Abstract
Functional selection of genetic suppressor elements (GSEs), engineered gene fragments that interfere with the function of a particular gene product, was used to identify regulators of FAS-induced apoptosis. Chicken DF-1 cells expressing human FAS receptor and susceptible to FAS-induced apoptosis were infected with a GSE library consisting of randomly fragmented normalized chicken cDNAs in a replication-competent avian retroviral vector. Virus-producing cells were subjected to several rounds of selection using FAS agonistic antibodies, resulting in isolation of a set of GSEs conferring resistance to FAS-induced apoptosis. Surprisingly, one of the isolated GSEs encoded a 42 amino acid-long polypeptide derived from the C-terminal half of cytochrome b (Cyt b) encoded by the mitochondrial genome. Subsequent experiments showed that caspase 8-dependent cleavage of mitochondrial Cyt b and translocation of its C-terminal half into the cytoplasm occurred during FAS-induced apoptosis in both chicken and human cells. Ectopic cytoplasmic expression of either full-length Cyt b or its C-terminal half in several human cell lines induced apoptosis, which could be suppressed by the isolated GSE, but not by Bcl2 over-expression or Apaf-1 or cytochrome c knock-down. These results reveal a cytochrome c-independent branch of FAS-induced apoptosis involving cleavage and cytoplasmic release of mitochondrial Cyt b.
Keywords: avian retroviral vector, Bcl-2, capsase, cytochrome c, genetic suppressor element
FAS is a member of the death domain-containing tumor necrosis factor receptor family of proteins that regulate apoptosis (1–3). FAS-induced apoptosis occurs upon binding of its physiological ligand, FAS ligand (FASL), or experimental FAS agonistic antibodies (4). The major physiological function of this conserved apoptotic pathway is to support homeostasis of the immune system, directing elimination of self-reacting lymphocytes and proliferation and maturation of some lymphocyte populations (5). Deregulation of the FAS pathway has been implicated in various malignancies and diseases (4). Tumor cells often escape immune attack by suppressing the FAS pathway or by using it to their advantage (3, 6, 7). For example, some tumors up-regulate expression of FASL to induce apoptosis in infiltrating lymphocytes (4). Some viruses promote their propagation by expressing FAS-inhibitory proteins (8, 9). Mutations in FAS and some haplotypes of FAS splice variants are associated with human diseases (4).
Binding of FASL or FAS agonistic antibodies to FAS on the cell surface triggers the extrinsic pathway of apoptosis through formation of a death-inducing signaling complex (DISC) (10). DISC contains a number of adaptor proteins, caspases 8 and 10 and c-Flip, a negative regulator of caspase 8 (11) that is used by some tumor cells and viruses to acquire FAS resistance (12). DISC formation is the main mechanism of FAS-mediated killing in cells designated “type I” (H9, SKW6.4, human lymphocytes). However, in “type II” cells (Jurkat, CEM, HeLa, human hepatocytes), FAS-mediated apoptosis occurs via the intrinsic pathway (13). Type II cells do not form enough DISC to induce apoptosis directly; instead, DISC-activated caspase 8 cleaves the Bcl2 family member Bid (14), which triggers the mitochondrial pathway of apoptosis (15) involving cytochrome c (Cyt c) release and activation of the Apaf1/procaspase 9 apoptosome (16). Other mitochondrial components have recently been defined as apoptosis mediators, including AIF, SMAC/Diablo, and Omi (16–20); however, their relative impact on FAS-mediated death remains unclear.
In this study we sought to identify different components and modulators of the FAS pathway through an unbiased genetic suppressor element (GSE) (21) library screen. This method allows identification of genes associated with specific cellular phenotypes by functional selection of cells expressing GSEs, engineered gene fragments constructed to encode either antisense RNAs or dominant negative partial proteins. Developed in 1992 (22), GSE technology has been used to identify unique genes involved in tumor suppression, drug sensitivity, apoptosis, and growth regulation (21, 23, 24). In past studies, GSE libraries were constructed in mammalian retroviral vectors that required packaging cells to produce viral stocks of the GSE library and selected clones and a second cell type for functional selection. Because clone rescue between rounds of selection was laborious and somewhat unreliable, we have modified the method to use a GSE library constructed in an RCAS avian retroviral vector (25). Because RCAS vectors are replication competent, functional selection can be performed directly on the virus-producing cells.
We used this method to isolate GSEs conferring resistance to FAS apoptosis in chicken cells expressing human FAS. Several of the isolated GSEs corresponded to genes with known relevance to apoptosis. However, one corresponded to the cytochrome b (Cyt b), a mitochondrial DNA-encoded component of complex III of the mitochondrial electron-transport chain (26–28), which has not previously been linked to apoptosis or any other nonmitochondrial activity. We were intrigued by how a GSE-encoded fragment of a mitochondrial protein could affect FAS apoptosis when expressed in the cytoplasm. This apparent paradox was resolved by our work demonstrating a different role for a processed Cyt b protein as a cytoplasmic mediator of FAS-induced apoptosis.
Results
Identification of GSEs Suppressing FAS-Mediated Apoptosis.
We designed a modified GSE screening approach that allows direct phenotypic selection of cells producing GSE-encoding retroviruses (see Materials and Methods for details). This was made possible by the use of a GSE library prepared in a replication-competent avian retroviral vector [RCAS series (25); see supporting information (SI) Fig. S1] comprised of randomly fragmented and normalized chicken cDNAs ligated to an adaptor enabling translation of the inserts in all three reading frames (a gift of E. Feinstein, see SI Text for details).
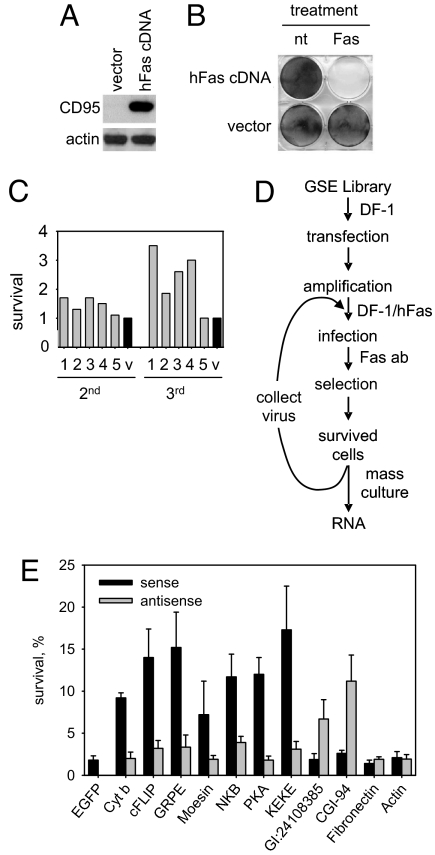
To use this library for identification of mediators of FAS-dependent apoptosis, we established a chicken cell line permissive to RCAS replication that is susceptible to apoptosis induced by human FAS agonistic antibodies. Chicken DF-1 cells transduced with an RCAS construct directing expression of human FAS (CD95) (Fig. 1A) became uniformly susceptible to FAS agonistic antibodies (Fig. 1B). This indicates that FAS was expressed in all of the DF-1FAS cells and that human FAS can function with chicken DISC components.
Fig. 1.
Identification of mediators of FAS apoptosis through GSE selection in a chicken system. (A) Chicken DF-1 cells transduced with a retrovirus encoding human FAS receptor (CD95) or an empty vector control were analyzed by Western blot for CD95 and actin (control) expression. (B) Chicken cells expressing human FAS undergo apoptosis upon exposure to FAS agonistic antibodies. Control DF-1 cells (vector) or DF-1 cells expressing human FAS (hFAS cDNA) were left untreated (nt) or treated with anti-FAS antibody (Fas). Cell viability was assessed 2 days later by methylene-blue staining. (C) Flow diagram of RCAS-based GSE selection for identification of modulators of FAS apoptosis (details in text). (D) Resistance to FAS-induced apoptosis conferred by enriched pools of GSEs. Virus collected from GSE library-transduced (five independent pools, numbered 1–5, gray bars) and empty vector-transduced (v, black bar) DF-1FAS cells after secondary (2′) and tertiary (3′) selection was used to infect DF-1FAS cells. Cells were exposed to FAS agonistic antibody and stained with methylene-blue 1 week later. Survival is shown relative to the number of colonies in the empty vector control, set at one unit for each selection. (E) GSEs recovered following tertiary selection were expressed in DF-1FAS cells in sense and antisense orientations. Cell viability was measured by methylene-blue staining 4 h after addition of FAS agonistic antibody. EGFP was used as a negative control. The gene names corresponding to each GSE are indicated. Survival of cells expressing each construct is given relative to the same cells in the absence of FAS antibody (set at 100%). Three independent experiments were performed; error bars indicate standard deviations.
The scheme of isolation of GSEs conferring FAS resistance is shown in Fig. 1D. Briefly, DF-1FAS cells were infected with virus produced in DF-1 cells transfected with library DNA. Superinfection of DF-1FAS cells with the library was possible because the FAS cDNA and GSE library were cloned into RCAS vectors differing in their env genes (29–31). The protocol used for library transduction ensured delivery of the library in its full complexity with each of the 1 × 106 clones delivered to at least 10 cells.
The library-transduced DF-1FAS cells were treated with a low concentration of FAS antibody that allowed survival of one out of 105 untransduced cells. These conditions were chosen to allow isolation of relatively weak GSEs and quantitative assessment of their biological effects. The primary selection was performed on five plates of 107 library-transduced cells and a single plate of 107 cells transduced with insert-free virus. Secondary and tertiary rounds of selection were performed similarly. Each of the five branches of library selection was processed separately to reduce the risk of losing weak GSEs because of competition with stronger ones.
The first round of selection revealed an equivalent frequency of FAS resistance in the library- and insert-free vector-transduced populations (≈500 clones were isolated from the five branches of the library experiment and ≈100 clones were isolated in the control experiment). However, in the secondary and tertiary rounds of selection, the selected viruses produced an increase in FAS resistance as compared to the empty vector (Fig. 1C). The degree of FAS resistance in each branch remained at the same level after further rounds of selection, suggesting that a plateau had been reached that presumably reflects the biological efficacy of the selected viruses. It is possible that the strength of the effects of the isolated virus populations might be reduced by competition of FAS- and GSE-encoding RNAs during viral packaging. However, our successful isolation of confirmed bioactive GSEs (see below) indicates that such competition did not affect selection. Presumably GSE-encoding RNA has a packaging advantage because it is significantly shorter than the FAS-transcript.
Identification of the Genes Corresponding to FAS-Inhibitory GSEs.
To identify the genes corresponding to selected GSEs, total RNA was isolated from the FAS-resistant populations of cells obtained after tertiary selection. RT-PCR was performed with a GSE-specific primer corresponding to the sense strand of the universal GSE adaptor (see Fig. S1B). The PCR products were cloned back into the RCAS(A) vector and sequenced. The isolated sequences are listed in Table S1. Some of them represented fragments of genes with known relevance to FAS-mediated apoptosis, including c-FLIP (12, 32), NK receptor KIR/NKB (33, 34), and protein kinase A (PKA) (35). The remainder corresponded to genes that have not been previously associated with apoptosis. A subset of these represented highly abundant transcripts (fibronectin and actin) or RNA sequences with no recognized relation to mammalian genes.
Because the same universal adaptor flanked each library insert at both ends, the orientation of the recovered GSEs that was effective in conferring FAS resistance was not known. We therefore prepared two constructs for each GSE differing in the orientation of the insert in the RCAS(A) vector and assessed their effect on the FAS sensitivity of DF-1FAS cells. Viruses grown in DF-1 cells were used to infect DF-1FAS cells, which were then treated with FAS-agonistic antibodies. For the majority of tested sequences, one of the two tested constructs was effective in conferring FAS resistance (Fig. 1E). No FAS resistance was associated with isolated fibronectin- and actin-derived sequences cloned in either orientation, confirming that they represent background (data not shown).
Seven out of nine GSEs with confirmed activity demonstrated the FAS protective effect only when cloned in their sense orientation (see Fig. 1E), suggesting that they likely act as protein fragments. FAS resistance conferred by a truncated protein could be to the result of dominant negative inhibition of a corresponding endogenous protein required for apoptosis, or represent a functional portion of an apoptosis inhibitor. For example, GSE F3 corresponds to the inhibitory subunit of PKA and presumably causes inactivation of this regulatory subunit and subsequent activation of PKA, which has been implicated in prosurvival signaling (35). On the other hand, GSE F25 derived from c-FLIP presumably acts as a functional equivalent of the full-length c-FLIP protein, a known suppressor of caspase 8 and inhibitor of FAS apoptosis (12, 32, 36).
One of the GSEs with confirmed sense orientation-dependent activity was GSE F21, which encodes a 42 amino acid-long portion of Cyt b, derived from its C-terminal half (see SI Text, Fig. S2). Cyt b is encoded by the mitochondrial genome, synthesized inside mitochondria and localized to the inner membrane of mitochondria where it functions as a subunit of the mitochondrial electron transport complex III. Cyt b has not been previously implicated in apoptosis.
Cleavage and Release of a C-Terminal Portion of Cyt b into the Cytoplasm During FAS-Mediated Apoptosis.
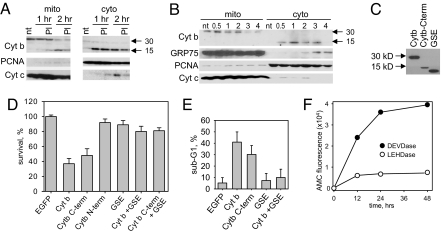
While Cyt b resides inside mitochondria, its corresponding GSE (F21) is expressed in the cytoplasm. To investigate whether GSE F21 has a chance to act against its endogenous target, we monitored the integrity and intracellular localization of Cyt b during FAS apoptosis (Fig. 2 A and B).
Fig. 2.
Cyt b is cleaved during FAS-induced apoptosis and its C-terminal part is released into the cytoplasm. (A) Intracellular distribution of Cyt b in DF-1 cells left untreated (control) or treated with FAS-agonistic antibody in the absence or presence on the proteosomal inhibitor MG-132 (PI). Cytoplasmic and mitochondrial fractions prepared either 1 or 2 h after treatment were analyzed by Western blotting with antibodies against Cyt b, proliferating cell nuclear antigen (PCNA) (cytoplasmic marker) and Cyt c. Full-length (30 kDa) and cleaved (15 kDa) Cyt b are indicated by arrows. (B) Mitochondrial and cytoplasmic fractions were prepared from HeLa cells left untreated (control) or treated with FAS antibody at the indicated times after treatment (0.5–4 h). Fractions were analyzed by Western blotting for Cyt b (full-length, 30 kDa, and cleaved, 15 kDa), GRP75 (mitochondrial marker), PCNA (cytoplasmic marker), and Cyt c. (C) Western blot analysis of HeLa cells expressing full-length Cyt b, Cyt b512–1146 (C-term), or the portion of Cyt b corresponding to GSE F21 in the cytoplasm. All recombinant proteins contained a C-terminal FLAG tag and were detected using anti-FLAG antibodies. (D) Full-length Cyt b and Cyt b512–1146 are toxic when expressed in the cytoplasm. HeLa cells were infected with lentiviruses directing expressing of EGFP (negative control), full-length Cyt b, the C-terminal half of Cyt b (Cyt b512–1146), the N-terminal half of Cyt b or the portion of Cyt b corresponding to GSE F21 as indicated below each bar. Cell survival was measured by methylene-blue staining 72 h after infection and is shown relative to the EGFP control (set at 100%). Here and in (E), the error bars indicate standard deviation of three independent experiments. (E) Quantitation of cells with subG1 DNA content. FACS analysis was performed on HeLa cells 72 h after infection with lentiviral Cyt b constructs as in (D). (F) Time course of caspase induction in HeLa cells expressing cytoplasmic Cyt b. Activation of caspases 3/7 and 9/6 were measured using the fluorogenic substrates DEVD-AMC (filled circles) and LEHD-AMC (open circles), respectively. AMC fluorescence resulting from substrate cleavage was measured at the indicated times after substrate addition.
Mitochondrial and cytoplasmic fractions were prepared from DF-1FAS cells and HeLa cells expressing endogenous FAS at various times after treatment with FAS agonistic antibodies. The quality of separation was assessed by detection of fraction-specific marker proteins (e.g., PCNA as a cytoplasmic marker and GRP75 as a mitochondrial marker). Cyt c was used as a classic example of a protein that undergoes mitochondria-to-cytoplasm translocation during apoptosis. Western blot analysis using P49 antibodies (37) generated against a C-terminal peptide of Cyt b (location of epitope is illustrated in Fig. S2) showed that in untreated (nonapoptotic) cells, Cyt b (30 kD) was restricted to the mitochondrial fraction. However, 0.5 to 1 h after FAS antibody treatment, a new band corresponding to a 15-kDa protein was detected primarily in the cytoplasmic fraction. Appearance of the 15-kDa Cyt b band in the cytoplasmic fraction was accompanied by a decrease in the intensity of the 30-kDa Cyt b band in the mitochondrial fraction. This profile of Cyt b expression and localization was observed in both chicken (DF-1FAS) and human (HeLa) cells.
Antibodies recognizing an N-terminal epitope of Cyt b also revealed the decrease in full-length Cyt b in the mitochondrial fraction of FAS antibody treated cells, but failed to identify the 15-kDa cytoplasmic band (data not shown). This result suggests that the 15-kDa protein is the result of cleavage of mitochondrial Cyt b and export of a C-terminal portion (roughly half) to the cytoplasm. The sequence corresponding to GSE F21 lies within this C-terminal portion (see Fig. S2). Neither cleavage nor translocation of Cyt b to the cytoplasm were detected in the same cells (DF-1FAS and HeLa) undergoing apoptosis because of treatment with staurosporin (data not shown), indicating that this phenomenon is not an essential part of any mitochondria-mediated apoptosis and might be specific for FAS.
The identity of the protease involved in Cyt b cleavage during FAS-induced apoptosis, as well as the exact cleavage site, remain to be determined; however, it should be noted that cleavage of Cyt b was not blocked by the proteasomal inhibitor MG-132 (see Fig. 2A).
Cytoplasmic Expression of Cyt b or the C-Terminal Fragment Cyt b512–1146 Induces Apoptosis.
Because Cyt b is encoded by the mitochondrial genome and translated by mitochondrial ribosomes from mitochondrial RNA, we could not use gene knock-out or knock-down techniques to test the role of Cyt b in FAS-induced apoptosis directly. As an alternative, we tested whether ectopic expression of full-length Cyt b or a C-terminal portion predicted to correspond to the 15-kDa Cyt b fragment (Cyt b512–1146) in the cytoplasm could initiate apoptosis in the absence of FAS activation.
Because of differences in the nuclear and mitochondrial genetic codes, we could not use the Cyt b coding sequence taken from mitochondrial DNA for cytoplasmic expression of the protein. The mitochondrial sequence of the Cyt b ORF contains 11 tryptophan residues encoded by codons that are interpreted as stop codons in the nuclear code. Therefore, we generated an artificial human Cyt b gene sequence encoding the same amino acids but using codons of the nuclear code that are most frequently used in the mammalian genome to optimize translation (see Fig. S2A). It should be noted that GSE F21 does not contain any tryptophan codons.
In addition to full-length Cyt b, three Cyt b derivatives were also generated from the recoded synthetic gene (see Fig. S2A). Two of these corresponded to the N- and C-terminal halves of the protein, roughly corresponding to the putative cleavage products of Cyt b generated during FAS-mediated apoptosis (Cyt b1–511 and Cyt b512–1146, respectively). The third encoded the 42 amino acid-long portion of Cyt b corresponding to GSE F21. To simplify delivery of these constructs into avian and mammalian cells, they were cloned into a lentiviral vector. The ability of the constructs to drive expression of FLAG-tagged proteins of the expected sizes in HeLa cells was analyzed by Western blot (Fig. 2C).
The effect of the generated Cyt b expression constructs on the viability of HeLa cells is shown in Fig. 2D. Similar results were obtained in DF-1 cells (data not shown). No cytotoxicity was associated with expression of either the N-terminal portion of Cyt b or the GSE F21-derived sequence. In contrast, expression of either full-length Cyt b or Cyt b512–1146 caused significant cell death, which could be blocked by coexpression of the GSE F21-derived sequence. This confirms that the apoptosis-inhibiting effect of GSE F21 is because of specific interference of the GSE with the biological activity of its cognate gene product, Cyt b. The fact that the original GSE F21 (mitochondrial sequence) and the recoded version (nuclear sequence) were equally effective in suppressing FAS-induced apoptosis indicates that they act as dominant negative peptides rather than inhibitory RNA molecules.
The apoptotic nature of the observed cell death was confirmed by FACS analysis of cellular DNA content, showing that ectopic expression of Cyt b or Cyt b512–1146 in HeLa cells results in appearance of a large population of cells with subG1 DNA content that can be blocked by coexpression of GSE F21(Fig. 2E, Fig. S3A). The proportion of cells with subG1 DNA content in cell cultures infected with different Cyt b constructs correlated with their cytotoxicity (see Fig. 2 D and E). In addition, ectopic expression of Cyt b resulted in caspase activation as illustrated by caspase enzymatic activity in cell lysates (Fig. 2F) and poly(ADP-ribose) polymerase (PARP) cleavage (Fig. S3B). The proapoptotic activity of Cyt b and Cyt b512–1146 was evident in several different cell lines (including PC3 and CWR22R, Fig. S3 C and D).
Caspase 8 and Bid Mediate Cyt b Cleavage During FAS Apoptosis.
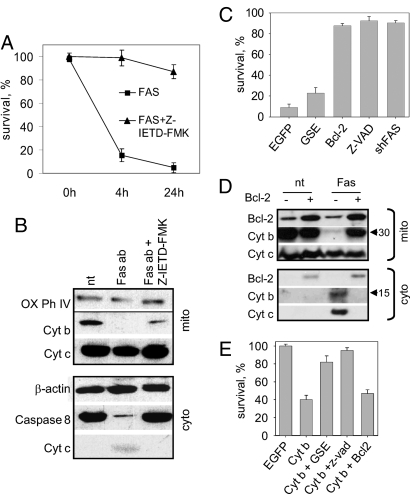
Caspase 8 activation is known to occur immediately downstream of DISC assembly during FAS-mediated apoptosis (38). To determine whether Cyt b release from mitochondria depends upon caspase 8 activation, we monitored Cyt b content in the mitochondria and cytoplasm of cells treated with FAS agonistic antibodies in the presence or absence of the specific caspase 8 inhibitor Z-IETD-FMK. As expected, Z-IETD-FMK treatment of HeLa cells resulted in inhibition of FAS-induced cell death (Fig. 3A). In addition, we found that the presence of Z-IETD-FMK resulted in reduced Cyt b cleavage (Fig. 3B).
Fig. 3.
Place of Cyt b in the FAS apoptosis pathway. (A) Caspase 8 is required for FAS-induced apoptosis. Cell survival was measured in HeLa cells by methylene-blue staining following treatment (at time 0 h) with FAS agonistic antibody or FAS antibody plus Z-IETD-FMK (specific caspase 8 inhibitor). Three independent experiments were performed; error bars indicate standard deviations. (B) Inhibition of Caspase 8 during FAS-induced apoptosis prevents Cyt b processing. Mitochondrial (Top) and cytoplasmic (Bottom) fractions were prepared from untreated HeLa cells (nt) or 8 h after treatment with FAS antibody plus cycloheximide (CHI) (Fas ab) in the absence or presence of Z-IETD-FMK. Western blotting used the indicated antibodies including β-actin and Cytochrome Oxidase Subunit IV as cytoplasmic and mitochondrial markers, respectively. (C) FAS apoptosis can be blocked by expression of GSE F21 or Bcl-2 and requires caspase and FAS activity. HeLa cells expressing EGFP (control), GSE F21, Bcl-2, or shFAS were treated with FAS antibody. Uninfected HeLa cells were treated with FAS antibody plus Z-VAD. Survival was measured by methylene-blue staining 4 h after treatment and is shown for each cell type relative to the same cells in the absence of FAS antibody. Here and in (E), three independent experiments were performed: error bars indicate standard deviations. (D) Western blot analysis of HeLa mitochondrial (left) and cytoplasmic (right) fractions 12 h after treatment with FAS antibody (nt, untreated). Cells were infected before treatment with control or Bcl-2 lentiviruses. (E) Cyt b-induced cell death can be blocked by GSE F21 or Z-VAD, but not Bcl-2. HeLa cells were infected with EGFP (control), Cyt b, GSE F21, and Bcl-2 lentiviral constructs. Z-VAD was added 24 h after infection and methylene-blue staining was performed 48 h after infection.
Caspase 8 mediates the mitochondrial branch of FAS apoptosis by proteolytic activation of Bid (1). We found that Bid knock-down by specific shRNA not only makes HeLa cells resistant to FAS agonistic antibody treatment, but also prevents Cyt b cleavage (Fig. S4B). Thus, activation of both caspase 8 and Bid is required for Cyt b-dependent apoptosis.
Bcl-2 and Cyt b-Mediated Apoptosis.
Blockade of caspase activation by the pan-caspase inhibitor Z-VAD resulted in inhibition of Cyt b-mediated apoptosis, as well as FAS-mediated apoptosis (Fig. 3 C and E). This provides further evidence that Cyt b mediates an apoptotic pathway and suggests that it functions in a manner similar to Cyt c, through induction of the caspase cascade upon release into the cytoplasm. To investigate how Cyt b fits into the current paradigm of mitochondrial apoptosis, we analyzed the effects of known regulators of mitochondrial apoptosis, Bcl-2 and Cyt c, on apoptosis induced by FAS-agonistic antibodies or cytoplasmic Cyt b expression. Bcl-2 is an antiapoptotic member of a large family of proteins that control apoptosis, at least in part, by regulating mitochondrial outer membrane permeability. Activated Bcl-2 counteracts proapoptotic family members and prevents mitochondrial release of apoptogenic factors such as Cyt c, which activate caspases in the cytoplasm (39). Bcl-2 over-expression effectively blocks FAS-induced apoptosis in HeLa cells (see Fig. 3C), indicating that, as for other Type II cells, they die following FAS ligation predominantly via a mitochondria-mediated mechanism. Analysis of endogenous Cyt b in cytoplasmic and mitochondrial fractions of cells overexpressing Bcl-2 following treatment with FAS agonistic antibodies showed that Cyt b cleavage and cytoplasmic release of its C-terminal portion were completely blocked (Fig. 3D). These data suggest that Bcl-2 might block a specific signal originating in the cytoplasm upon FAS activation that is required to trigger mitochondrial events leading to Cyt b cleavage. Bcl-2 could presumably prevent such a signal from passing through the mitochondrial membrane. This hypothesis is supported by our finding that over-expression of Bcl-2 did not alleviate apoptosis induced by ectopic cytoplasmic expression of full-length Cyt b or Cyt b512–1146 (see Fig. 3E).
Cyt c in Cyt b-Mediated Apoptosis.
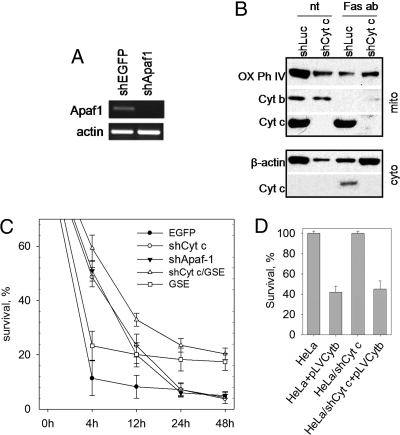
The failure of Bcl-2 over-expression to prevent Cyt b-induced apoptosis suggests that FAS apoptosis mediated by Cyt b might involve a different intrinsic pathway independent of Cyt c/Apaf-1. To test this hypothesis, specific shRNAs were used to knock down expression of Apaf-1 (Fig. 4A) or Cyt c (Fig. 4B) in HeLa cells. Cells were then treated with FAS agonistic antibodies. Knock-down of either component of the apoptosome resulted in a similar significant delay in the rate of FAS-induced cell death; however, the overall extent of cell death 48 h after FAS antibody treatment was the same, regardless of Cyt c or Apaf-1 presence or absence (Fig. 4C). At 4 h after FAS antibody treatment, nearly 90% of cells with intact Cyt c/Apaf1 were dead, as compared to only ≈50% of cells lacking Cyt c or Apaf-1. This could not be accounted for by incomplete gene knock-down, because shCyt c completely blocked apoptosis induced by staurosporine, a known inducer of intrinsic apoptosis (Fig. S5A). These observations indicate that the Cyt c/Apaf-1 pathway can accelerate but is not essential for FAS-mediated apoptosis. In addition, FAS-induced disappearance of Cyt b from mitochondria (see Fig. 4B) and apoptosis induced by cytoplasmic expression of Cyt b were unaffected by knock-down of Cyt c (Fig. 4D). Moreover, expression of GSE F21 increased the proportion of surviving cells to a level that did not depend upon Cyt c knock-down (see Fig. 4C). Taken together, these data suggest that Cyt b acts independently of Cyt c during FAS apoptosis (Fig. S6).
Fig. 4.
Limited dependence of FAS apoptosis on Cyt c and Apaf-1. (A) RT-PCR detection of Apaf-1 and actin RNA in HeLa cells expressing shRNAs targeting Apaf-1 or EGFP (control). (B) Suppression of Cyt c does not affect Cyt b processing during FAS apoptosis. HeLa cells expressing shCyt c or shLuc (negative control) were left untreated (nt) or treated with FAS agonistic antibody and CHI (Fas ab). Western blot analysis of Cyt b and Cyt c was performed on mitochondrial (Upper) and cytoplasmic (Lower) fractions 8 h after treatment. Cytochrome Oxidase Subunit IV (OK Phos Compl IV) and β-actin were used as mitochondrial and cytoplasmic markers, respectively. (C) Time course of FAS apoptosis in control HeLa cells and HeLa cells expressing the indicated constructs. Methylene-blue staining was performed at the indicated times after exposure to FAS antibody plus CHI. Here and in (D), three independent experiments were performed; error bars indicate standard deviations. (D) Knock-down of Cyt c does not prevent Cyt b-induced apoptosis. Methylene-blue staining was performed in control HeLa cells and HeLa cells expressing cytoplasmic Cyt b (pLVCytb), shRNA targeting Cyt c (shCytc) or both. Survival was measured 3 days after lentiviral infection.
Discussion
In this study, we used a previously unusedGSE method based upon RCAS replication competent avian retroviral vectors that allow immediate rescreening of enriched virus subpopulations after each round of functional selection. In addition to expedited screening, this system also offers improved safety, as chicken retroviruses cannot replicate in mammalian cells. The concern of biological differences between avian and mammalian cells is not a serious drawback when the method is applied to evolutionarily conserved pathways.
The RCAS-based GSE method was validated by our isolation of a number of GSEs capable of protecting both chicken and human cells from FAS-mediated apoptosis. The known involvement of several of the identified genes in apoptosis provided confidence in our method and suggested that isolation of a GSE corresponding to Cyt b was meaningful. Our investigation into the relevance of Cyt b to FAS-induced apoptosis revealed that cleavage and cytoplasmic release of a C-terminal portion of Cyt b occurs during FAS-induced apoptosis and that cytoplasmic expression of Cyt b is sufficient to induce apoptosis.
Several lines of evidence indicate that FAS-induced cleavage of Cyt b occurs inside the mitochondria before its release into the cytoplasm. First, ectopically expressed cytoplasmic Cyt b was not cleaved upon treatment with FAS agonistic antibodies (data not shown). Second, in our cell fractionation experiments full-length Cyt b was only observed in mitochondrial fractions. In addition, our finding that ectopic cytoplasmic expression of either full-length Cyt b or Cyt b512–1146 induced apoptosis supports the idea that intra-mitochondrial cleavage of Cyt b is needed not for activation of its proapoptotic activity, but rather for its release from the mitochondria.
If cleavage of Cyt b is a prerequisite for its release to the cytoplasm, then the antiapoptotic activity of GSE F21 might be because of its acting as an inhibitor of Cyt b cleavage. However, although GSE F21-expressing cells are resistant to FAS-dependent apoptosis, FAS antibody treatment still results in cleavage and cytoplasmic release of Cyt b in these cells (data not shown). This, together with the proapoptotic effect of cytoplasmic Cyt b and inhibition of this effect by GSE F21, indicate that the GSE blocks some proapoptotic cytoplasmic function of Cyt b (see below).
Our data suggest the following hypothetical sequence of events occurring upon FAS activation (see Fig. 4): Caspase 8 activation following DISC formation results in generation of a signal of unknown nature that triggers intra-mitochondrial cleavage of Cyt b. This signal is Bid-dependent and can be blocked by over-expression of Bcl-2, suggesting that it involves a factor passing through the mitochondrial pore. It is unlikely that Bcl-2 acts at the point of the Cyt b C terminus leaving the mitochondria, because Bcl-2 over-expression prevents Cyt b cleavage. The C-terminal half of cleaved Cyt b then appears in the cytoplasm. Given the high hydrophobicity of Cyt b and its tight binding to the inner mitochondrial membrane (27), it will be important to identify mediators of this process.
One hypothesis regarding the mechanism of action of the C-terminal Cyt b peptide is that it induces activation of the caspases that execute apoptosis through interaction with a cellular factor, a process which might be blocked by GSE F21. The identity of such cellular mediators of Cyt b-induced caspase activation and of the protease responsible for Cyt b cleavage remains to be determined. These are likely to be unique factors, as (i) the classic Cyt c/Apaf-1 apoptosome is not responsible for Cyt b-induced apoptotic function, and (ii) there are no conventional protease cleavage sites recognizable within Cyt b (although there is a cluster of three overlapping imperfect caspase-recognition sites in the middle of Cyt b) (see Fig. S2). Complete elucidation of the Cyt b apoptosis pathway will be important to resolve the apparent discrepancy between the near complete block of FAS apoptosis observed with Bcl-2 over-expression (see Fig. 3C) and the lack of complete inhibition seen upon knock-down of Cyt c or Apaf-1 (see Fig. 4), suggesting that FAS signaling only partially goes through the canonical Cyt c path.
Preliminary observations (see Fig. S4 A and B) suggest that a similar Cyt b-mediated mechanism is involved in apoptosis initiated by TRAIL and TNFα, thus extending the generality of the described phenomenon beyond FAS apoptosis to all death receptor-mediated programmed cell death.
Materials and Methods
GSE Library Screening.
To convert library DNA into viral stock, 2.5 × 106 DF-1 cells were plated on each of five 15-cm plates and transfected the next day with 50-μg library DNA per plate using Lipofectamine Plus (Invitrogen). A plate of DF-1 cells was transfected with empty RCASBP(A) vector for use as a negative control. Transfection conditions were optimized using an RCASBP(A)-based GFP expression construct to achieve virus titers of at least 106 IU/ml. The virus from each of the five library-transfected plates was kept separate, generating five parallel experimental branches. DF-1FAS cells at ≈20% confluency on 15-cm plates (≈2 × 106 cells per plate) were infected with library virus (five plates) or negative control virus (one plate) overnight with 4-μg/ml polybrene. Two days later, when the cells were nearly confluent (107 cells per plate), they were treated overnight with FAS agonistic antibody (1 μg/ml). Cells were allowed to recover for 1 week and those surviving formed clonal colonies (dead cells were removed by aspiration during medium changes). Virus-containing media collected from the pooled clones surviving from each plate was used to infect fresh DF-1FAS cells. The secondary and tertiary selections were performed as above, except in a six-well plate format (2 × 105 cells per well). Five wells were each infected with one of the five independent virus subpopulations generated by the primary selection and one well was infected with negative control virus (original virus stock). Two days later, the nearly confluent cells (about 106 cells per well) were again subjected to FAS antibody selection. Following tertiary selection, surviving clones from each well were pooled and expanded. Total RNA was isolated and used for RT-PCR with a primer specific for the adaptor flanking each library insert: 5′-AATCATCGATCATGGGTCATGGGTCATGG-3′. Recovered GSEs were sequenced and identified using NCBI BLAST.
Supplementary Material
Acknowledgments.
We thank Elena Feinstein for providing the chicken GSE library and helpful discussions, Stephen Hughes for RCAS vectors, Yuri Lazebnik for the shRNA construct against Cyt c, and Patti Baker for help in manuscript preparation. This work was funded by National Institutes of Health Grants CA60730 and CA75179 (to A.V.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807549105/DCSupplemental.
References
- 1.Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281(5381):1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 2.Chen G, Goeddel DV. TNF-R1 signaling: A beautiful pathway. Science. 2002;296(5573):1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 3.Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 2003;10(1):26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- 4.Krammer PH. CD95's deadly mission in the immune system. Nature. 2000;407(6805):789–795. doi: 10.1038/35037728. [DOI] [PubMed] [Google Scholar]
- 5.Gati A, et al. CD158 receptor controls cytotoxic T-lymphocyte susceptibility to tumor-mediated activation-induced cell death by interfering with Fas signaling. Cancer Res. 2003;63(21):7475–7482. [PubMed] [Google Scholar]
- 6.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1(1):19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 7.Rathmell JC, Thompson CB. Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell. 2002;109(Suppl):S97–S107. doi: 10.1016/s0092-8674(02)00704-3. [DOI] [PubMed] [Google Scholar]
- 8.Thome M, et al. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386(6624):517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 9.Bertin J, et al. Death effector domain-containing herpes virus and pox virus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94(4):1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wajant H. The Fas signaling pathway: More than a paradigm. Science. 2002;296(5573):1635–1636. doi: 10.1126/science.1071553. [DOI] [PubMed] [Google Scholar]
- 11.Kataoka T, et al. The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Curr Biol. 2000;10(11):640–648. doi: 10.1016/s0960-9822(00)00512-1. [DOI] [PubMed] [Google Scholar]
- 12.Irmler M, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388(6638):190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 13.Boatright KM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11(2):529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 14.Yin XM, et al. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature. 1999;400(6747):886–891. doi: 10.1038/23730. [DOI] [PubMed] [Google Scholar]
- 15.Wei MC, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292(5517):727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90(3):405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 17.Verhagen AM, et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102(1):43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 18.Joza N, et al. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410(6828):549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 19.Saelens X, et al. Toxic proteins released from mitochondria in cell death. Oncogene. 2004;23(16):2861–2874. doi: 10.1038/sj.onc.1207523. [DOI] [PubMed] [Google Scholar]
- 20.Susin SA, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397(6718):441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 21.Gudkov AV, et al. Cloning mammalian genes by expression selection of genetic suppressor elements: association of kinesin with drug resistance and cell immortalization. Proc Natl Acad Sci USA. 1994;91(9):3744–3748. doi: 10.1073/pnas.91.9.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holzmayer TA, Pestov DG, Roninson IB. Isolation of dominant negative mutants and inhibitory antisense RNA sequences by expression selection of random DNA fragments. Nucleic Acids Res. 1992;20(4):711–717. doi: 10.1093/nar/20.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudkov AV, et al. Isolation of genetic suppressor elements, inducing resistance to topoisomerase II-interactive cytotoxic drugs, from human topoisomerase II cDNA. Proc Natl Acad Sci USA. 1993;90(8):3231–3235. doi: 10.1073/pnas.90.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roninson IB, et al. Genetic suppressor elements: new tools for molecular oncology. Thirteenth Cornelius P. Rhoads Memorial Award Lecture. Cancer Res. 1995;55(18):4023–4028. [PubMed] [Google Scholar]
- 25.Hughes S, Kosik E. Mutagenesis of the region between env and src of the SR-A strain of Rous sarcoma virus for the purpose of constructing helper-independent vectors. Virology. 1984;136(1):89–99. doi: 10.1016/0042-6822(84)90250-2. [DOI] [PubMed] [Google Scholar]
- 26.Iwata S, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281(5373):64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 27.Xia D, et al. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277(5322):60–66. doi: 10.1126/science.277.5322.60. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392(6677):677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 29.Young JA, Bates P, Varmus HE. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67(4):1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith EJ, Brojatsch J, Naughton J, Young JA. The CAR1 gene encoding a cellular receptor specific for subgroup B and D avian leukosis viruses maps to the chicken tvb locus. J Virol. 1998;72(4):3501–3503. doi: 10.1128/jvi.72.4.3501-3503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adkins HB, Brojatsch J, Young JA. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J Virol. 2000;74(8):3572–3578. doi: 10.1128/jvi.74.8.3572-3578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274(3):1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- 33.Chwae YJ, et al. Molecular mechanism of the activation-induced cell death inhibition mediated by a p70 inhibitory killer cell Ig-like receptor in Jurkat T cells. J Immunol. 2002;169(7):3726–3735. doi: 10.4049/jimmunol.169.7.3726. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima H, Yamada N, Takiguchi M. Fas-independent apoptosis of T cells via killer cell inhibitory receptors. Int Immunol. 1998;10(1):85–90. doi: 10.1093/intimm/10.1.85. [DOI] [PubMed] [Google Scholar]
- 35.Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89(3):413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 36.Chang DW, et al. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21(14):3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He DY, Yu L, Yu CA. Ubiquinone binding domains in bovine heart mitochondrial cytochrome b. J Biol Chem. 1994;269(3):2292–2298. [PubMed] [Google Scholar]
- 38.Chang DW, Xing Z, Capacio VL, Peter ME, Yang X. Interdimer processing mechanism of procaspase-8 activation. EMBO J. 2003;22(16):4132–4142. doi: 10.1093/emboj/cdg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science. 1997;275(5303):1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.