Abstract
Hepatitis C virus (HCV) NS3-4A is a membrane-associated multifunctional protein harboring serine protease and RNA helicase activities. It is an essential component of the HCV replication complex and a prime target for antiviral intervention. Here, we show that membrane association and structural organization of HCV NS3-4A are ensured in a cooperative manner by two membrane-binding determinants. We demonstrate that the N-terminal 21 amino acids of NS4A form a transmembrane α-helix that may be involved in intramembrane protein–protein interactions important for the assembly of a functional replication complex. In addition, we demonstrate that amphipathic helix α0, formed by NS3 residues 12–23, serves as a second essential determinant for membrane association of NS3-4A, allowing proper positioning of the serine protease active site on the membrane. These results allowed us to propose a dynamic model for the membrane association, processing, and structural organization of NS3-4A on the membrane. This model has implications for the functional architecture of the HCV replication complex, proteolytic targeting of host factors, and drug design.
Keywords: HCV, NMR, replication complex, serine protease, nonstructural protein
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide. HCV contains a 9.6-kb positive-strand RNA genome encoding a polyprotein precursor of ∼3,000 amino acids, which is co- and post-translationally processed by cellular and viral proteases to yield the structural and nonstructural proteins (reviewed in ref. 1). Nonstructural protein 3 (NS3) is a multifunctional protein, with a serine protease located in the N-terminal one-third (amino acids 1–180) and an RNA helicase in the C-terminal two-thirds (amino acids 181–631). The protease domain adopts a chymotrypsin-like fold with two β-barrel subdomains (reviewed in ref. 2). The catalytic triad is formed by His 57, Asp 81, and Ser 139. The 54-aa NS4A polypeptide functions as a cofactor for the NS3 serine protease. Its central portion comprises a β-strand that is incorporated into the N-terminal β-barrel of NS3 (3–5) whereas the hydrophobic N-terminal segment is required for membrane association (6) and the C-terminal acidic domain was recently shown to modulate HCV RNA replication (7).
NS3-4A has emerged as a prime target for antiviral intervention (reviewed in ref. 8). In addition, it has recently been shown that the NS3-4A protease cleaves, and thereby inactivates, two crucial adaptor proteins in innate immune sensing, namely Trif (9) and Cardif (10) (also known as MAVS, IPS-1, and VISA), thereby blocking IFN production. Thus, the NS3-4A complex plays an essential role in HCV replication and pathogenesis.
As in all positive-strand RNA viruses investigated thus far, the HCV nonstructural proteins form a membrane-associated replication complex. Here, we examined the determinants for membrane association of the NS3-4A complex through biochemical assays, site-directed mutagenesis, and CD and NMR structural analyses. We demonstrate that the N-terminal 21 amino acids of NS4A form a transmembrane α-helix required for integral membrane association of the NS3-4A complex. Moreover, we demonstrate that NS3 helix α0 represents a second essential determinant for membrane association, allowing proper positioning of the serine protease active site on the membrane. These results allowed us to propose a dynamic model for the membrane association, processing, and structural organization of NS3-4A on the membrane.
Results
The N-Terminal Segment of NS4A Forms a Transmembrane α-Helix.
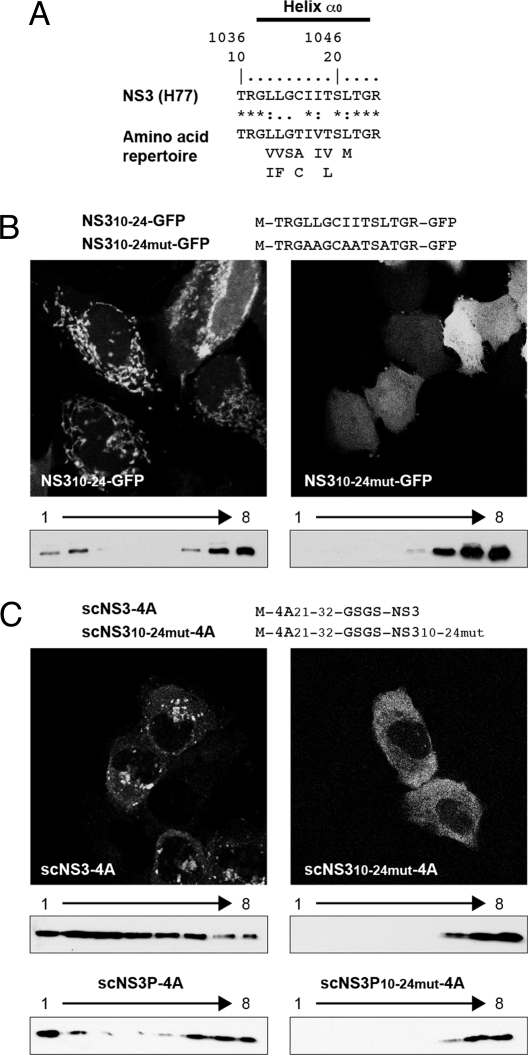
The amino acid repertoire derived from ClustalW alignment of 26 reference sequences representative of all major HCV genotypes and subtypes revealed that the N-terminal predicted membrane segment (amino acids 1–20) and the C-terminal acidic domain (amino acids 40–54) are highly conserved (Fig. 1A). By contrast, the central portion of NS4A, including the NS3 cofactor (amino acids 21–32) and kink (amino acids 33–39) segments, appears much more variable. However, this apparent variability is limited at most positions because the observed residues exhibit similar physico-chemical properties.
Fig. 1.
Identification of the membrane segment of NS4A. (A) Amino acid sequence analyses. The NS4A sequence from the HCV H77 consensus clone (GenBank accession no. AF009606) is shown at the top. Amino acids are numbered with respect to NS4A and the HCV polyprotein (top row). The amino acid repertoire of 26 reference sequences representative of all major HCV genotypes and subtypes (http://euhcvdb.ibcp.fr; ref. 21) is given below, with the observed amino acids listed in decreasing order of frequency and the similarity index according to ClustalW convention (asterisk, invariant; colon, highly similar; dot, similar). (B) Subcellular localization of NS4A-GFP fusion constructs. U-2 OS cells transiently transfected with pCMVNS4A-GFP (NS4A) or C- and N-terminal NS4A deletion constructs fused to GFP (see SI Text for details) were examined by fluorescence microscopy. (C) Membrane topology of NS4A. Lysates of U-2 OS cells transfected with pCMVgt-NS4A-GFP or pCMVgtmut-NS4A-GFP (see SI Text for details) were digested with endoglycosidase H (Endo H) or N-glycosidase F (N-glyc F), followed by 15% SDS/PAGE and immunoblot with monoclonal antibody JL-8 against GFP (Clontech). The position of glycosylated gt-NS4A-GFP is indicated by an asterisk.
We and others have previously shown that NS4A mediates membrane association of the NS3-4A complex (6). A panel of NS4A deletion constructs fused to the GFP was prepared to determine the segment required for membrane association. As shown in Fig. 1B, constructs bearing 21 or more N-terminal amino acid residues of NS4A showed a membrane-associated fluorescence pattern. By contrast, the other constructs showed a diffuse fluorescence pattern. These observations were confirmed by membrane flotation analyses (data not illustrated). Taken together, these data indicate that the N-terminal 21 amino acids of NS4A mediate membrane association.
Next, an NST consensus motif for N-linked glycosylation, followed by a 14-aa spacer sequence, was added to the N terminus of NS4A to determine its membrane topology (gt-NS4A-GFP, Fig. 1C). A construct harboring Gln instead of Asn within the glycosylation acceptor site served as control (gtmut-NS4A-GFP). As shown in Fig. 1C, an additional +3-kDa band was observed for construct gt-NS4A-GFP, indicating that the added Asn residue was glycosylated by oligosaccharyltransferase present in the endoplasmic reticulum (ER) lumen. Digestion with both endoglycosidase H and N-glycosidase F led to disappearance of this band, with the remaining band migrating at the same position as gtmut-NS4A-GFP. These results indicate that the N-terminal segment of NS4A traverses the phospholipid bilayer as a transmembrane segment and that, given the endoglycosidase H-sensitive nature of the transferred carbohydrate moiety, NS4A is retained in the ER. The transmembrane topology of the N-terminal segment of NS4A was confirmed by insertion of a glycosylation acceptor site in the context of a functional NS3-4A complex [supporting information (SI) Fig. S1]. As expected, studies involving a proteolytically inactive NS3-4A mutant and differential membrane extraction and flotation analyses indicated that processing between NS3 and NS4A is required for integral membrane association of the NS3-4A complex (Fig. S1 and data not illustrated).
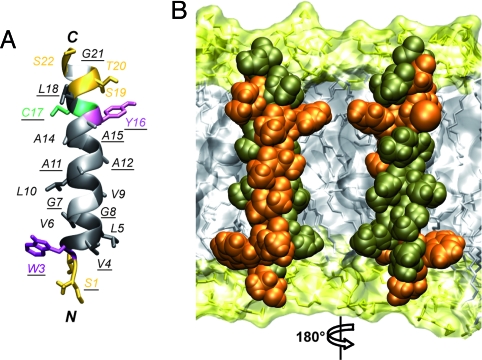
The structure of the transmembrane segment of NS4A was examined by CD and NMR by using a synthetic peptide with KKGG and GGKK solubilization tags at the N- and C-terminal ends, respectively, designated as NS4A[1-22]*. After solubilization in various membrane mimetics NS4A[1-22]* exhibited CD spectra typical for an α-helix (Fig. S2). For NMR, the peptide was in 50% TFE-d2. An overview of nuclear Overhauser enhancement (NOE) connectivities and the deviation of 1Hα chemical shifts from random coil values are shown in Fig. S3. These analyses clearly showed that the main body of the peptide (Thr 2–Thr 20) forms an α-helix. On the basis of the NOE-derived interproton distance constraints, a structural model of NS4A[1-22]* was calculated that fully satisfied the experimental NMR data (Fig. 2A and Table S1). The NS4A transmembrane α-helix displays several remarkable features, including an unusually high number of small Gly and Ala residues, an absolutely conserved side (Fig. 2B, residues highlighted in orange), and typical interface residues (Trp 3 and Tyr 16) at both ends. As discussed below, the numerous well-conserved small amino acid residues may play a role in the post-translational membrane insertion process and their conservation on one side of the helix may point to intramembrane protein–protein interactions.
Fig. 2.
Structure of the NS4A transmembrane segment. (A) Ribbon representation of the best representative structure of NS4A[1-22]* selected from the final set of 26 calculated NMR structures (BMRB entry 15580). Residue side chains are shown as sticks and are colored on the basis of the chemical properties of their side chains (hydrophobic, dark gray; polar, yellow). Gly are light gray. Tyr, Trp, and Cys are violet, magenta, and green, respectively. Fully conserved residues are underlined. (B) Amino acid van der Waals representation and tentative position of NS4A amino acids 1–22 within a phospholipid bilayer. Fully and less conserved residues are colored orange and bronze, respectively. Orientation of the left structure is the same as in image A. Note that the majority of fully conserved residues are located on one side of the transmembrane α-helix. The membrane is represented as a simulated model of a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer (http://moose.bio.ucalgary.ca). Polar heads and hydrophobic tails of phospholipids (surface and stick structures) are light yellow and gray, respectively.
Mutational Analysis of the NS4A Membrane Anchor.
The high degree of amino acid sequence conservation within the NS4A transmembrane segment suggests that it may have additional functions apart from serving as membrane anchor. To further explore this idea, we performed site-directed mutagenesis of selected conserved amino acid residues and examined the polyprotein processing, membrane association, and RNA replication of these mutants. In addition, the formation of cytoplasmic dot-like structures as a correlate of replication complex assembly (1) was investigated by immunofluorescence microscopy. Mutations were designed to preserve the α-helical fold of the NS4A transmembrane segment. All mutants were properly processed and membrane-associated when expressed in the context of the HCV polyprotein (data not illustrated). When analyzed in the context of selectable subgenomic replicons, mutants G8L and G21V did not yield any colonies whereas RNA replication of mutants L13A/A14L and C17A was only moderately impaired (Table 1 and Fig. S4). Mutant Y16F yielded only a few viable clones. Interestingly, mutant G21V revealed a striking loss of dot-like structures, suggesting a defect in replication complex assembly, whereas the replication defect of mutants G8L and Y16F may be more subtle, possibly resulting from disturbed intramembrane protein–protein interactions within the replication complex (Fig. S4).
Table 1.
Analysis of NS4A membrane anchor mutants
NS3 Helix α0 Mediates Membrane Association.
The previously stated results define the N-terminal transmembrane segment of NS4A as a key determinant for membrane association of NS3-4A. However, close inspection of available NS3-4A structures revealed a peculiar amphipathic α-helix at the N terminus of NS3, represented by amino acid residues 12–23 and designated α0, which forms a hydrophobic patch on the protein surface and was previously hypothesized to interact with membranes (4).
As shown in Fig. 3A, the NS3 amino acid 10–24 segment is highly conserved. In particular, hydrophobic residues at positions 13, 14, 17, 18, and 21 are conserved among all HCV genotypes, forming a strongly hydrophobic helix side on the protein surface whereas the polar helix side interacts with the rest of the protein. A construct harboring the NS3 amino acid 10–24 segment fused to GFP, NS310–24-GFP, was prepared in order to examine the potential of helix α0 to associate with membranes. Construct NS310–24mut-GFP, in which the 5 hydrophobic residues Leu 13, Leu 14, Ile 17, Ile 18, and Leu 21 were replaced by Ala, served as control (Fig. 3B). As shown by CD analyses of the corresponding synthetic peptide, these changes preserved the α-helical fold propensity although abrogating the hydrophobic character of amphipathic helix α0 (Fig. S2). As shown in Fig. 3B, NS310–24-GFP displayed a membrane-associated fluorescence pattern whereas NS310–24mut-GFP was distributed diffusely. Membrane flotation analyses confirmed that NS310–24-GFP, but not NS310–24mut-GFP, associates with membranes (Fig. 3B, bottom).
Fig. 3.
NS3 helix α0 mediates membrane association. (A) Sequence analyses of the NS3 amino acid 10–24 segment. See Fig. 1 legend for details. (B) Membrane association of NS3 helix α0-GFP fusion construct. Constructs NS310–24-GFP or NS310–24mut-GFP (see SI Text for details) were expressed in U-2 OS cells and examined by fluorescence microscopy and membrane flotation analyses. Monoclonal antibody JL-8 against GFP was used for immunoblot. Membranes float to the upper, low-density fractions (left portion of the blots). (C) Membrane association of NS3-4A single-chain constructs. Constructs scNS3–4A, scNS310–24mut-4A, scNS3P-4A, and scNS3P10–24mut-4A (see SI Text for details) were examined by immunofluorescence microscopy and membrane flotation by using monoclonal antibody 1B6 against NS3 (3).
To explore the role of NS3 helix α0 in the membrane association of the NS3-4A complex, single-chain (sc) constructs were prepared in which the central portion of NS4A was fused to the N terminus of either full-length NS3 (scNS3–4A) (5) or the protease domain alone (scNS3P-4A). Constructs scNS310–24mut-4A and scNS3P10–24mut-4A harbor the Ala substitutions described above. As shown in Fig. 3C, scNS3–4A displayed a membrane-associated staining pattern with formation of granular structures in the cytoplasm. By contrast, diffuse staining with sparing of the nucleus was observed for scNS310–24mut-4A. These results were confirmed by membrane flotation analyses (Fig. 3C). Similar observations were made for scNS3P-4A and scNS3P10–24mut-4A (Fig. 3C, bottom). The broad distribution of scNS3–4A and, to a lesser extent, scNS3P-4A in the density gradient may be explained by the formation of micellar protein aggregates, mainly because of the hydrophobic Leu and Ile residues in helix α0.
Structural Analyses of NS3 Helix α0 and Implications for the Membrane Topology of NS3-4A.
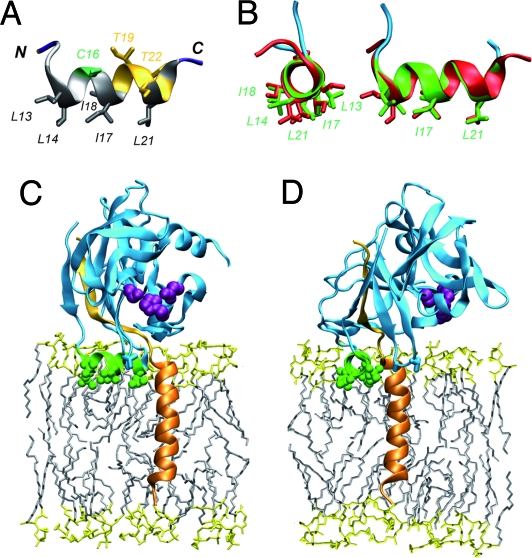
The lipophilic properties and structure of NS3 helix α0 were examined by CD and NMR in membrane-mimetic media by using a synthetic peptide representing the NS3 amino acid 10–24 segment, NS3[10–24] (Fig. S2). This peptide is unfolded in water but folds into an α-helix in all membrane mimetics tested, indicating the lipophilic properties of helix α0. NMR analyses of NS3[10–24] in 50% TFE-d2 and 100 mM SDS-d25 yielded well-resolved spectra that allowed complete sequential attribution and, together with the 1Hα and 13Cα chemical shift variation (Fig. S3), revealed an amphipathic α-helix between Leu 13 and Thr 22 (Fig. 4A). Importantly, the structure resolved by NMR in membrane-mimetic media was perfectly superposable to the structure of helix α0 as present in the X-ray structure of NS3-4A (Fig. 4B). Taken together, the CD and NMR data indicate that amphipathic helix α0 likely folds upon interaction with the membrane interface. Importantly, the positioning of amphipathic helix α0 in an in-plane topology at the membrane interface, with the hydrophobic residues oriented toward the hydrophobic membrane core, and of the transmembrane α-helix of NS4A dictate the topology of the NS3 serine protease domain on the membrane (Fig. 4 C and D).
Fig. 4.
Structural analyses of NS3 helix α0. (A) Ribbon representation of the best representative structure of NS3[10–24] selected from the final set of 26 calculated NMR structures in 100 mM SDS-d25 (BMRB entry 15582). Residue side chains are colored as in Fig. 2A. (B) Comparison of NS3 amino acid 10–24 structures obtained by NMR (red) and by X-ray crystallography (ref. 5; PDB entry 1CU1; green and cyan). The two structures were superimposed from residues 12–23. Left and right images correspond to axial and perpendicular views. (C and D) Model of the membrane-associated NS3 serine protease domain complexed with NS4A (perpendicular and axial views relative to NS3 helix α0). This model was constructed by using the coordinates reported by Yao et al. (ref. 5; PDB entry 1CU1) and the structure of NS4A[1-22]* reported in Fig. 2. The NS3 serine protease domain is cyan, with side chain atoms of the catalytic triad (His 57, Asp 81, and Ser 139) highlighted as purple spheres. NS3 helix α0 is green and the five hydrophobic residues are represented by sticks and balls. The N-terminal transmembrane (amino acids 1–20) and central (amino acids 21–32) segments of NS4A are orange and light orange, respectively. The membrane is represented as a simulated model of POPC bilayer (see Fig. 2 legend for details).
Discussion
Here, we show that membrane association of NS3-4A is conferred by two determinants, amphipathic helix α0, formed by NS3 residues 12–23, which interacts in-plane with the membrane interface, and the N-terminal 21 amino acids of NS4A, which form a transmembrane α-helix. These observations and available structural data allowed us to propose a dynamic model for the membrane association, processing, and structural organization of NS3-4A on the membrane (Fig. 5; video available at http://www.ibcp.fr/en/gallery/49/gallery.php).
Fig. 5.
Mechanistic model of the membrane association process of NS3-4A. See Discussion for comments. The cytosolic side of the membrane bilayer is on the top. (1) N-terminal portion of NS3 (amino acids 13–23 highlighted in green) before folding of helix α0 (constructed by using the NS3 serine protease structure without the central segment of NS4A solved by X-ray crystallography (ref. 24; PDB entry 1A1Q). (2) Membrane association of uncleaved NS3-NS4A by NS3 helix α0. The NS4A structure is undefined at this step. It is represented as sticks with N-terminal segment 1–20, central segment 21–32, and the beginning of the C-terminal segment (amino acids 33–40) in medium, light, and dark orange, respectively. The NS3 structure was constructed by using: (i) The crystal structure of scNS3–4A (ref. 5; PDB entry 1CU1), where the C terminus of the helicase domain (silver) lies within the active site of the serine protease domain (cyan); (ii) The NS3 serine protease structure without the central segment of NS4A solved by NMR (ref. 25; PDB entry 1BT7) for the N-terminal β-barrel subdomain; and (iii) The NMR structure of NS3[10–24] comprising helix α0 (this study). Well folded structures (helix α0, C-terminal β-barrel subdomain, and helicase domain) are represented as ribbon diagrams whereas the less stable or unfolded structures are represented as sticks (NS3 segment 1–9 and N-terminal β-barrel subdomain). Side-chain atoms of the catalytic triad (His 57, Asp 81, and Ser 139) are highlighted as purple spheres. (3) Cleavage at the NS3/NS4A site allows membrane insertion of the N-terminal segment of NS4A, resulting in a transmembrane α-helix. (4) Cofolding of the central segment 21–32 of NS4A into the N-terminal β-barrel subdomain stabilizes the structure of the serine protease, which is locked onto the membrane by NS3 helix α0 and the NS4A transmembrane α-helix. Note that the hydrophilic helicase domain would be partially immersed into the membrane in the NS3-NS4A cis-cleavage conformation (5) (model in brackets). (5) Final topology of the NS3-4A complex on the membrane.
In the HCV polyprotein context, translation of NS3 likely occurs at the membrane (Fig. 5, step 1). Induced folding of amphipathic helix α0 upon interaction with the membrane interface may therefore represent a cotranslational event, followed by folding of the protease and helicase domains (Fig. 5, step 2). The zinc molecule likely plays a central role in the folding of the C-terminal β-barrel subdomain of the NS3 serine protease (11). The folding of uncleaved NS4A is not defined at this stage. As its central portion is required for cleavage at the NS3/NS4A site (12), it is expected that this segment interacts with the protease domain before cleavage. However, tight incorporation of the central NS4A β-strand into the N-terminal β-barrel subdomain of the protease would induce its final folding and, as subsequently discussed, would break the protease–helicase interaction. Thus, a low affinity interaction with NS3 serine protease β-strands A1 and A0 (see ref. 2 for the nomenclature of NS3-4A secondary structure elements) may be postulated at this point. At this stage, the in-plane membrane association of helix α0 does not impose any constraints on the positioning of NS3, which could thus participate in NS2–NS3 processing. Forward movement of NS3 could bring the hydrophobic N-terminal segment of NS4A into close contact with the membrane, thereby facilitating its post-translational insertion into the membrane after processing at the NS3/NS4A site (Fig. 5, step 3). Final incorporation of the central segment of NS4A induces the cofolding of composite β-sheet A0(NS3)-D1′(NS4A)-A1(NS3) within the N-terminal β-barrel which in turn stabilizes the interaction of helix α0 with the NS3 serine protease. This complete folding and membrane association by amphipathic helix α0 and the transmembrane segment of NS4A lock the protease in a strictly defined position onto the membrane (Fig. 5, step 4). As shown in the brackets, the hydrophilic helicase domain would be immersed into the membrane at this stage in the NS3-4A cis-cleavage conformation (5). Hence, the helicase domain has to move away from the protease in the final membrane-associated stage through a rotation of the linker segment connecting the two domains (Fig. 5, step 5). As a consequence, the helicase domain is free to interact with other components of the HCV replicase. Moreover, one should postulate a second conformation of the NS3-4A complex, with the protease and helicase domains interacting by contacts different from the ones identified in the cis-cleavage structure. Indeed, the recently reported crystal structure of NS3 from the related Dengue virus shows a relative orientation between the protease and helicase domains drastically different from the one known for the HCV NS3-4A cis-cleavage structure, resulting in an elongated shape of the molecule (13). Interestingly, a similar conformation on the membrane has recently been proposed for the NS2B-3 serine protease of the related flaviviruses (14, 15). In this case, hydrophobic residues within an N-terminal hairpin loop of NS3 are spatially homologous to the hydrophobic residues of helix α0, suggesting convergent evolution of the membrane binding elements.
Our model suggests a possible interaction between NS3 helix α0 and the transmembrane α-helix of NS4A around NS4A amino acids 19–22. This may explain the absolute conservation of Gly 21 and the dramatic phenotype of NS4A mutant G21V. As a further consequence of the positioning of helix α0 at the membrane interface, NS3 loop 38–40 is expected to contact the membrane interface, resulting in tripod-like positioning of the NS3-4A complex on the membrane surface (Fig. 4 C and D).
This positioning has implications for polyprotein processing by NS3-4A. The first trans-cleavage occurs rapidly and without absolute requirement for NS4A (12) at the NS5A/NS5B site. As this site is likely not located at the membrane surface, it is conceivable that cleavage already occurs before full incorporation of the central NS4A segment into the NS3 serine protease and its locking onto the membrane (Fig. 5, step 3). In this scenario, however, the helicase domain has to already move away from the protease to accommodate trans-cleavage between NS5A and NS5B. The resulting NS4A-5A precursor is cleaved first between NS4A and NS4B, yielding in a relatively stable NS4B-5A intermediate, and finally between NS4B and NS5A. As the NS4A/NS4B and NS4B/NS5A cleavage sites are predicted to be located at the membrane surface, it is likely that trans-cleavage at these sites is performed by the NS3-4A protease in its final membrane-associated conformation (Fig. 5, step 5).
Another important consequence of our model relates to the proteolytic targeting of host factors. In this context, strict positioning of the protease active site with respect to the membrane confers a high degree of selectivity to potential cellular trans-cleavage substrates. Indeed, NS3-4A cleavage of the RIG-I adaptor Cardif at Cys 508 occurs very close to its C-terminal transmembrane segment (amino acids 514–535), resulting in displacement from the outer mitochondrial membrane and inactivation of Cardif (10, 16). Of note, we have previously shown that a minor proportion of NS3-4A localizes to mitochondria (6).
Our results show that NS3 helix α0 is a key determinant in the membrane association process of NS3-4A. Accordingly, mutant constructs harboring Ala substitutions of the five hydrophobic residues in helix α0 showed major defects in the association between NS3 and NS4A, polyprotein processing, and cleavage of Cardif (data not illustrated). Interestingly, helix α0 overlaps with an important HLA-A2-restricted cytotoxic T lymphocyte epitope (NS3 amino acids 12–21) (17). The central role of helix α0 in NS3-4A function likely explains the conservation of this epitope which may render it an attractive candidate for immunotherapeutic interventions.
We also report here that certain NS4A membrane anchor mutants show a striking discordance between preserved polyprotein processing and membrane association on the one hand and RNA replication on the other hand. We have previously reported similar observations for the membrane anchors of HCV NS5A (18) and NS5B (19), suggesting that these segments have additional functions and are likely involved in protein–protein interactions essential for the assembly of a functional replication complex. Mutant G8L is particularly noteworthy in this regard, as glycines are frequently involved in transmembrane helix–helix interactions (20). In addition, the high degree of amino acid conservation within the NS4A transmembrane segment may be related to its unusual mechanism of post-translational membrane insertion.
In conclusion, we demonstrate that membrane association of HCV NS3-4A is conferred by two structural determinants, NS3 amphipathic helix α0 and the transmembrane segment of NS4A. On the basis of these results we propose a dynamic molecular model in which the sequential membrane association of both determinants plays an active role in the processing and structural organization of NS3-4A and its final topology on the membrane. This model has implications for the functional architecture of the HCV replication complex, proteolytic targeting of host factors, and drug design.
Materials and Methods
Sequence Analyses.
Sequence analyses were performed by using the European Hepatitis C Virus Database (http://euhcvdb.ibcp.fr; ref. 21).
Expression Constructs.
HCV sequences were derived from the HCV H77 consensus (22) and Con1 clones (23). Details are reported in SI Text and Table S2.
Immunofluorescence Microscopy, Membrane Flotation, and Immunoblot.
Immunofluorescence microscopy, membrane flotation, and immunoblot were performed as described (18).
Peptide Synthesis and Purification.
Peptides NS3[10–24] (TRGLLGCIITSLTGR), NS3[10–24]mut (TRGAAGCAATSATGR), and NS4A[1-22]* (KKGGSTWVLVGGVLAALAAYCLSTGSGGKK) were synthesized by Clonestar Biotech and purified by RP-HPLC (purity >98%).
Structure Determination by CD and NMR.
CD, NMR spectroscopy, NMR-derived constraints and structure calculation, and molecular modeling and structure representation were performed by standard approaches as described in SI Text.
Supplementary Material
Acknowledgments.
We thank Raffaele de Francesco for discussion and critical review of the manuscript; Elke Bieck and Anja Wahl for excellent technical assistance; Charles M. Rice, Ralf Bartenschlager, and Jan Albert Hellings for reagents; Christophe Combet for bioinformatics supports; and Emmanuel Bettler for video preparation. This work was supported by the Swiss National Science Foundation (3100A0–107831/1), the Swiss Cancer League (OCS-01762–08-2005), the Leenaards Foundation, the Deutsche Forschungsgemeinschaft (Mo 799/1–3 and Br 3440/2–1), the Bundesministerium für Bildung und Forschung (01 KI 9951), the European Commission (LSHM-CT-2004–503359, VIRGIL), and the French Centre National de la Recherche Scientifique and Agence Nationale de Recherches sur le SIDA et les Hépatites Virales.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates of have been deposited in the BioMagResBank (BMRB) (accession nos. 15580 (NS4A[1-22]*) and 15582 (NS3[10-24]).
This article contains supporting information online at www.pnas.org/cgi/content/full/0807298105/DCSupplemental.
References
- 1.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 2.De Francesco R, Steinkühler C. Structure and function of the hepatitis C virus NS3-NS4A serine protease. Curr Top Microbiol Immunol. 2000;242:149–169. doi: 10.1007/978-3-642-59605-6_8. [DOI] [PubMed] [Google Scholar]
- 3.Kim JL, et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 4.Yan Y, et al. Complex of NS3 protease and NS4A peptide of BK strain of hepatitis C virus: A 2.2 Å resolution structure in a hexagonal crystal form. Protein Sci. 1998;7:837–847. doi: 10.1002/pro.5560070402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao N, et al. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure Fold Des. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 6.Wölk B, et al. Subcellular localization, stability and trans-cleavage competence of the hepatitis C virus NS3–NS4A complex expressed in tetracycline-regulated cell lines. J Virol. 2000;74:2293–2304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindenbach BD, et al. The C-terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J Virol. 2007;81:8905–8918. doi: 10.1128/JVI.00937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Francesco R, Carfi A. Advances in the development of new therapeutic agents targeting the NS3-4A serine protease or the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Adv Drug Deliv Rev. 2007;59:1242–1262. doi: 10.1016/j.addr.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 9.Li K, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 11.Urbani A, et al. The metal binding site of the hepatitis C virus NS3 protease. A spectroscopic investigation. J Biol Chem. 1998;273:18760–18769. doi: 10.1074/jbc.273.30.18760. [DOI] [PubMed] [Google Scholar]
- 12.Tomei L, et al. A central hydrophobic domain of the hepatitis C virus NS4A protein is necessary and sufficient for the activation of the NS3 protease. J Gen Virol. 1996;77:1065–1070. doi: 10.1099/0022-1317-77-5-1065. [DOI] [PubMed] [Google Scholar]
- 13.Luo D, et al. Crystal structure of the NS3 protease-helicase from Dengue virus. J Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chernov AV, et al. The two-component NS2B–NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J Biol Chem. 2008;25:17270–17278. doi: 10.1074/jbc.M801719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XD, et al. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fournillier A, et al. An accelerated vaccine schedule with a poly-antigenic hepatitis C virus MVA-based candidate vaccine induces potent, long lasting, and in vivo cross-reactive T cell responses. Vaccine. 2007;25:7339–7353. doi: 10.1016/j.vaccine.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Penin F, et al. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem. 2004;279:40835–40843. doi: 10.1074/jbc.M404761200. [DOI] [PubMed] [Google Scholar]
- 19.Moradpour D, et al. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J Virol. 2004;78:13278–13284. doi: 10.1128/JVI.78.23.13278-13284.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senes A, Ubarretxena-Belandia I, Engelman DM. The Cα—H·O hydrogen bond: A determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Combet C, et al. euHCVdb: The European hepatitis C virus database. Nucleic Acids Res. 2007;35:D363–366. doi: 10.1093/nar/gkl970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolykhalov AA, et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 24.Love RA, et al. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 25.Barbato G, et al. The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism. J Mol Biol. 1999;289:371–384. doi: 10.1006/jmbi.1999.2745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.