Abstract
Target of rapamycin (TOR) kinases control cell growth through two functionally distinct multiprotein complexes. TOR complex 1 (TORC1) controls temporal cell growth and is sensitive to rapamycin, whereas TOR complex 2 (TORC2) is rapamycin resistant and regulates spatial cell growth. Here, we identified two TOR orthologues, TbTOR1 and TbTOR2, in the protozoan parasite Trypanosoma brucei, as well as orthologues of the well-known TORC1 and TORC2 partners, KOG1/raptor and AVO3/rictor. TbTOR proteins differ in their functions, subcellular localization, and rapamycin sensitivity. TbTOR1 controls cell growth by regulating cell cycle, nucleolus structure, and protein synthesis, whereas TbTOR2 coordinates cell polarization and cytokinesis. Rapamycin treatment of bloodstream trypanosomes resulted in a pronounced reduction of cell proliferation, with an EC50 of 152 nM. Unique for a eukaryote, we observed that rapamycin acted exclusively by preventing TORC2 formation, with no effect on TORC1. Our findings on TOR signaling in this protozoan, which is located in a distal position in the eukaryotic cell lineage, highlight the clinical possibilities of rapamycin derivates and provide valuable insights into understanding rapamycin-mediated inhibition of TORC2.
Keywords: FKBP12, PIKK, target of rapamycin, Trypanosoma brucei
The protozoan parasite Trypanosoma brucei is the etiological agent of African trypanosomiasis, which causes sleeping sickness in humans, affecting up to half a million people. This disease is fatal without medical treatment and development of an effective vaccine is hampered by antigenic variation of the surface coat of this extracellular parasite. Current drugs used to treat African trypanosomiasis either show high toxicity or are not effective at the late neurological stage of the disease.
T. brucei is among the most ancient and evolutionarily divergent eukaryotes, and provides a unique distant reference point from which to investigate crucial biological processes (1). Indeed, trypanosomes possess many unique biological features: marked polarization of the endocytic/exocytic system, metabolic compartmentalization, a single mitochondrion, RNA editing, transsplicing, and transcription of protein-coding genes by RNA polymerase I (2).
We wished to investigate the function of the target of rapamycin (TOR) protein in trypanosomes because of its role as a major regulator of cell growth and proliferation. TOR is a serine/threonine kinase of the phosphatidylinositol kinase-related kinase (PIKK) family that controls cell growth in eukaryotes in response to nutrients, energy conditions, and growth factors (for reviews, see refs. 3 and 4). TOR controls two distinct aspects of cell growth via two different TOR-containing complexes: TOR complex 1 (TORC1) controls temporal aspects of cell growth through processes such as ribosome biogenesis, transcription, and translation, whereas TORC2 controls spatial aspects of cell growth by actin cytoskeleton remodeling. Rapamycin, a highly specific inhibitor of TOR, is a macrolide with a potent immunosuppressant and antitumoral activity. Rapamycin selectively inhibits TORC1 signaling, whereas TORC2 is resistant to the action of this drug (5, 6). Recently, TORC2 was reported to be inhibited in certain human cell lines upon prolonged rapamycin treatment (7).
We have identified two TOR orthologues, named TbTOR1 and TbTOR2, and two other proteins with significant homology to yeast or mammalian TORs, named TbTOR-like 1 and TbTOR-like 2. We demonstrate that control of cell growth in T. brucei is achieved by two functionally distinct TOR kinases through signaling by two distinct TOR complexes. TbTOR1, regulates temporal aspects of cell growth by binding to TORC1. In contrast, TbTOR2 binds exclusively to TORC2 and regulates cell polarization. In contrast to other eukaryotes, rapamycin potently inhibited T. brucei cell proliferation by exclusive inhibition of TORC2 signaling.
Results
Rapamycin Inhibits T. brucei Cell Proliferation.
We first wished to investigate whether the antifungal drug rapamycin can function as a trypanocidal drug. After 72 h of treatment in culture medium, rapamycin showed a potent inhibition effect on bloodstream trypanosomes growth in vitro, with an EC50 of 152 nM (Fig. 1A). In eukaryotes, rapamycin action is mediated by the FKBP12 prolyl isomerase, which binds to and inhibits TOR, blocking protein synthesis and cell cycle progression (8–10). However, protein synthesis was not affected in rapamycin-treated trypanosomes (Fig. 1C), but instead exhibited defects in cytokinesis leading to nonviable multinucleated cells with a prominent enlarged flagellar pocket (FP) (Fig. 1B). Interestingly, treatment of bloodstream trypanosomes with rapamycin did not arrest cell cycle in G1/S phase, which is characteristic of TORC1 inhibition. Thus, we investigated the molecular mechanism of rapamycin in T. brucei, by characterizing the proteins involved.
Fig. 1.
Rapamycin inhibits T. brucei cell proliferation, producing defects in cytokinesis. (A) Rapamycin has potent antiproliferative effects on BSF T. brucei. T. brucei BSF (Molteno Institute Trypanozoon antigenic type 1.2, MITat 1.2, clone 221a) was cultured in HMI-9 medium with the indicated concentrations of rapamycin for 72 h. Culture density was measured as described in Experimental Procedures. The results are expressed as the mean ± standard deviation of three determinations. (B) Rapamycin produces defects in FP structure, cell polarization, and cytokinesis. The effects of rapamycin on the cellular morphology of BSF cells grown in the presence or absence of 1 μM rapamycin for 24 h were monitored by using Nomarski optics and transmission electron microscopy (TEM). (C) Rapamycin treatment does not inhibit protein synthesis in T. brucei. Total protein synthesis was estimated in cells incubated with the indicated concentrations of rapamycin for 24 h.
Four TOR orthologues containing characteristic domains of the PIKK superfamily were identified in the T. brucei genome database. Two of these proteins displayed features not found in other TORs described to date [supporting information (SI) Fig. S1]. These structural features suggested a division between TOR proteins and TOR-like proteins: two sensu stricto TOR orthologues, TbTOR1 and TbTOR2, and two related proteins, TbTOR-like 1 and TbTOR-like 2. The FRB domain was significantly conserved in TbTOR1, TbTOR2, and TbTOR-like 1, but not in TbTOR-like 2. This feature of TOR-like 2 led us to focus on the TbTOR1, TbTOR2, and TbTOR-like 1 kinases. We developed affinity-purified antisera raised against the carboxyl-terminal region of TbTOR1, TbTOR2, and TbTOR-like 1 that specifically recognized proteins of 250–270 kDa in both the bloodstream and procyclic developmental forms (Fig. S2).
TbTOR1 and TbTOR2 Act Through Two Distinct Multiprotein Complexes in T. brucei.
Yeast KOG1 and AVO3, raptor and rictor in mammals, define two distinct TOR-containing complexes, TORC1 and TORC2, which participate in different signaling cascades. KOG1/raptor and AVO3/rictor homologues can be identified in the T. brucei genome (Fig. S3). We expressed myc-tagged TbRaptor and TbAVO3 in bloodstream trypanosomes to investigate whether the TbTOR proteins interacted with any of the two conserved partners necessary for TOR complex function. To analyze specific interactions within TbTOR complexes, we performed coimmunoprecipitation (co-IP) assays by using anti-myc monoclonal antibody and affinity-purified antisera against TbTOR1, TbTOR2, or TbTOR-like 1 using conditions that preserve TOR complex integrity (see Materials and Methods).
Co-IP experiments revealed that TbTOR1 predominantly interacts with TbRaptor, although a weak but reproducible interaction with TbAVO3 was detected (Fig. 2A). We performed the reciprocal experiment by using anti-myc antibody, which confirmed a robust TbTOR1-TbRaptor interaction, but did not reveal a TbTOR1-TbAVO3 interaction. These results suggested that TbTOR1 primarily localizes to TORC1, although there may be a weak association with TORC2. Conversely, co-IP experiments using TbTOR2 anti-serum demonstrated that TbAVO3, but not TbRaptor, coprecipitates with TbTOR2 (Fig. 2B), indicating that TbTOR2 is a TORC2-specific protein. No interaction between TbTOR-like 1 and TbRaptor or TbAVO3 was detected (Fig. 2C), suggesting that TbTOR-like 1 is not involved in either of the two previously described TOR signaling cascades.
Fig. 2.
Identification of two distinct TOR complexes in T. brucei. (A) TbTOR1 coimmunoprecipitates with TbRaptor and TbAVO3. Immunoblotting for TbTOR1 and myc-tagged proteins was performed on TbTOR1 immunoprecipitates prepared from 109 221 BSF trypanosomes expressing TbRaptor-myc, TbAVO3-myc, or TbRaptor-like-myc (see Fig. S4). Anti-myc immunoprecipitates were analyzed in parallel by immunoblotting with the antibodies mentioned above. The arrow indicates the weak association of TbTOR1 with TbAVO3. (B) TbTOR2 specifically interacts with TbAVO3. Lysates expressing TbRaptor-myc, TbAVO3-myc, or TbRaptor-like-myc were subjected to IP with anti-TbTOR2. Immunoprecipitates were analyzed by immunoblotting using anti-myc or anti-TbTOR2. (C) TbTOR-like 1 does not interact with TbRaptor or TbAVO3. (D) Modification of TOR complex stability confirms TbTOR1 interaction with TbAVO3 and the detergent-sensitive association of TbTOR proteins with TOR-interacting proteins. Immunoprecipitates prepared from cells lysed in buffer containing either 0.3% CHAPS or 1% Triton X-100 were analyzed by immunoblotting to detect TbTOR1, TbTOR2, and myc-tagged proteins. Cells were treated as indicated with the cross-linker DSP before (DSP) or after (TX100+DSP) cell lysis with buffer containing 1% Triton X-100. The arrow indicates the weak association of TbTOR1 with TbAVO3.
To further investigate the specificity of these interactions, we used more stringent lysis conditions (1% Triton X-100) to disrupt protein interactions (11), or a protein-cross-linker to stabilize them [reversible cross-linker dithiobis(succinimidyl)propionate (DSP)]. As expected, TbTOR1-TbRaptor and TbTOR2-Tb AVO3 interactions were disrupted in the presence of 1% Triton X-100 (Fig. 2D). Conversely, DSP increased the amount of coprecipitated proteins, confirming the weak TbTOR1-TbAVO3 interaction (see Fig. 2D).
TbTOR1 and TbTOR2 Have Differential Subcellular Localizations.
Localization of signaling molecules plays a critical role in regulating their function and specificity. TOR proteins have been predominantly localized to endoplasmic reticulum (ER) and Golgi membranes, consistent with roles in translational machinery (TORC1) and actin cytoskeleton organization (TORC2). In recent years, localization to mitochondria has revealed that TOR plays a role in sensing osmotic stress and ATP levels (12). TOR has also been localized to the nucleus in mammals and yeasts (13–15). Using three-dimensional (3D) microscopy, we found that TbTOR1 (TORC1) was predominantly nuclear (Fig. 3A). Unlike TbTOR1, TbTOR2 (TORC2) was distributed within the cell cytoplasm, but was excluded from the nucleus (Fig. 3B). Double indirect immunofluorescence (IF) using anti-TbTOR2 plus anti-BiP (an ER marker) or mitotracker for mitochondrial staining showed that TbTOR2 was associated with the ER and mitochondria (Fig. 3C).
Fig. 3.
Distinct subcellular localization of TbTOR proteins in T. brucei. (A) TbTOR1 localizes to the nucleus of T. brucei. The subcellular localization of TbTOR1 was detected by indirect immunofluorescence in bloodstream trypanosomes by using TbTOR1 affinity-purified antiserum. Cells were counterstained with DAPI to locate the nuclear (N) and kinetoplast mitochondrial (K) DNA. (B) TbTOR2 localizes to the cytosol in a punctate pattern and is excluded from the nucleus. (C) TbTOR2 associates with ER membranes and mitochondria as revealed by colocalization with BiP and the mitotracker stain.
TbTOR1 and TbTOR2 Control Distinct Aspects of Cell Growth in T. brucei.
In eukaryotes, TOR kinases regulate temporal and spatial cell growth by their association with distinct protein complexes. To determine the role of TbTOR1 and TbTOR2 in T. brucei growth regulation, we investigated the phenotypic effects of protein depletion by using a tetracycline-inducible RNA interference (RNAi) system (16).
Depletion of each of the RNAi-targeted TbTOR pathway components has a considerable effect on cell proliferation. TbTOR1 protein knockdown (KD), in a tetracycline-inducible manner, resulted in a significant reduction of cell proliferation compared to noninduced cells (Fig. 4A). Similarly, incomplete TbTOR2 depletion upon RNAi induction showed a reduction in cell proliferation (Fig. 4 B and C). This result suggests TbTOR1 does not complement the loss of TbTOR2 in TORC2, as might be inferred from the weak interaction detected between TbTOR1 and TbAVO3 (see Fig. 2A). These data strongly suggest that only TbTOR2, and not TbTOR1, is involved in TORC2 signaling in T.brucei.
Fig. 4.
TbTOR1 and TbTOR2 independently control two distinct aspects of cell growth. (A, B, D, and E) RNAi knockdown of TbTOR1, TbTOR2, TbRaptor, and TbAVO3 affects cell proliferation. Growth curves of bloodstream trypanosome cell lines containing inducible RNAi constructs (as indicated) were obtained after RNAi induction with doxycycline or uninduced as a control. Both induced and uninduced cultures were diluted daily to 2.5 × 104 cells per ml to maintain cell density between 2.5 × 104 and 106 cells per ml. (C) Western blot analysis of TbTOR1 and TbTOR2 in total cell extracts (5 × 106 cells per lane) upon RNAi induction and during the following 4 days. Tubulin was used as a loading control. (F) Analysis of the cellular morphology in bloodstream trypanosome living cells after knockdown of TbTOR1, TbRaptor, TbTOR2, and TbAVO3. TbTOR1 and TbRaptor (TORC1)-depleted cells do not show major changes in morphology. TbTOR2 and TbAVO3 (TORC2) depleted trypanosomes display cytokinesis defects leading to the formation of large abnormal cells. Cells were examined by using Nomarski optics after 2 days of RNAi induction as indicated. (Scale bar: 2 μm.) (G) TORC1 and TORC2 disruption by RNAi have opposite effects on cell size. FACS analysis showing forward scatter (FSC) of RNAi cell lines targeting components of both TOR-containing complexes 2 days after RNAi induction. Kolmogorov–Smirnov statistical analysis of the FSC data showing that changes in cell volume upon protein depletion are statistically significant (P < 0.001) is labeled by asterisks. (H) TbTOR1 regulates nucleolar structure and RNA pol I localization to the nucleolus. Exponentially growing RNAi cell lines were analyzed 2 days after RNAi induction. Nucleolar structure was visualized by DAPI stain as a spherical dark region in the DAPI-stained nucleus, and RNA pol I localization was analyzed by IF using anti-TbRPA1 antibody. Note the presence of multiple nuclei and kinetoplasts in TbTOR2 KD cells without major changes in nucleolus structure. (I) TbTORC1 (TbTOR1 and TbRaptor) positively regulates protein synthesis in T. brucei. Total protein synthesis was estimated in TbTOR1, TbTOR2, TbAVO3, and TbRaptor RNAi KD cell lines after 24 and 48 h of RNAi induction as described in SI Materials and Methods. A significant reduction in protein synthesis was detected upon TbTOR1 and TbRaptor depletion, but not upon TbTOR2 and TbAVO3 depletion experiments. Plots of the means ± SD of four assays are shown.
Observation of live cells by wide field microscopy revealed that TbAVO3 and TbTOR2 depletion both resulted in abnormal cells, whereas TbRaptor or TbTOR1 KD reduced cell growth without altering cell morphology (Fig. 4F). As expected, reduction in the proliferation of TbTOR1- and TbRaptor-depleted cells was because of halted cell cycle progression in G1 phase, revealed by nuclear staining analyzed by FACS analysis (see Fig. S4). In contrast, reduction of cell proliferation by TbTOR2 and TbAVO3 RNAi yield to multinucleated and multiflagellated cells (see Fig. 4F and Fig. S4). These results suggest that cytokinesis is dramatically affected by TbTOR2 and TbAVO3 depletion. Taken together, loss-of-function studies revealed that TbTOR1 and TbTOR2 are functionally independent and control two separate pathways for regulation of cell growth.
TbTOR1 (TORC1) Regulates Protein Synthesis and Cell Size.
Cell size, determined by forward light scatter using flow cytometry, was reduced upon TbTOR1/TbRaptor depletion compared with uninduced cells (Fig. 4G). Thus, TbTOR1 controls cell size in trypanosomes, confirming its positive role in TORC1 signaling. The rate of cell growth correlates with ribosome number and translation initiation efficiency. Because RNA pol I plays a key role in ribosome biogenesis, we analyzed RNA pol I localization to the nucleolus, the region where rRNA transcription and ribosome biogenesis occurs. IF analysis by 3D microscopy revealed clear modification of nuclear DAPI staining and delocalization of TbRPA1 from the nucleolus in TbTOR1-depleted cells (Fig. 4H). To further investigate TbTOR1 function, protein synthesis was assayed upon knockdowns (Fig. 4I). The incorporation of 35S-labeled amino acids into proteins was decreased in TbTOR1- and TbRaptor-depleted cells, indicating that TbTOR1, but not TbTOR2, positively regulates protein synthesis in trypanosomes through TORC1 signaling.
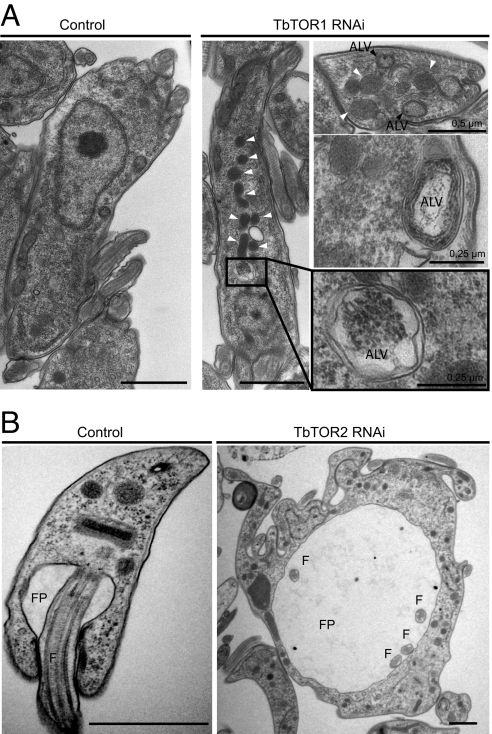
Autophagy is not well studied in T. brucei and no molecular markers have been described so far, although it was suggested to be involved in differentiation processes including glycosomes turnover (17). However, TORC1-mediated regulation of autophagy in other eukaryotes is well known. Analysis of TbTOR1-depleted cells by electron microscopy revealed an increase in the number of electrodense vacuoles and double-membraned vacuoles containing cytoplasmic material reminiscent of autophagosomes (Fig. 5A).
Fig. 5.
Ultrastructural analysis of cells after depletion of TbTOR1 and TbTOR2. (A) TbTOR1 loss-of-function induces autophagic-like vacuoles. TEM of the TbTOR1 RNAi cells reveals the appearance of electrodense vacuoles (white arrowheads) and double-membraned vacuoles (black arrowheads) containing cytoplasmic material. ALV, autophagic-like vacuoles. (B) FP enlargement upon TbTOR2 RNAi induction. Note the size of the lumen of the FP in TbTOR2 RNAi cells compared with that in control cells. Multiple flagella (F) are visible in the TbTOR2 RNAi cells.
TbTOR2 (TORC2) Is Essential for Polarized Cell Growth.
Initial visualization of TbTOR2- and TbAVO3-depleted cells showed profound morphological alterations including large multinucleated cells, in contrast to depletion of TbTOR1 or TbRaptor. Flow cytometry analysis confirmed a dramatic increase in cell size upon TbTOR2 and TbAVO3 depletion (see Fig. 4G). Ultrastructural analysis of TbTOR2-depleted bloodstream trypanosomes showed profound alterations, including a prominent enlarged FP and multinucleated cells (Fig. 5B). In trypanosomes, cell growth is highly polarized toward the FP, the only region where endo- and exocytosis processes take place. Actin is highly polarized toward this region and its depletion in bloodstream trypanosomes results in endocytic defects, producing cells with an enlarged FP (18). Upon TbTOR2 depletion, actin localization by IF in the proximity of the FP was altered resulting in a punctuated pattern throughout the cell (Fig. S5A), suggesting TbTORC2 functions in regulating actin polarization in T. brucei, as proposed for other eukaryotes (6, 19). Furthermore, a reduction of endocytosis upon TbTOR2 depletion was detected by fluorescence-labeled Con A (Fig. S5 B and C).
Importantly, TbTOR2/TbAVO3 RNAi phenotype was similar to that obtained in cells treated with rapamycin (see Fig. 1). These data suggest that rapamycin inhibition of trypanosome growth occurs via inhibition of TbTOR2 (TORC2) signaling, not TbTOR1 (TORC1) signaling.
TbFKPB12-Rapamycin Prevents TbTORC2 Formation.
Rapamycin action is mediated by the FKBP12 prolyl-isomerase through the formation of the FKBP12-rapamycin complex, which binds to and inhibits TOR function. We identified a trypanosome FKBP12 homolog in the T. brucei GeneDB (Fig. S6). To determine whether TbFKBP12 is involved in the formation of the FKBP12-rapamycin-TOR complex, we performed a S. cerevisiae complementation assay. TbFKBP12 complemented yeast FKBP12 function and restored rapamycin sensitivity (Fig. 6A), demonstrating that TbFKBP12 inhibits yeast TOR proteins by binding to rapamycin. To precisely determine the binding specificity of the TbTORs, we investigated whether the TbFKBP12-rapamycin complex was able to bind to each TbTOR protein in vitro. We performed GST pull-down assays by using recombinant FRB domains from the TbTOR1, TbTOR2, and TbTOR-like 1 proteins and incubated with soluble extracts of bloodstream trypanosomes in the presence of rapamycin or drug carrier. Interestingly, only the FRB domain of TbTOR2 interacted with TbFKBP12 in the presence of rapamycin (Fig. 6B). As a control, the TbFKBP12-rapamycin complex binds efficiently to the more conserved FRB domain of Chlamydomonas TOR (see Fig. 6B).
Fig. 6.
Rapamycin inhibits TORC2 signaling solely by preventing TORC2 formation. (A) T. brucei FKBP12 functionally complements a yeast FKBP12 mutant. WT JK9–3da cells and the fpr1 mutant strain lacking the FKBP12 protein were transformed with empty vector or with plasmids expressing WT TbFKBP12. Cultures were normalized, subjected to 10-fold serial dilutions, and spotted onto either SD plates without rapamycin or on plates containing the indicated concentrations of rapamycin. Plates were incubated at 30°C for 3 days. (B) Only the FRB domain of TbTOR2 interacts with TbFKBP12 in the presence of rapamycin. Five micrograms of purified GST-fusion protein containing the FRB domain of TbTOR1, TbTOR2, and TbTOR-like 1 were incubated with 2.5 mg of BSF T. brucei cell extract in the presence of 4 μM rapamycin (+rap) or the same volume of drug vehicle (−rap). The conserved FRB domain of C. reinhardtii was used as a positive control. GST-fusion proteins and TbFKBP12 were detected by Coomassie staining and immunoblotting with the anti-TbFKBP12 antibody, respectively. (C) TbFKBP12-rapamycin complex interacts with free TbTOR2 exclusively. Total BSF cell extract was lysed in CHAPS- or TX-100-containing buffers that preserved or disrupt TORC stability, respectively. TbTOR2 immunoprecipitates were analyzed by immunoblotting for the presence of TbFKBP12. Immunoprecipitates and cell lysates were analyzed by immunoblotting for the levels of TbTOR1, TbTOR2, and TbFKBP12. (D) TbFKBP12-rapamycin complex binds to TbTOR2 in vivo. Trypanosome cells grown in the presence of 1 μM rapamycin were lysed in CHAPS-containing buffers and subjected to TbTOR2 IP. Immunoprecipitates and cell lysates were analyzed by immunoblotting for the levels of TbTOR2 and TbFKBP12. (E) Rapamycin affects the TbAVO3-TbTOR2 interaction (TORC2) progressively, but not TbTOR1-TbRaptor interaction (TORC1). BSF cells expressing TbRaptor or TbAVO3 myc-tagged proteins were treated with rapamycin for 2 or 16 h, resuspended in lysis buffer, and subjected to co-IP using anti-myc antibodies. Myc-tagged immunoprecipitates and cell lysates were analyzed by immunoblotting for the levels of TbTOR1, TbTOR2, or TbRaptor/TbAVO3.
To investigate the inhibition mechanism of TbTORC2 by rapamycin, we analyzed the association of the TbFKBP12-rapamycin complex to TbTOR2 by in vitro co-IP assays by using conditions that either maintain (CHAPS) or destabilize (Triton X-100) the complexes, as shown in Fig. 2D. No interaction between TbFKBP12 and TbTOR1 was observed in the presence of rapamycin (Fig. 6C), in agreement with the recombinant FRB domains binding studies (see Fig. 6B). Interestingly, in vitro co-IP experiments revealed binding of TbFKBP12-rapamycin to TbTOR2 only under conditions that compromised TORC2 integrity, as binding was not observed to intact TORC2 (see Fig. 6C). These results indicate that TbTOR2 in TORC2 is not readily accessible to TbFKBP12-rapamycin, suggesting that other proteins in the complex may block access of TbFKBP12-rapamycin to the FRB domain of TbTOR2. Next, we examined the in vivo binding of TbFKBP12-rapamycin to TbTOR2 by co-IP assays in rapamycin-treated cells. Under these conditions a TbFKBP12-rapamycin-TbTOR2 ternary complex indeed occurs in cells treated with rapamycin for 2 h (Fig. 6D). Together, our in vitro and in vivo co-IP data suggest that TbFKBP12-rapamycin binds exclusively to TbTOR2.
In yeast and humans, the interaction of TOR proteins with raptor or AVO3 is necessary for TORC1 or TORC2 signaling, respectively (11, 19–21). We next asked whether rapamycin affects the stability of trypanosome TOR complexes. Trypanosomes were grown in the presence of rapamycin for either 2 or 16 h, and the amount of TbRaptor and TbAVO3 bound to TbTOR1 and TbTOR2 was monitored (Fig. 6E). The TbTOR1-TbRaptor interaction was not disrupted, consistent with the inability of TbTOR1 to bind to FKBP12-rapamycin (see Fig. 6 B and C). However, rapamycin treatment progressively reduced the TbTOR2-TbAVO3 interaction, which was nearly undetectable after 16 h of rapamycin treatment (see Fig. 6E). In sum, these results demonstrate that rapamycin inhibition in trypanosomes is mediated through the binding of TbFKBP12-rapamycin to TbTOR2, which prevents the formation of TORC2 complexes and signaling.
Discussion
TOR is an essential and conserved protein that governs the spatial and temporal regulation of cell growth in eukaryotes. This study provides a detailed characterization of TOR proteins and rapamycin action in protozoa, and several new findings have emerged. T. brucei represents the earliest eukaryotic branch in which the role of TOR proteins in growth regulation has been reported.
Our data show that TbTOR1 positively controls cell growth in trypanosomes through TORC1 signaling. We demonstrated that TORC1 basic functions are conserved in T brucei. Cells with reduced levels of TbTOR1 showed disperse nucleolar localization of RNA pol I, protein synthesis inhibition, and reduced cell size, as previously shown for other eukaryotes (22–24).
TbTOR2 seems to act exclusively through TORC2 signaling. TbTOR2-depleted cells are not able to correctly segregate their organelles during cytokinesis, and therefore degenerate into large rounded cells containing nuclear aggregates, as well as multiple kinetoplasts and flagella (see Fig. 4). To exclude a possible off-targeting effect, we generated an additional TbTOR2 RNAi cell line, confirming this phenotype (Fig. S7). In trypanosomes, exocytic and endocytic systems are highly polarized toward a discrete growth site, the FP, and determine the spatial growth pattern of trypanosome cells. Interestingly, actin plays an essential role in endocytosis in trypanosome as actin depletion produces an enlarged FP (18) similar to TbTOR2/TbAVO3 RNAi phenotype (see Fig. 5B). Typical actin filaments in T. brucei have not yet been described; however, the enriched area of actin in the proximity of the FP was delocalized and endocytosis was affected upon TbTOR2 depletion (see Fig. S5). Thus, similar to TORC2 function in yeast and mammalian cells (6, 19, 21), our findings strongly suggest that TbTOR2 controls actin cytoskeleton organization and endocytosis through TORC2 signaling.
Most eukaryotes coordinate cell growth via a single TOR, but other organisms, such as yeast and trypanosomes, possess additional TOR genes. A common feature of these organisms is the presence of specialized TOR proteins controlling two distinct aspects of cell growth (25, 26). S. cerevisiae TOR2 is found almost exclusively bound to TORC2, although it is able to bind to TORC1 and to functionally substitute for TOR1 in a tor1 mutant strain (25, 27). Similarly, two functionally distinct TOR proteins control different aspects of T. brucei cell growth (see Fig. 4). Moreover, in organisms with two TOR proteins, polarization is a key aspect of cell growth and viability that is controlled by actin cytoskeleton remodeling through TORC2 signaling. Our results demonstrated that partial decreases in TORC2 signaling produce abnormal growth, leading to nonviable trypanosomes and illustrating the importance of tightly controlled regulation of cell growth polarization for maintaining cell viability.
The functional independence of TbTOR1 and TbTOR2 in trypanosome allowed us to study the subcellular localization of TbTORC1 and TbTORC2. Studies in mTOR localization are controversial, with significant differences depending on the cell line tested or the tissue analyzed (12, 14, 15, 28, 29), and S. cerevisiae TOR1 is the subject of discussion (30). TbTOR proteins localize to distinct subcellular compartments. TbTOR1 (TORC1) is localized to the nucleus (see Fig. 3 A and B). In contrast, TbTOR2 (TORC2) staining shows a cytosolic pattern associated with cytoplasmic organelles, such as the mitochondria and, to a lesser extent, ER (see Fig. 3C). Therefore, localization of TbTOR2 may be essential for interactions with downstream effectors involved in vesicular traffic regulation through actin cytoskeleton organization.
Our results show that trypanosomes are sensitive to rapamycin. Surprisingly, treatment of bloodstream form (BSF) trypanosomes with rapamycin resulted in nonviable multinucleated cells with an enlarged FP (see Fig. 1B), similar to TbTORC2-depleted cells, but did not resemble cells with reduced TbTORC1 function. Contrary to the classical rapamycin action in eukarya, co-IP assays demonstrated that TbFKBP12-rapamycin binds exclusively to TbTOR2, although it is not capable of binding to TbTOR1 (see Fig. 6). TORC2 inhibition by rapamycin has been reported recently in mammalian cells and in fission yeast (7, 31). However, TORC1 signaling is potently inhibited during long-term treatments, and potentially, TORC2 disruption might be an indirect effect because of TORC1 inhibition. In contrast, the rapamycin insensitiveness of TbTOR1 (TORC1) in trypanosomes, together with the functional independence of TbTOR proteins, establishes that TORC2 inhibition by rapamycin occurs in cells with normal TORC1 signaling.
It has been previously reported that FKBP12-rapamycin cannot bind to mTORC2 (19, 21). We demonstrate that FKBP12-rapamycin complex does not bind to TbTOR2 when complexed with other proteins in TORC2, but does bind to free TbTOR2, suggesting that insensitivity of TORC2 toward rapamycin may be because of steric effects caused by TOR binding proteins. Because trypanosome TORC1 is not inhibited by rapamycin, protein synthesis is not affected. In this context, newly synthesized TbTOR2 may be able to bind to TbFKBP12-rapamycin, preventing its association with TORC2 proteins, and thus affecting TORC2 signaling (see Fig. 6). T. brucei is a unique example of an organism in which rapamycin molecular mechanism of action is prevention of functional TORC2 formation exclusively.
These observed differences in the molecular mechanism of rapamycin action, together with the essential role of TORC2 in trypanosomes, make TOR suitable as a drug target. The nonconserved FRB of TbTOR proteins is particularly interesting because multiple residues involved in FKBP12-rapamycin binding differ from human FRB. These differences may be the basis for developing rapamycin analogs with decreased immunosuppressive activity that selectively inhibit TOR proteins in T. brucei.
Materials and Methods
Bioinformatics, Plasmids Constructs, Cell Lines, Antibodies, Recombinant Proteins, Metabolic Labeling, Electron Microscopy, and IF.
Details are provided in SI Materials and Methods.
IP and Cross-linking Assays.
These assays were performed as previously described (11). Anti-myc monoclonal antibody or polyclonal antibodies against different TOR homologs were used to immunoprecipitate TOR complexes in cell lines expressing recombinant myc-tagged TbRaptor or TbAVO3. Cleared extract (5 mg) was used for each IP. To assess the specificity of the co-IP, purification of a raptor-like protein and mock purification of the control cell line extract were also carried out.
Pull-Down Assays.
TbTOR1, TbTOR2, TbTOR-like 1, and crTOR FRB domains (residues 1871–1966, 1893–1987, 2012–2096, and 1961–2055, respectively) were expressed in bacteria as GST-fusion proteins (see above). Purified GST-fusion proteins were immobilized on glutathione Sepharose 4B beads and incubated with 2 mg of T. brucei BSF protein extracts (soluble fraction) and 4 μM rapamycin (LC Labs) or with an equal volume of drug vehicle for 4 h in lysis buffer at 4°C on a rotary incubator. Beads were washed three times with the same buffer, once without CHAPS, resuspended in 30 μl of 2X Laemmli buffer, and resolved by 15% SDS/PAGE.
Yeast Complementation Studies.
Growth of yeast cells on media containing the indicated concentration of rapamycin was assayed by spotting 2.5 μl of 10-fold dilutions of normalized cultures of exponentially growing cells. Plates were incubated at 30°C for 3 days.
Supplementary Material
Acknowledgments.
We thank D. Landeira and X. Peñate for helpful discussions, I. Vidal for technical support, J. Bangs for anti-BiP, and J.A. Garcia-Salcedo for anti-Actin antibodies. This work was funded by Spanish Ministry of Science Grants SAF2006-01763 and BFU2007-60805 (to J.L.C.), and Red de Investigación de Centros de Enfermedades Tropicales Grant (RD06/0021/0010) (to M.N.). M.N. is a Howard Hughes Medical Institute International Research Scholar (HHMI-55005525). A.B. was supported by a Programa de Formación del Profesorado Universitario PhD fellowship (AP-2004-4414).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802668105/DCSupplemental.
References
- 1.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 2.Barry D, McCulloch R, Mottram L, Acosta-Serrano A, editors. Trypanosomes—after the genome. Norfolk, U.K.: Horizon Scientific Press; 2007. [Google Scholar]
- 3.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Zheng XF, et al. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt A, Kunz J, Hall MN. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 8.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 9.Barbet NC, et al. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabatini DM, et al. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 11.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 12.Desai BN, Myers BR, Schreiber SL. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc Natl Acad Sci USA. 2002;99:4319–4324. doi: 10.1073/pnas.261702698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, et al. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 14.Kim JE, Chen J. Cytoplasmic-nuclear shuttling of FKBP12-rapamycin-associated protein is involved in rapamycin-sensitive signaling and translation initiation. Proc Natl Acad Sci USA. 2000;97:14340–14345. doi: 10.1073/pnas.011511898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J Biol Chem. 2002;277:28127–28134. doi: 10.1074/jbc.M202625200. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, et al. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 17.Herman M, et al. Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy. 2008;4:294–308. doi: 10.4161/auto.5443. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Salcedo JA, et al. A differential role for actin during the life cycle of Trypanosoma brucei. EMBO J. 2004;23:780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 20.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 21.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 22.Honma Y, et al. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO J. 2006;25:3832–3842. doi: 10.1038/sj.emboj.7601262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsang CK, et al. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, et al. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 26.Matsuo T, et al. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol Cell Biol. 2007;27:3154–3164. doi: 10.1128/MCB.01039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunz J, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 28.Drenan RM, et al. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J Biol Chem. 2004;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- 29.Bachmann RA, et al. A nuclear transport signal in mammalian target of rapamycin is critical for its cytoplasmic signaling to S6 kinase 1. J Biol Chem. 2006;281:7357–7363. doi: 10.1074/jbc.M512218200. [DOI] [PubMed] [Google Scholar]
- 30.Martin DE, Powers T, Hall MN. Regulation of ribosome biogenesis: Where is TOR? Cell Metab. 2006;4:259–260. doi: 10.1016/j.cmet.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Petersen J, Nurse P. TOR signaling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol. 2007;9:1263–1272. doi: 10.1038/ncb1646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.