Abstract
Antigen specific T cell migration to sites of infection or cancer is critical for an effective immune response. In mouse models of cancer, the number of lymphocytes reaching the tumor is typically only a few hundred, yet technology capable of imaging these cells using bioluminescence has yet to be achieved. A combination of codon optimization, removal of cryptic splice sites and retroviral modification was used to engineer an enhanced firefly luciferase (ffLuc) vector. Compared with ffLuc, T cells expressing our construct generated >100 times more light, permitting detection of as few as three cells implanted s.c. while maintaining long term coexpression of a reporter gene (Thy1.1). Expression of enhanced ffLuc in mouse T cells permitted the tracking of <3 × 104 adoptively transferred T cells infiltrating sites of vaccination and preestablished tumors. Penetration of light through deep tissues, including the liver and spleen, was also observed. Finally, we were able to enumerate infiltrating mouse lymphocytes constituting <0.3% of total tumor cellularity, representing a significant improvement over standard methods of quantitation including flow cytometry.
Keywords: bioluminescence, immunology, molecular biology
Complete resolution of infection or cancer often depends on an effective T cell mediated immune response. In many cases, in particular that of a tumor setting, only hundreds of tumor-specific T cells traffic to the tumor. To date, the signal intensity generated by bioluminescent (BLI) reporter genes has been insufficient to track fewer than tens of thousands of T cells in living animals. This has made it very difficult to study strategies for improving tumor homing.
In addition to BLI, positron emission tomography (PET) (1) and intravital microscopy (IVM) (2, 3) offer dynamic imaging but are less applicable to routine preclinical studies because of their expense and laborious nature. PET also requires the use of radioactive substrates and IVM is highly invasive and generally requires that studied animals be euthanized. Recently, IVM was used to image tumor specific T cells at tumor sites indicating that tumor cell killing by an individual T cell occurs slowly (> 6 h) (4).
Pioneered by Contag et al. and using bioluminescent bacteria (5), BLI is among the most commonly used modalities for noninvasive imaging of small animals. To date, the luciferase genes from Photinus pyralis (American firefly; ffLuc), Pyrophorus plagiophthalamus (CB-Luc), Renilla reniformis (rLuc), and Gaussia princes (gLuc) are the most commonly used light emitting reporters. Of these, only ffLuc and CB-Luc catabolize a substrate (d-Luciferin) that is relatively stable in vivo. gLuc and rLuc catabolize coelenterazine but their use is limited by background luminescence (6), rapid clearance of their substrate, and flash kinetics and blue/green emission, respectively (7, 8).
Mouse T cells expressing ffLuc have been obtained from transgenic (tg) mice (9–11), bone marrow (BM) chimeric mice (obtained after lentiviral transduction of donor BM) (12), and direct retroviral transduction of T cells in vitro (13, 14). Unfortunately, ffLuc is expressed poorly in mouse T cells resulting in poor sensitivity of detection (at the level of thousands of cells), requiring investigators to acquire images with high binning densities [8–16 pixels (px) grouped together) to capture weak signals in general areas (9, 13–15). In digital imaging, a px is the smallest piece of data in an image. For review, see ref. 16.
Experience from human clinical trials employing the adoptive transfer of expanded autologous tumor-infiltrating T cells into cancer patients indicates that very small numbers of lymphocytes (<0.005% per gram of tumor) reach the tumor (17). To date, most reports studying T cell trafficking to tumors have been performed in xenograft models because expression of ffLuc in human T cells provides sufficient sensitivity for BLI (18–23). These models, however, do not consider the complex interplay of T cells with an autologous immune system. Several investigators have thus attempted to use BLI for T cell trafficking in immunocompetent models of cancer, but because of the weak signal intensity generated by ffLuc-expressing T cells, images had to be acquired using high binning density (ρ) (≤256 × 256 px) (15, 24). Comparison of xenogeneic and syngeneic systems has been recently performed in a model of mammary carcinoma in which BLI was clearly superior in the xenogeneic system (24). Other models investigating the trafficking of ffLuc-expressing mouse T cells have also necessitated high px ρ, thus limiting spatial analysis. These include collagen-induced arthritis (CIA) (14), experimental autoimmune encephalitis (EAE) (13) and graft versus host disease (GVHD) (10, 12, 25).
We engineered an enhanced version of ffLuc (effLuc) encoded within a retroviral vector to facilitate detection of <10, 000 T cells at a given site in living animals. Retroviral transduction was chosen because it is a rapid procedure (3 days) for achieving stable expression of a given gene, which facilitates subsequent experimentation within days to weeks. Compared with mouse T cells expressing ffLuc, those expressing effLuc generated >100 times as much detectable light. Further, the augmented signal intensity provided the means to image as few as three effLuc-expressing T cells and to examine the limits of detection of cells expressing effLuc in models of vaccination and adoptive immunotherapy of established tumors.
Results
Construction of an Enhanced Firefly Luciferase and Expression in Mouse T Cells.
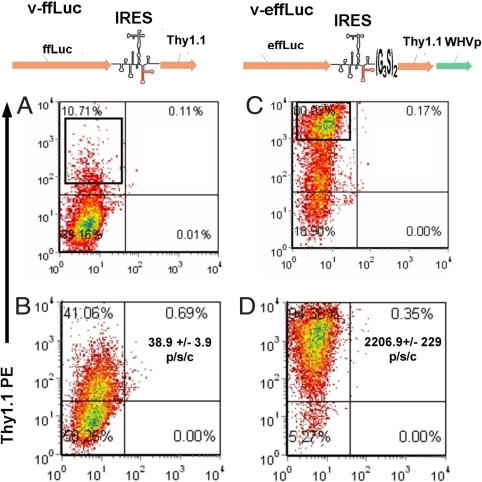
We made modifications to an ffLuc encoding retroviral vector (pMSCV-ffLuc-pIRES2-Thy1.1 [v-ffLuc]) to improve expression of the reporter transgene, in particular for mouse T cells. Thy1.1 was chosen as the coreporter because it differs from its allelic variant Thy1.2 sufficiently to be detected by a specific monoclonal antibody but is not immunogenic in Thy1.2+ strains. Mouse CD8 OT-1 TCR tg T cells specific for Ovalbumin peptide 257–264 in the context of H2-kb (26) transduced with v-ffLuc demonstrated weak Thy1.1 (flow cytometry) and luciferase activity (in vitro). (Fig. 1 A and B). To generate an improved construct, effLuc, we added a (G3S)2 linker to the 3′ end of the pIRES2 [supporting information (SI) Fig. S1], codons were optimized to the highest frequency of Mus musculus and a Woodchuck Hepatitis Virus (WHV) pre element was added downstream of Thy1.1 to augment export of the viral mRNA into the cytosol (27). Construct illustrations are shown in Fig. 1. Codon optimization lead to the modification of three consensus acceptor splice sites (positions 131, 894, and 1004), 9 cryptic acceptor sites (positions 194, 305, 551, 639, 669, 801, 1011, 1112, and 1403) and five cryptic donor sites (positions 334, 349, 1083, 1169, and 1173). One consensus (position 1433) and two cryptic donor splice sites (positions 334 and 1173) from the ffLuc ORF were eliminated by the GeneOptimizer software. Removal of cryptic splice sites was considered important for mouse T cells because excessive aberrant splicing has been reported as a protective tactic used by mouse T cells against retroviral infection (28).
Fig. 1.
Comparison of coreporter expression and in vitro luminescent activity of T cells expressing standard versus enhanced retroviral firefly luciferase constructs. OT.1 T cells were transduced with the retroviruses v-ffLuc (A and B) or v-effLuc (C and D) (illustrations of vector coding regions are shown above each column) and assessed for Thy1.1 expression 3 days later via flow cytometry (A and C). The boxed areas represent the regions chosen for sorting. 7 days after sorting, transduced T cells were reassessed for longevity of Thy1.1 expression. (B and D) OT.1 T cells (1 × 105) transduced with the indicated retroviruses were assessed for bioluminescent activity before sorting and after addition of d-Luciferin. Signal intensity (photons per second per cell) is in the top right quadrant of B and D.
These modifications resulted in >80% of effLuc-transduced T cells expressing Thy1.1 (a 6-fold increase) and a >100-fold increase in Thy1.1 mean fluorescent intensity in the unsorted population. Less than 25% of effLuc OT-I lost Thy1.1 expression over 7 days of culture compared with >85% for ffLuc. During this time, relative to ffLuc-expressing T cells, luciferase acitivity from effLuc-expressing OT-I T cells increased from 60% to >250% (Fig. S2B). After sorting, we found that effLuc-transduced (compared with ffLuc-transduced) T cells demonstrated a >55-fold increase in intensity [photon flux (the radiance (photons per second) in each px integrated over the ROI area (cm2) × 4π at a given cell number]. The signal intensity of OT-I effLuc versus OT-1 ffLuc was 2,206.9 ± 229 photons per second per cell and 38.9 ± 3.9 photons per second per cell, respectively (Fig. 1 B and D). We also observed a 100- to 110-fold increase in sensitivity (number of cells detected at the same photon flux) (Table S1). Viral integration, measured on days 3 and 5 after transduction, was 4- to 6-fold better for v-effLuc versus v-ffLuc (Fig. S3), thus the increase in luciferase acitivity is a function of both integration efficiency and expression efficiency.
Because the levels of firefly luciferase expression reported here are superior to previously observed, we compared several parameters of in vitro immunological function between sorted v-ffLuc, v-effLuc, and untransduced OT-I cells. We found no differences in proliferation, cytotoxicity, or IFN-γ production (Fig. S4). Interestingly, in the unsorted population, we found that, at 24 h after transduction, the number of cells in the v-ffLuc population was approximately half that in the v-effLuc. The proliferation curves normalized after 24 h (Fig. S5). Because there was no difference in cell viability, we speculate that the initial difference was due to diminished proliferation of ffLuc OT-I cells that were forced to handle the expression of ffLuc, which utilizes many rare codons.
effLuc was designed for mouse T cells. Nevertheless, we hypothesized that effLuc should provide enhanced sensitivity when transduced into a variety of tissues. We transduced a panel of human and mouse cell lines to investigate the universal applicability of this technology. We found that enhanced intensity and sensitivity of effLuc versus ffLuc ranged from 10- to 250-fold and 10- to >400-fold, respectively (Table S1).
Imaging effLuc-Expressing T Cells in Vivo.
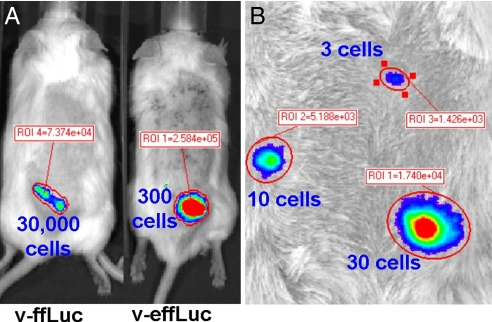
C57BL/6 T cells were transduced with each of the two constructs, and T cell numbers ranging from 3 to 30,000 were injected s.c. into C57BL/6 (B6) Albino mice. The limit of detection for T cells transduced with ffLuc was between 10,000 and 30,000 cells. We found that <300 T cells expressing effLuc were required to match the photon flux of 30,000 T cells expressing ffLuc (2.7 × 105 ± 1.3 × 104 versus 8 × 104 ± 8.3 × 103 photons/sec/ROI). Thus, effLuc resulted in an increase in sensitivity of 200–400 times (Fig. 2A). Strikingly, we found that as few as three T cells expressing effLuc could be imaged even when acquisition was performed using a binning setting of 4 (512 × 512 px) (Fig. 2B). These results suggest that effLuc should permit the study of detailed trafficking patterns of even small populations of lymphocytes. Moreover, although there was 40% loss in signal, ≤300 effLuc-expressing T cells could still be detected through the pigmented skin of wild type C57BL/6 mice. This will allow for imaging studies in a wide variety of tg and knockout animals not available on albino strains (Fig. S6).
Fig. 2.
In vivo imaging of s.c. implanted T cells transduced with optimized firefly luciferase. (A) B6 T cells transduced with v-ffLuc or v-effLuc (as indicated) were injected s.c. at the indicated numbers into B6 Albino mice, and photon emissions were measured on an IVIS 200. (B) T cells transduced with v-effLuc were injected s.c. at the level of 30, 10, or 3 cells as indicated.
Our in vivo images of effLuc T cells were acquired using a small px ρ (binning of 4), allowing us to pinpoint T cell location with better confidence than has been historically possible with ffLuc T cells, which requires larger px ρ settings (binning of 8–16 corresponding to 256 × 256 and 128 × 128 px, respectively) to raise the signal above the noise (9, 13, 14).
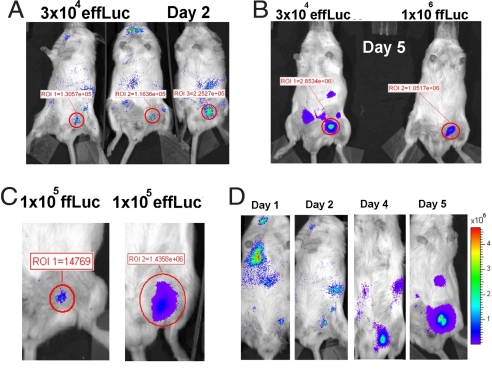
We examined the difference in sensitivity between T cells transduced with v-ffLuc versus v-effLuc in two different immunocompetent animal models. The first was a model of vaccination whereby T cells migrate into the s.c. tissue of the vaccine site. Ova peptide-pulsed or DCs (105 cells) were injected s.c. into the left or right inguinal region, respectively. On the same day, ffLuc- or effLuc-expressing OT-I T cells ranging from 3 × 104 to 106 in number, were injected retro-orbitally. Imaging was performed on days 1–6. For both groups, we found that T cells were detected at the site of the Ova vaccination but not the mock vaccination. Results were highly reproducible between animals even when as few as 3 × 104 T cells expressing effLuc were transferred (Fig. 3A). The number of adoptively transferred T cells required for detection at the vaccination site for ffLuc and effLuc was 105 and ≤3 × 104 T cells, respectively. We found that light emission at the vaccination site was ≈3-fold higher for animals that received 3 × 104 versus 106 ffLuc T cells. A representative day 5 image is shown in Fig. 3B. This result was highly reproducible at all of the imaging time points. A side by side comparison of luminescent activity of equal numbers of transferred ffLuc- versus effLuc-expressing T cells that migrated to the vaccination site indicated a >100-fold difference in sensitivity, similar to our in vitro data (Fig. 3C; day 5 is shown). Photon flux produced by T cells transduced with v-effLuc and injected s.c. into the ventral skin of a mouse (n = 3), was used to establish a standard curve (Fig. S7) to back-calculate the approximate number of effLuc-expressing T cells in the s.c. tissue in the group receiving 3 × 104 T cells. This approach indicated that ≤450 T cells had migrated to the vaccination site. Bioluminescence was also observed emanating from deeper tissues (Fig. 3D; 105 transferred T cells are shown), which allowed us to perform simple kinetic analysis (Fig. 4D). These images capture the trafficking of as few as 0.003% (3 × 104 T cells) of the total lymphocyte pool (≈109 cells).
Fig. 3.
Comparison of migration of OT.1 T cells transduced with v-ffLuc versus v-effLuc to Ova peptide-pulsed DCs. 1 × 105 Ova-peptide-pulsed or unpulsed dendritic cells were injected into the left or right inguinal region, respectively. Simultaneously, T cells transduced with v-effLuc or v-ffLuc were injected intravenously. (A) Mice receiving 3 × 104 v-effLuc-transduced T cells on day 2. (B) Comparison of mice receiving 3 × 104 v-effLuc-transduced T cells (Left) versus 1 × 106 v-ffLuc-transduced T cells (Right), imaged on day 5. (C) Comparison of mice that received 1 × 105 v-ffLuc-transduced T cells (Left) versus 1 × 105 v-effLuc-transduced T cells (Right) imaged on day 5. (D) Sequential imaging on days 1–6 (days 1, 2, 4, and 5 are shown) of an animal that received 1 × 105 v-effLuc-transduced T cells. The y axis represents photons per second per square centimeter per sr. In each case, a representative experiment is shown.
Fig. 4.
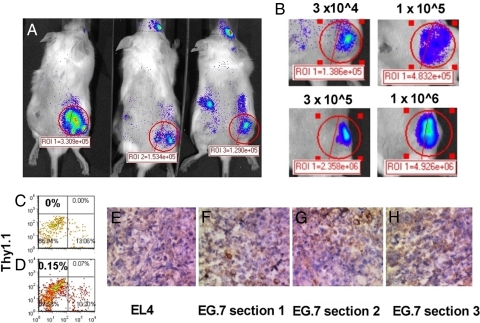
Small numbers of adoptively transferred tumor specific T cells can be detected at preexisting tumors. B6 Albino mice were sublethally irradiated (300 cGy) and implanted s.c. with 1 × 106 Ova-expressing EL4 (EG.7) tumor cells in the right flank. The same number of control EL4 cells was implanted on the left flank. Seven days later, different numbers of OT.1 T cells transduced with v-effLuc were intravenously injected into the animals. Five days after adoptive transfer, animals were imaged. (A) A group of animals injected with 3 × 104 OT.1 T cells. (B) A representative example of tumor infiltration on day 5 after transfer of effLuc-expressing OT.1 T cells (as indicated). (C–G) EL4 (C and E) and EG.7 (D, F, and G) tumors were resected 5 days after adoptive transfer of 1 × 106 effLuc-expressing OT-I T cell and examined for Thy1.1 expression by either flow cytometry (C and D) or immunohistochemistry (E and F).
Imaging Mouse T Cells in an Established Immunocompetent Tumor Model.
Finally, we evaluated our ability to image effLuc T cells within large preestablished tumors (area >50 mm2) in a well established model of adoptive immunotherapy. B6 Albino mice bearing EL4 (left flank) and Ova-expressing EL4 (EG.7; right flank) tumors were injected retro-orbitally with effLuc-transduced OT-1 T cells ranging in number from 3 × 104 to 106. The preferential therapeutic efficacy against EG.7 tumors is well described in this model after the transfer of ≥3 × 105 OT.1 T cells (29), a result we duplicated (data not shown). T cells were detected at the site of EG.7 but not EL4 tumors after the transfer of as few as 3 × 104 T cells (Fig. 4A; day 5 is shown). Bioluminescent intensity correlated with the number of transferred T cells (Fig. 4B). The observed photon flux at the EG.7 tumor site was used to calculate the number of infiltrating T cells. We investigated whether the photon flux of effLuc T cells injected intratumorally (IT) was lower than those injected s.c.. No statistically significant difference was observed (Fig. S8). Consequently, even when 106 OT.1 T cells were transferred, our calculations estimate an infiltrate on day 5 of only 3–5 × 103 cells. We excised the tumors and subjected them to Thy1.1 detection by flow cytometry (Fig. 4 C and D) or IHC (Fig. 4 E–H). We found that Thy1.1+ cells were observed at a frequency of <0.3%. In our opinion, flow cytometry cannot accurately quantitate cell numbers at these levels. Using IHC, the number of positive cells varied wildly between tissue sections. This indicates that BLI of mouse T cells expressing effLuc is a more accurate method of cell number quantitation and subject to less handling error, variation, and human interpretation.
Discussion
In this report we addressed the long-standing problem of how to express ffLuc in mouse T cells at sufficient magnitude to achieve a signal intensity suitable to detect <10,000 T cells at a given location and track ≤0.003% of the lymphoid pool within living mice. We decided to focus on retroviral vectors and transduction because this procedure is rapid, simple, and less expensive than crossing ffLuc-tg mice onto immunologically relevant backgrounds.
We made several changes to the standard retro-viral construct encoding (1) ffLuc, (2), an EMCV IRES and (3) Thy1.1. We added a (G3S)2 flexible linker at the 3′ end of the IRES to facilitate wild type RNA folding (RE-IRES). Next, we proceeded to codon optimize, remove cryptic splice sites from ffLuc and add a WHV pre element. These modifications resulted in >100-fold increase in luciferase activity and Thy1.1 expression intensity. Our data indicate that tRNA availability and cryptic splicing are the major factors limiting expression of ffLuc in mouse T cells after transduction. We found that, compared with ffLuc, transduction of effLuc into human PBMC-derived T cells resulted in only a 10-fold increase in light emission (Table S1). This interspecies difference in ffLuc expression is important because it has allowed for the study of T cell trafficking to solid tumors in xenogeneic models and validation of the therapeutic efficacy of chimeric activation receptors (18–23). Such models, however, are difficult to evaluate from a dynamic immunogical perspective because they necessitate the use of immuno-deficient mice.
Our data suggest that the use of effLuc will provide sufficient sensitivity to investigate differences in trafficking and efficacy between different populations of only 3–10 × 104 adoptively transferred, phenotypically distinct, or genetically modified, tumor-specific T cells (0.003–0.005% of the lymphoid pool). Using a standard curve of effLuc-expressing T cells injected s.c. or IT, we were able to back calculate the approximate number of T cells at a s.c. or IT site. Although three replicate mice were used to establish these curves for each experiment, we feel that the calculated T cell numbers should be considered semiquantitative. Further, compared with s.c., IT injection resulted in what appeared to be a trend toward a 25% loss in signal, but we could not establish statistical significance. Current adoptive immunotherapeutic regimens involve the transfer between 1 and 40 billion heterogeneous tumor specific T cells. In the setting of metastatic melanoma, tumor infiltration has been measured as <0.005% per gram of tumor (17, 30–32) and therapeutic activity is not predictable (33, 34). The injection of large cell numbers has many drawbacks. The procedure is expensive and labor intensive, results in the transfer of later passage cells that are likely less potent effectors and is complicated by T cell trapping in the lungs that can cause respiratory distress (35) and vascular leak syndrome (36). The use of effLuc in models of adoptive immunotherapy will hopefully provide the technology necessary to resolve these side effects and allow the identification of a small subset(s) of T cells, which are responsible for anti-tumor activity.
In summary, we have developed an optimized version of ffLuc that can be transduced into primary mouse T cells. As few as three T cells expressing effLuc injected s.c., can be imaged in living mice. Our data suggests that this technology is better suited for quantification of ≤0.3% of a population of cells compared with flow cytometry and eliminates variation introduced by tissue disruption. IHC appeared to visualize small cell numbers, but considerable variation was observed between sections at different tissue depths. Crossing ffLuc-tg mice onto the multiple strains currently used as models for studying T cells is a valuable technique for various applications including the response of undisturbed populations to multiple stimuli. It will be interesting to see whether tg mice expressing effLuc will result in superior detection of transgene-expressing cell populations versus ffLuc tg mice. The approach described here offers investigators the ability to address questions more quickly than with tg models. From a therapeutic perspective, it should now be possible to visualize and identify subsets of lymphocytes constituting ≤0.003% of the lymphoid pool that are responsible for different aspects of anti-tumor activity and autoimmunity.
Methods
Animals.
C57BL/6J-Tyr-2J/J Albino mice were purchased from The Jackson Laboratory. C57BL/6 OT-I TCR tg mice were bred in the vivarium at M. D. Anderson Cancer Center. The M. D. Anderson Cancer Center Animal Care and Use Committee approved all protocols.
Construction of Retroviral Vector Encoding Optimized Firefly Luciferase and Thy1.1.
Plasmids were derived from the pMSCV-ffFLuc-pIRES-Thy1.1 encoding ffLuc (pGL3; Promega), EMCV pIRES2 (Clontech) and Thy1.1 (H.I.L., unpublished data). The reengineered EMCV IRES (RE-IRES) includes a 3′ linker (G3S)2 synthesized by Genscript and cloned between HpaI and Xho-I sites. Codon optimization, removal of cryptic splice sites and negative cis-acting motifs were performed using GeneOptimizer and synthesized by GeneArt. effLuc was cloned between Bgl-II and Hpa-I sites. The optimized construct was generated by cloning a Sal-I and Cla-I flanked WHVpre element (generated via PCR) downstream of Thy1.1 between Sal-I and Cla-I. All constructs were sequence verified.
Packaging of Retrovirus.
Retroviral vectors were packaged into ecotropic virus as described in ref. 37 with the noted addition of pCLEco (38) (Addgene) to the transfection mix. Supernatants were concentrated between 25× and 100×, using Centricon Plus-20 Centrifugal Filter Units (Millipore), and then titered on NIH 3T3 cells.
Transduction of Mouse T Cells.
Splenocytes from C57BL/6 wild type mice or OT-I TCR tg mice were cultured in X-Vivo-15 (Lonza), Normocin (Invivogen), 200 IU Proleukin (Chiron), and 0.1 μg/ml anti-mouse CD3 (clone 2C11; BD Biosciences). After 22–24 h, retrovirus was added at an MOI of 5 in the presence of polybrene (Sigma) and Lipofectamine 2000 (Invitrogen) at 1.6 μg/ml and 2 μg/ml, respectively, and spinfected at 850 × g for 2 h. The following day, cells were washed and expanded in Alpha-MEM, 10% FBS, Normocin, and 200 IU/ml Proleukin. Cells were stained three days after transduction with anti-mouse Thy1.1 PE (BD-PharMingen) and sorted using a FACSVantage (BD Biosciences).
In Vitro Bioluminescence Assay.
Transduced T cells were washed in PBS and 1 × 105 cells tested for light emission in a 96-well format in triplicate in 200 μl OPTI-MEM/150 μg/ml d-Luciferin (Xenogen), using a Topcount apparatus (Perkin–Elmer).
Ova Vaccination Model.
To generate CD11b+ DCs, Bone marrow was harvested from C57BL/6 wild type mice and seeded in 6-well plates at 1 × 106/ml (5 ml) in Alpha-MEM, 10% FBS, Normocin, and 200 μg/ml Flt3-L and cultured at 37°C in 7.5% CO2. On day 8, GM-CSF was added to 20 μg/ml. On day 10, LPS was added at to 100 ng/ml. On day 11, DCs were pulsed with 1 μg/ml Ova peptide 257–264 (Ovap) and washed, and 1 × 105 cells were injected into the left inguinal region of C57BL/6 Albino mice. The same number of nonpulsed DCs was injected on the right side. Immediately after DCs injection, 3 × 104 − 1 × 106 OT-I TCR tg T cells expressing ffLuc or effLuc were injected i.v. via retro-orbital injection. Imaging was performed on days 1, 2, 4, and 5.
Model of Adoptive Immunotherapy for Cancer.
EL4 or EG.7 tumor cells (2 × 106) were injected into the left and right flank of C57BL/6 Albino mice. Seven days later, 3 × 104 −106 OT-I TCR tg T cells expressing effLuc were injected IV via retro-orbital injection. Imaging was performed on days 4 and 5. In some cases, tumors were resected and examined for Thy1.1 expression either via standard IHC of formalin fixed paraffin embedded sections or via flow cytometry.
In Vivo Bioluminescence Imaging.
Isofluorane-anesthetized animals were imaged using an IVIS 200 system (Xenogen) 8 min after i.p. injection of 2 mg of d-luciferin according to the manufacturer's specifications. Living Image software was used to analyze the data.
Supplementary Material
Acknowledgments.
The authors thank Sunny D. Armstrong for supporting this work and Will Hauser for his assistance with Living Image 3.0. This work was supported by National Institutes of Health Grants RO1 CA116206, CA120956, and R21 CA129390; M. D. Anderson Cancer Center Grant CA16672; Netherlands Organisation for Scientific Research Veni Grant 916.046.014, and M. D. Anderson Cancer Center Specialized Programs of Research Excellence Grant P50 CA093459.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804105105/DCSupplemental.
References
- 1.Hardy J, et al. Bioluminescence imaging of lymphocyte trafficking in vivo. Exp Hematol. 2001;29:1353–1360. doi: 10.1016/s0301-472x(01)00756-1. [DOI] [PubMed] [Google Scholar]
- 2.Sumen C, Mempel TR, Mazo IB, von Andrian UH. Intravital microscopy: Visualizing immunity in context. Immunity. 2004;21:315–329. doi: 10.1016/j.immuni.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Jain RK, Munn LL, Fukumura D. Dissecting tumour pathophysiology using intravital microscopy. Nat Rev Cancer. 2002;2:266–276. doi: 10.1038/nrc778. [DOI] [PubMed] [Google Scholar]
- 4.Breart B, Lemaitre F, Celli S, Bousso P. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J Clin Invest. 2008;118:1390–1397. doi: 10.1172/JCI34388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contag CH, et al. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 6.Otto-Duessel M, et al. In vivo testing of Renilla luciferase substrate analogs in an orthotopic murine model of human glioblastoma. Mol Imaging. 2006;5:57–64. [PubMed] [Google Scholar]
- 7.Zhao H, et al. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10:41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 8.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Azadniv M, Dugger K, Bowers WJ, Weaver C, Crispe IN. Imaging CD8+ T cell dynamics in vivo using a transgenic luciferase reporter. Int Immunol. 2007;19:1165–1173. doi: 10.1093/intimm/dxm086. [DOI] [PubMed] [Google Scholar]
- 10.Beilhack A, et al. In vivo analyses of early events in acute graft-versus-host disease reveal sequential infiltration of T-cell subsets. Blood. 2005;106:1113–1122. doi: 10.1182/blood-2005-02-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka M, et al. In vivo visualization of cardiac allograft rejection and trafficking passenger leukocytes using bioluminescence imaging. Circulation. 2005;112:I105–I110. doi: 10.1161/CIRCULATIONAHA.104.524777. [DOI] [PubMed] [Google Scholar]
- 12.Edinger M, et al. CD4+CD25+ regulatory T cells preserve graft-versus-tumor activity while inhibiting graft-versus-host disease after bone marrow transplantation. Nat Med. 2003;9:1144–1150. doi: 10.1038/nm915. [DOI] [PubMed] [Google Scholar]
- 13.Costa GL, et al. Adoptive immunotherapy of experimental autoimmune encephalomyelitis via T cell delivery of the IL-12 p40 subunit. J Immunol. 2001;167:2379–2387. doi: 10.4049/jimmunol.167.4.2379. [DOI] [PubMed] [Google Scholar]
- 14.Nakajima A, et al. Antigen-specific T cell-mediated gene therapy in collagen-induced arthritis. J Clin Invest. 2001;107:1293–1301. doi: 10.1172/JCI12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D, Hung CF, Wu TC. Monitoring the trafficking of adoptively transferred antigen- specific CD8-positive T cells in vivo, using noninvasive luminescence imaging. Hum Gene Ther. 2007;18:575–588. doi: 10.1089/hum.2007.038. [DOI] [PubMed] [Google Scholar]
- 16.Graph RF. Modern Dictionary of Electronics. Burlington, MA: Elsevier Science & Technology; 1999. [Google Scholar]
- 17.Pockaj BA, et al. Localization of 111indium-labeled tumor infiltrating lymphocytes to tumor in patients receiving adoptive immunotherapy. Augmentation with cyclophosphamide and correlation with response. Cancer. 1994;73:1731–1737. doi: 10.1002/1097-0142(19940315)73:6<1731::aid-cncr2820730630>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 18.Kowolik CM, et al. CD28 costimulation provided through a CD19-specific chimeric antigen receptor enhances in vivo persistence and antitumor efficacy of adoptively transferred T cells. Cancer Res. 2006;66:10995–11004. doi: 10.1158/0008-5472.CAN-06-0160. [DOI] [PubMed] [Google Scholar]
- 19.Vera J, et al. T lymphocytes redirected against the κ light chain of human immunoglobulin efficiently kill mature B lymphocyte-derived malignant cells. Blood. 2006;108:3890–3897. doi: 10.1182/blood-2006-04-017061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savoldo B, et al. Epstein Barr virus specific cytotoxic T lymphocytes expressing the anti-CD30ζ artificial chimeric T-cell receptor for immunotherapy of Hodgkin disease. Blood. 2007;110:2620–2630. doi: 10.1182/blood-2006-11-059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quintarelli C, et al. Co-expression of cytokine and suicide genes to enhance the activity and safety of tumor-specific cytotoxic T lymphocytes. Blood. 2007;110:2793–2802. doi: 10.1182/blood-2007-02-072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown CE, et al. Tumor-derived chemokine MCP-1/CCL2 is sufficient for mediating tumor tropism of adoptively transferred T cells. J Immunol. 2007;179:3332–3341. doi: 10.4049/jimmunol.179.5.3332. [DOI] [PubMed] [Google Scholar]
- 23.Singh H, et al. Combining adoptive cellular and immunocytokine therapies to improve treatment of B-lineage malignancy. Cancer Res. 2007;67:2872–2880. doi: 10.1158/0008-5472.CAN-06-2283. [DOI] [PubMed] [Google Scholar]
- 24.Thorne SH, Negrin RS, Contag CH. Synergistic antitumor effects of immune cell-viral biotherapy. Science. 2006;311:1780–1784. doi: 10.1126/science.1121411. [DOI] [PubMed] [Google Scholar]
- 25.Zeiser R, et al. Inhibition of CD4+CD25+ regulatory T-cell function by calcineurin-dependent interleukin-2 production. Blood. 2006;108:390–399. doi: 10.1182/blood-2006-01-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 27.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumann JG, et al. Murine T cells potently restrict human immunodeficiency virus infection. J Virol. 2004;78:12537–12547. doi: 10.1128/JVI.78.22.12537-12547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helmich BK, Dutton RW. The role of adoptively transferred CD8 T cells and host cells in the control of the growth of the EG7 thymoma: Factors that determine the relative effectiveness and homing properties of Tc1 and Tc2 effectors. J Immunol. 2001;166:6500–6508. doi: 10.4049/jimmunol.166.11.6500. [DOI] [PubMed] [Google Scholar]
- 30.Economou JS, et al. In vivo trafficking of adoptively transferred interleukin-2 expanded tumor-infiltrating lymphocytes and peripheral blood lymphocytes. Results of a double gene marking trial. J Clin Invest. 1996;97:515–521. doi: 10.1172/JCI118443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goedegebuure PS, et al. Adoptive immunotherapy with tumor-infiltrating lymphocytes and interleukin-2 in patients with metastatic malignant melanoma and renal cell carcinoma: A pilot study. J Clin Oncol. 1995;13:1939–1949. doi: 10.1200/JCO.1995.13.8.1939. [DOI] [PubMed] [Google Scholar]
- 32.Griffith KD, et al. In vivo distribution of adoptively transferred indium-111-labeled tumor infiltrating lymphocytes and peripheral blood lymphocytes in patients with metastatic melanoma. J Natl Cancer Inst. 1989;81:1709–1717. doi: 10.1093/jnci/81.22.1709. [DOI] [PubMed] [Google Scholar]
- 33.Mackensen A, et al. Phase I study of adoptive T-cell therapy using antigen-specific CD8+ T cells for the treatment of patients with metastatic melanoma. J Clin Oncol. 2006;24:5060–5069. doi: 10.1200/JCO.2006.07.1100. [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg SA, Dudley ME. Cancer regression in patients with metastatic melanoma after the transfer of autologous antitumor lymphocytes. Proc Natl Acad Sci USA. 2004;101:14639–14645. doi: 10.1073/pnas.0405730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glauser FL, et al. Cardiopulmonary toxicity of adoptive immunotherapy. Am J Med Sci. 1988;296:406–412. doi: 10.1097/00000441-198812000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Bechard DE, et al. Nonspecific cytotoxicity of recombinant interleukin-2 activated lymphocytes. Am J Med Sci. 1989;298:28–33. doi: 10.1097/00000441-198907000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Rabinovich BA, et al. Activated, but not resting, T cells can be recognized and killed by syngeneic NK cells. J Immunol. 2003;170:3572–3576. doi: 10.4049/jimmunol.170.7.3572. [DOI] [PubMed] [Google Scholar]
- 38.Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: Rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.