Levels (BAGEL) (34). FDR were estimated based on the variation observed when randomized versions of the original dataset were analyzed in BAGEL. This procedure showed that at Bayesian Posterior Probability (BPP) >0.999, <10–20 genes are expected to be found differentially expressed by chance between any contrast, whereas ≈1,233 were found between PS1/PS1 and PS2/PS2 (FDR <0.02). We have adopted this threshold for all of the following contrasts: PS1/PS1 vs. PS2/PS2, PS1/PS3 vs. PS2/PS3, PS1/CS vs. PS2/CS, and PS1/Z53 vs. PS2/Z53. To select genes disrupted by the heterozygous effects of chromosomes PS3, CS, and Z53 we performed contrasts between PS1/PS1, vs. each of the following three relevant genotypes (PS1/PS3, PS1/CS, and PS1/Z53), and between PS2/PS2 vs. each of the following three relevant genotypes (PS2/PS3, PS2/CS, and PS2/Z53). The effect of each chromosome (PS3, CS, and Z53) was defined as those that are common to both backgrounds (PS1 and PS2). Carried out at BPP >0.99, this resulted in an FDR <0.01. Finally, third-chromosome substitution data were obtained from Hughes et al. (17). Normalization was carried out as in Hughes et al. (17), and differential expression was assessed with standard t tests.
Supplementary Material
Acknowledgments.
We thank J. Birchler, G. Gibson, N. Johnson, C. Meiklejohn, H.-Y. Wang, and P. Wittkopp for helpful comments on an early version of the manuscript; and K. Hughes (University of Illinois at Urbana-Champaign), L. McIntyre (University of Florida), and M. Wayne (University of Florida) for making their data available. This research was funded by National Institutes of Health Grant GM068465 (to D.L.H.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE12191).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805160105/DCSupplemental.
References
- 1.Clark TA, Townsend JP. Quantifying variation in gene expression. Mol Ecol. 2007;16:2613–2616. doi: 10.1111/j.1365-294X.2007.03354.x. [DOI] [PubMed] [Google Scholar]
- 2.Denver DR, et al. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- 3.Lemos B, Meiklejohn CD, Caceres M, Hartl DL. Rates of divergence in gene expression profiles of primates, mice, and flies: Stabilizing selection and variability among functional categories. Evol Int J Org Evol. 2005;59:126–137. [PubMed] [Google Scholar]
- 4.Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- 5.Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science. 2007;317:118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- 6.Lemos B, Bettencourt BR, Meiklejohn CD, Hartl DL. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol Biol Evol. 2005;22:1345–1354. doi: 10.1093/molbev/msi122. [DOI] [PubMed] [Google Scholar]
- 7.Voolstra C, Tautz D, Farbrother P, Eichinger L, Harr B. Contrasting evolution of expression differences in the testis between species and subspecies of the house mouse. Genome Res. 2007;17:42–49. doi: 10.1101/gr.5683806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol. 2004;21:1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- 9.Beldade P, Brakefield PM. The genetics and evo-devo of butterfly wing patterns. Nat Rev Genet. 2002;3:442–452. doi: 10.1038/nrg818. [DOI] [PubMed] [Google Scholar]
- 10.Hansen TF. The evolution of genetic architecture. Annu Rev Ecol Evol Syst. 2006;37:123–157. [Google Scholar]
- 11.West MA, et al. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics. 2007;175:1441–1450. doi: 10.1534/genetics.106.064972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- 13.Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104:8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura M. On the probability of fixation of mutant genes in a population. Genetics. 1962;47:713–719. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazzaro BP, Sceurman BK, Clark AG. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science. 2004;303:1873–1876. doi: 10.1126/science.1092447. [DOI] [PubMed] [Google Scholar]
- 16.Wu CI, et al. Sexual isolation in Drosophila melanogaster: A possible case of incipient speciation. Proc Natl Acad Sci USA. 1995;92:2519–2523. doi: 10.1073/pnas.92.7.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes KA, et al. Segregating variation in the transcriptome: Cis regulation and additivity of effects. Genetics. 2006;173:1347–1355. doi: 10.1534/genetics.105.051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wayne ML, Pan YJ, Nuzhdin SV, McIntyre LM. Additivity and trans-acting effects on gene expression in male Drosophila simulans. Genetics. 2004;168:1413–1420. doi: 10.1534/genetics.104.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown KM, Landry CR, Hartl DL, Cavalieri D. Cascading transcriptional effects of a naturally occurring frameshift mutation in Saccharomyces cerevisiae. Mol Ecol. 2008;17:2985–2997. doi: 10.1111/j.1365-294X.2008.03765.x. [DOI] [PubMed] [Google Scholar]
- 20.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. New York: Prentice Hall; 1996. [Google Scholar]
- 21.Wayne ML, et al. Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:18577–18582. doi: 10.1073/pnas.0705441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang HY, et al. Complex genetic interactions underlying expression differences between Drosophila races: Analysis of chromosome substitutions. Proc Natl Acad Sci USA. 2008;105:6362–6367. doi: 10.1073/pnas.0711774105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crow JF. Mutation, mean fitness, and genetic load. Oxford Surv Evol Biol. 1993;9:3–42. [Google Scholar]
- 24.Simmons MJ, Crow JF. Mutations affecting fitness in Drosophila populations. Annu Rev Genet. 1977;11:49–78. doi: 10.1146/annurev.ge.11.120177.000405. [DOI] [PubMed] [Google Scholar]
- 25.Charlesworth B, Charlesworth D. The genetic basis of inbreeding depression. Genet Res. 1999;74:329–340. doi: 10.1017/s0016672399004152. [DOI] [PubMed] [Google Scholar]
- 26.Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 27.Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: Biological implications. Trends Genet. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Dopman EB, Hartl DL. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:19920–19925. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Repping S, et al. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 30.Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–682. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- 31.Berger J, et al. Genetic mapping with SNP markers in Drosophila. Nat Genet. 2001;29:475–481. doi: 10.1038/ng773. [DOI] [PubMed] [Google Scholar]
- 32.Hild M, et al. An integrated gene annotation and transcriptional profiling approach towards the full content of the Drosophila genome. Genome Biol. 2003;5:R3. doi: 10.1186/gb-2003-5-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth GK. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 34.Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: Statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 2002;3:R71. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.