Abstract
Due to their low immunogenicity in patients, humanized or fully human mAbs are becoming increasingly important for the treatment of a growing number of diseases, including cancer, infections, and immune disorders. Here, we describe a technology allowing for the rapid isolation of fully human mAbs. In contrast to previously described methods, B cells specific for an antigen of interest are directly isolated from peripheral blood mononuclear cells (PBMC) of human donors. Recombinant, antigen-specific single-chain Fv (scFv) libraries are generated from this pool of B cells and screened by mammalian cell surface display by using a Sindbis virus expression system. This method allows isolating antigen-specific antibodies by a single round of FACS. The variable regions (VRs) of the heavy chains (HCs) and light chains (LCs) are isolated from positive clones and recombinant fully human antibodies produced as whole IgG or Fab fragments. In this manner, several hypermutated high-affinity antibodies binding the Qβ virus like particle (VLP), a model viral antigen, as well as antibodies specific for nicotine were isolated. All antibodies showed high expression levels in cell culture. The human nicotine-specific mAbs were validated preclinically in a mouse model. Thus, the technology presented here allows for rapid isolation of high-affinity, fully human antibodies with therapeutic potential from human volunteers.
Monoclonal antibodies (mAbs) have proven their usefulness for a wide spectrum of research, diagnostic, and therapeutic applications (1). mAbs generated by the conventional hybridoma technology from mice comprise nonhuman sequences, giving rise to an undesired immune response against the foreign sequence when administered therapeutically. Such anti-immunoglobuline responses can interfere with therapy (2) or cause allergic or immune complex hypersensitivity (3). Humanized antibodies (4, 5) and even more so fully human antibodies (6–9) are, therefore, becoming increasingly important for therapeutic applications.
Given the enormous therapeutic and commercial potential of human mAbs, a lot of effort has been put into the development of screening platforms allowing for the isolation of human mAbs with predetermined selectivity. The numerous strategies available for isolation of recombinant antibodies have been reviewed recently (10). In each case, a number of consecutive steps are involved. First, cloning of the immunological diversity contained in the VRs of antibodies by recombinant DNA technology. Second, expression of such antibody libraries by using an expression system suitable for coupling of phenotype with genotype (i.e., binding properties of expressed antibody with its encoding nucleic acid). Third, application of an appropriate selective pressure, typically selection for binding to antigen. And forth, amplification of the selected antibody-encoding clones, leading to an enrichment of specific binders. Typically, antibody libraries are enriched by several rounds of selection before individual clones are analyzed.
The most frequently used screening methods for the isolation of recombinant antibodies are phage display (11–13), ribosome/mRNA display (14, 15), and microbial cell display (16). Whereas each of these screening platforms has its specific advantages, they share the drawback of involving expression of antibodies in a nonnatural environment. Selection not only occurs for desired binding properties but also for physicochemical properties advantageous under the respective screening conditions, leading to a bias in the set of antibodies isolated. In contrast, a selection platform based on the expression of antibodies in the secretory pathway of mammalian cells ensures that all of the cellular components normally involved in antibody synthesis and processing are available, and is likely to yield a set of antibodies less biased by properties other than binding to the desired antigen.
Here, we describe a Sindbis virus-mediated mammalian cell display, a screening platform for the isolation of human antibodies that benefits from the advantages of a mammalian cell-based expression system and is completed in a single round of selection. As a proof of principle, we isolated fully human high-affinity antibodies against the VLP Qβ from an immunized human volunteer. Toward a therapeutic application of the screening strategy, we also isolated a panel of high affinity, fully human antibodies against nicotine, the principle addictive component in tobacco. Preventing the entry of nicotine into the brain by means of active or passive immunization is a promising strategy to aid in smoking cessation (17, 18). As a preclinical proof-of-concept, the therapeutic potential of nicotine-specific antibodies was demonstrated in vivo by showing their ability to inhibit nicotine entry into the brain in mice.
Results
Construction of a Qβ-Specific Antibody Library.
To establish a method for isolation of high-affinity human mAbs (outlined in Fig. 1), the VLP derived from the coat protein of the bacteriophage Qβ, a carrier well suited for vaccine production (18, 19), was used as a model antigen. We based our screening strategy on the generation of libraries from antigen-specific rather than total B cells to ascertain high representation of specific antibodies. To enrich for Qβ-specific, isotype switched B cells, a staining procedure previously established in mice was adapted to humans (20). In this manner, 230 isotype switched Qβ-specific B cells were isolated (Fig. 2A). Total RNA was isolated from these cells, and the immunoglobin VRs were amplified and assembled to scFvs. To allow for the rapid isolation of scFvs by mammalian cell display we adapted our DELphi Sindbis expression cloning system (21, 22) to the screening of surface-displayed antibody libraries. To allow for cell surface display and subsequent screening by FACS, assembled scFvs were cloned into pDel-SP-TM in frame with a mouse Igκ-signal sequence (SP) and the transmembrane domain (TM) of the human PDGFR beta chain. The resulting library made from the 230 Qβ-specific isotype switched B cells consisted of approximately 1 million independent transformants, a number sufficiently high to cover the 52′900 (230 × 230) possible combinations of HCVRs and LCVRs.
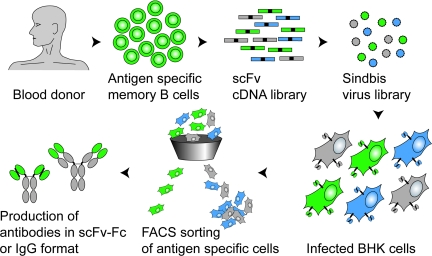
Fig. 1.
Isolation of human antibodies by mammalian cell display. Antigen-specific, isotype switched B cells are isolated from the PBMC of a human donor by FACS by using fluorescently labeled antigen as a bait. RNA isolated from these specific B cells is used to generate a random combinatorial scFv library. This enriched library, typically consisting of high-affinity binders (functional pairing of HCVR and LCVR, green), low affinity binders (suboptimal pairing, blue), and nonbinders (nonfunctional pairing, gray), is then converted to a high-titer Sindbis virus expression library. Infection of BHK cells at a low MOI generates a pool of infected cells, each expressing at their surface one specific antibody. Single cells expressing a functional antibody are then isolated by flow cytometry and sorted onto a monolayer of BHK cells. Once the virus has spread and expression of antigen-specific antibody is verified, the VRs are cloned and the antibody can be expressed in any desired format.
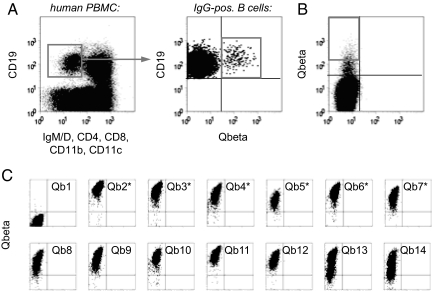
Fig. 2.
Screening for Qβ-specific antibodies. (A) Isolation of Qβ-specific B cells by FACS. Human PBMC were stained for the presence or absence of the indicated surface markers and analyzed for expression of Qβ-specific antibodies. IgM-IgD-negative, isotype switched B cells (as gated in Left) specifically binding to Qβ (as gated in Right) were sorted for library construction. (B) Sorting of BHK cells displaying Qβ-specific antibodies. BHK cells were infected at a low MOI with a Sindbis virus library encoding scFv antibodies constructed from Qβ-specific B cells. After staining with Alexa 647 nm-labeled Qβ, cells displaying specific antibodies were sorted onto semiconfluent BHK feeder cells (1 cell per well). (C) Rescreening for Qβ-specific antibodies. Two to three days after sorting, BHK cells showing signs of viral infection were analyzed for binding to Qβ. The first 14 of the 276 wells analyzed are shown. Antibodies expressed by those clones indicated with an asterisk were selected for gene rescue and sequencing.
To establish the quality, diversity, and overall structural organization of the generated library, six randomly picked clones were sequenced. Each clone corresponded to a different scFv with unique HCVR and LCVR. This observation indicates that even though the library was generated from few antigen-specific B cells, a certain level of diversity was maintained. The HCVRs comprised sequences belonging to the VH3 and VH4 families of VR gene segments (three each). The LCVRs comprised three κ (one Vκ1 and two Vκ3) and three λ LCVRs (two Vλ1 and one Vλ2), indicating that both classes of LCs were represented similarly. Importantly, all six clones were fused in-frame to both signal peptide and transmembrane region. Only one of the analyzed clones had an in-frame stop codon in the HCVR, presumably as a result of a point mutation resulting from the PCR amplification during library construction. Also, all antibodies showed signs of hypermutation if compared with germ-line sequences. The HCVR gene segments had on average 12 mutations leading to amino acid substitutions. Similarly, the κ and λ LCVR gene segments carried on average nine nonsilent point mutations. In conclusion, the human scFv cell-surface display library was diverse, hypermutated, and predominantly consisted of functional antibodies that can be expected to be displayed on the cell surface.
Isolation and Characterization of Qβ-Specific Antibodies.
The pDel-SP-TM scFv library was used to generate replication-competent Sindbis virus with a titer of ≈107 pfu/ml, essentially as described in ref. 21. BHK cells were infected at a low multiplicity of infection (MOI) to ascertain expression of a single antibody species per infected cell. After staining with fluorescently-labeled Qβ VLPs, a total of 480 Qβ-positive BHK cells were single-cell sorted by FACS onto a semiconfluent monolayer of BHK cells to allow for the expansion of the corresponding Sindbis virus clone (Fig. 2B). Two to 3 days later, wells showing typical signs of viral infection were reanalyzed for Qβ binding by FACS. In total, the cells of 276 wells were screened, of which 238 showed binding to Qβ (Fig. 2C).
The scFv coding regions from the first six positive wells were amplified by RT-PCR from virus-containing supernatants and subjected to sequencing. The sequence diversity of these clones was significantly reduced, compared with the one of the original library [supporting information (SI) Fig. S1]. Whereas none of the scFvs were identical, many of the analyzed clones were closely related and contained identical VRs. For instance, scFv-Qb2, scFv-Qb3, scFv-Qb4, and scFv-Qb6 shared the same HCVR. With the exception of scFv-Qb3, all antibodies contained λ LCs. In addition, the LCVRs of scFv-Qb2, scFv-Qb5, and scFv-Qb7 were almost identical and differed by only one to three amino acids.
Based on their sequences, the three antibody clones Qb2, Qb3, and Qb5 were selected for further analysis. To characterize their binding properties, each antibody was produced in three formats: as scFv-Fc fusion protein, as human IgG2, and as human Fab. Antibodies were expressed in HEK 293T cells by using an Epstein–Barr virus (EBV)-based episomal expression vector and purified by affinity chromatography. Notably, all isolated antibodies were highly expressed with yields of ≈30 mg per liter for all three clones in all three formats (Table 1).
Table 1.
Expression levels and affinities of Qβ-specific antibodies
| Format | Clone | Yield, mg/liter | EC50; ELISA, nM | Kd; Friguet, nM |
|---|---|---|---|---|
| scFv-Fc | Qb2 | 30 | 0.05 | ND |
| Qb3 | 27 | 0.04 | ND | |
| Qb5 | 30 | 0.05 | ND | |
| Fab | Qb2 | ND* | 0.9 | 169 |
| Qb3 | ND* | 0.5 | 16.9 | |
| Qb5 | ND* | 0.5 | 0.9 | |
| IgG2 | Qb2 | 37 | 0.06 | 11.7 |
| Qb3 | 29 | 0.04 | 1.5 | |
| Qb5 | 25 | 0.05 | 0.3 |
EC50, half maximal effective concentration; Kd, dissociation constant; ND, not determined.
*Expression levels similar to IgG and scFv-Fc as judged by SDS/PAGE.
The ability of each antibody to bind to the screening antigen (Qβ) was first confirmed by ELISA. Half-maximal binding of Qβ-specific antibodies at subnanomolar concentrations was observed for all investigated antibodies no matter whether the binding was bivalent, as with the scFv-Fc and IgG2, or monovalent, as with the Fab fragment (Table 1). These results suggest that the isolated antibodies are of high affinity. To determine the affinities of the antibody-Qβ interactions more precisely, the equilibrium dissociation constants (Kd) of antigen–antibody interactions were determined in solution by using the method of Friguet (23). As expected, the Kd values obtained in this manner differed significantly from the ELISA results. However, high-affinity interactions could be confirmed, with Kd values in the nanomolar to subnanomolar range (Table 1). The clone with the highest affinity was Qb5 which bound Qβ with a Kd <1 nM, both as Fab and IgG2. In general, the Kd values of the IgG2-Qβ interactions were significantly lower than those of the Fab-Qβ interactions, most likely due to the bivalency of the interactions caused by the multimeric VLP structure of Qβ. The range of affinities observed for the monovalent interactions is very similar to the one reported for tetanus toxoid-specific human antibodies isolated using the Symplex Technology (24).
Isolation and Characterization of Nicotine-Specific Antibodies.
Having demonstrated the potency of Sindbis-based cell display, we set out to screen for antibodies with therapeutic potential. To this end, nicotine, the principle addictive component in tobacco, was chosen as a target. We isolated 443 nicotine-specific B cells from the PBMCs of a volunteer who had been immunized with Qβ-nicotine (Fig. 3). RNA was isolated from these cells and used for construction of a scFv display library as described above. Two sublibraries were generated, one encoding scFv antibodies with κ, the other with λ LCVRs. The scFv-κ and scFv-λ sublibraries consisted of 1.5 × 106 and 1.1 × 106 independent transformants, respectively. Recombinant Sindbis virus libraries with titers of 3 × 106 pfu/ml and 4 × 106 pfu/ml, respectively, were then produced and used to infect BHK cells at low MOI. To avoid isolating carrier-specific antibodies, nicotine coupled to RNase rather than Qβ was used for screening. Thus, cells were stained using RNase-nicotine and appropriate secondary reagents and subjected to FACS. Infected cells which bound RNase-nicotine were single-cell sorted into wells containing BHK feeder cells to allow for amplification of the corresponding Sindbis virus. Two to 3 days later, cells from infected wells were reanalyzed for RNase-nicotine binding and supernatants from positive wells were used to clone the corresponding scFv coding regions by RT-PCR.
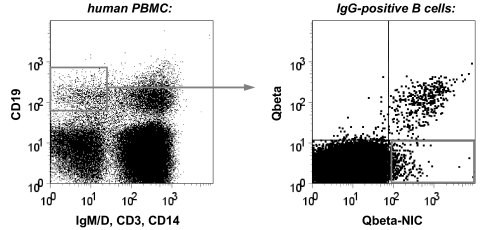
Fig. 3.
Isolation of nicotine-specific B cells by FACS. Human PBMC were stained for the presence or absence of the indicated surface markers and analyzed for expression of Qβ- and nicotine-specific antibodies. Isotype switched B cells (as gated in Left) specifically binding to nicotine but not Qβ (as gated in Right) were sorted for library construction.
Based on the FACS rescreening, 19 clones (Nic01-Nic19) were chosen for further analysis. Each of these scFv was cloned and produced as an Fc fusion protein. Nicotine-specificity was then verified in two ways: by analyzing binding to immobilized RNase-nicotine by ELISA, and by analyzing binding to free nicotine in an inhibition ELISA (data not shown). The seven best binders were chosen for further characterization. First, the dissociation constant of the antibody-nicotine interaction was determined for each of the scFv-Fc by equilibrium dialysis (18). All except clone Nic18 bound nicotine with Kd values in the low nanomolar range, ranging from 12 to 73 nM (Table 2). These Kd values compare favorably with the relatively modest affinities reported for nicotine-specific mAbs generated by standard hybridoma technology (25). In view of their potential use as therapeutic antibodies, the scFv-Fc were also converted to fully human IgG2. Similar to the Qβ-specific antibodies, nicotine-specific antibodies were highly expressed, typically yielding 30 to 50 mg per liter, in IgG2 as well as in scFv-Fc format (Table 2). The IgG2 antibodies had affinities comparable with the scFv-Fc, with Kd values that differed by no more than ≈2-fold. The IgG2-Nic12 had a dissociation constant of 7 nM, almost 2-fold lower than the one measured for scFv-Fc-Nic12 and at least four times lower than the Kd of the other IgG2. It is noteworthy that the affinity of polyclonal antibodies against nicotine is in the range of 30–70 nM, i.e., considerably lower than the affinity of clone Nic12 (18).
Table 2.
Expression levels and affinities of nicotine-specific antibodies
| Format | Clone | Yield, mg/liter | Kd; dialysis, nM |
|---|---|---|---|
| scFv-Fc | Nic12 | 39 | 12 |
| Nic14 | 35 | 71 | |
| Nic15 | 34 | 43 | |
| Nic16 | 48 | 33 | |
| Nic17 | 19 | 47 | |
| Nic18 | 8.6 | 686 | |
| Nic19 | 45 | 73 | |
| IgG2 | Nic12 | 47 | 7 |
| Nic14 | 17 | 31 | |
| Nic15 | 37 | 90 | |
| Nic16 | 36 | 65 | |
| Nic17 | 34 | 30 | |
| Nic18 | 32 | 744 | |
| Nic19 | 34 | 36 |
Efficacy of Passive Immunization in Mice.
To evaluate whether the human mAbs generated may be suitable for the treatment of nicotine addiction, they were tested in a well established animal model for their ability to inhibit entry of nicotine into the brain in mice (17, 18, 25, 26). Thus, mice were injected i.p. with 0.5 mg of each mAb or control IgG. One day later, 750 ng of tritium-labeled (−)-nicotine (≈0.03 mg/kg) were injected i.v. This dose leads to nicotine concentrations in serum similar to the ones found in regular smokers (see ref. 29). Mice were killed 5 min later and the nicotine concentrations in serum and brain determined. Compared with control mice, serum concentrations of nicotine were significantly increased in mice treated with each of the antibodies (data not shown), likely due to the formation of antibody-nicotine complexes, whereas brain nicotine concentrations were significantly reduced (Fig. 4A). In line with its high affinity for nicotine, mAb-Nic12 was the most efficient antibody and led to an almost 3-fold lower brain nicotine concentration. There appeared to be a clear correlation between affinity and efficacy, because mAb-Nic18 which bound nicotine only with a modest affinity displayed the lowest efficacy, whereas the other antibodies with intermediate affinities also showed intermediate efficacy.
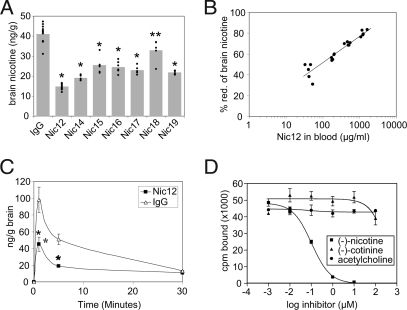
Fig. 4.
Characterization of nicotine-specific antibodies. (A) Inhibition of nicotine entry into brain. Mice were injected i.p. with 0.5 mg of the indicated human IgG2. One day later, mice were injected i.v. with 750 ng of tritium-labeled nicotine. Mice were killed after 5 min and nicotine concentrations in brains were measured. Individual mice are shown; bars indicate average values. IgG, control human IgG. *, P < 0.001 vs. IgG; **, P = 0.003 vs. IgG. (B) Dose-response of inhibition of nicotine entry into brain by IgG2-Nic12. Mice were injected i.p. with 0.1, 0.3, 1, or 3 mg of mAb-Nic12. One day later, mice were injected i.v. with 750 ng of tritium-labeled nicotine. Mice were killed after 5 min and nicotine concentrations in brains were measured. Antibody concentrations in sera were determined and plotted against the percentage nicotine-reduction in the brain. (C) Time course of inhibition of nicotine entry into brain by IgG2-Nic12. Mice were injected with 0.5 mg antibody, challenged with 750 ng of tritium-labeled nicotine and nicotine in the brain was measured at the indicated times. Averages of 4 to 5 mice are given with standard deviations. IgG, control human IgG. *, P < 0.001 vs. IgG. (D) Specificity of mAb-Nic12. The binding of tritium-labeled (−)-nicotine (56 nM) to mAb-Nic12 (50 nM) was measured by equilibrium dialysis in the presence of increasing concentrations of unlabeled (−)-nicotine, (−)-cotinine, or acetylcholine. Averages of two independent experiments are given with standard deviations.
Characterization of Nicotine-Specific mAb-Nic12.
Inhibition of nicotine entry into the brain by mAb-Nic12 was investigated in more detail. To this end, we first carried out an antibody dose titration. Mice were injected i.p. with 0.1, 0.3, 1, or 3 mg of mAb-Nic12 or with 3 mg control IgG. The next day, mice were challenged with 750 ng of nicotine and brain nicotine concentrations determined 5 min later. Compared with control mice, nicotine entry into the brain of mice injected with 0.1, 0.3, 1, or 3 mg of mAb-Nic12 was inhibited by 43%, 56%, 70%, or 80%, respectively (Fig. 4B). Thus, inhibition of nicotine distribution to the brain shows a clear dose-response.
The kinetics of nicotine entry into the brain was also analyzed in more detail. To this end, mice were injected i.p. with 0.5 mg of mAb-Nic12 or control IgG. The next day, the animals were challenged with 750 ng of nicotine and brain nicotine concentrations determined after 1, 5, or 30 min. Nicotine rapidly entered the brain in control animals, and highest amounts of nicotine were detected after 1 min. Concentrations rapidly decreased thereafter and were reduced by 48% after 5 min and by 87% after 30 min (Fig. 4C). This rapid decline in nicotine is best explained by the short half life of nicotine. Importantly, mAb-Nic12 inhibited the entry of nicotine into the brain by 55% and 64% after 1 and 5 min, respectively (Fig. 4C). Thus, mAb-Nic12 was able to efficiently blunt the early nicotine peak. This finding is of particular importance because the fast rise in brain nicotine levels on smoking is believed to provide the rewarding signal to the brain which is central to the nicotine addiction (26). Furthermore, complete blockage of nicotine entry to brain does not appear to be necessary, because passive transfer of polyclonal IgG, causing a 50% reduction of nicotine entry to brain, was shown to be sufficient to reduce nicotine-induced alleviation of nicotine abstinence syndrome in a rat model of nicotine addiction (17). In addition, active vaccination, causing a 40% reduction of nicotine entry to brain, was shown to prevent reinstatement of nicotine-seeking behavior in rats (27).
In view of the promising in vivo activity of mAb-Nic12, its binding properties were investigated in more detail. Thus, binding of tritium-labeled (−)-nicotine was measured by equilibrium dialysis in the presence of increasing concentrations of unlabeled (−)-nicotine, (−)-cotinine, or acetylcholine (Fig. 4D) (18). As expected, unlabeled (−)-nicotine readily displaced tritium-labeled (−)-nicotine. In contrast, acetylcholine, which is structurally unrelated to nicotine but binds to the same binding pocket of the nicotinic acetylcholine receptor (28), did not interfere with nicotine binding even at a 4,000-fold molar excess. Similarly, (−)-cotinine, which is the major metabolic product of (−)-nicotine and present in a 10- to 20-fold molar excess over nicotine in the blood of smokers (29), did not effectively compete with (−)-nicotine for binding to mAb-Nic12. Based on the EC50 observed, it can be estimated that mAb-Nic12 binds (−)-cotinine ≈1,000-fold less strongly than (−)-nicotine. Therefore, the presence of an excess of (−)-cotinine in the blood of a smoker is unlikely to interfere with the therapeutic efficacy of mAb-Nic12.
Discussion
Here, we describe a technology allowing to isolate therapeutically useful mAbs from humans by mammalian cell display. The method offers three key advantages. First, due to the fact that antibodies are isolated from humans, it makes full use of the affinity maturation machinery of the immune system, leading to high-affinity clones. Second, due to the mammalian cell display, isolation of antibodies which show high-expression levels during recombinant expression is favored. And third, because it is completed in a single round of selection, the time-lines are short and purified antigen-specific antibodies can be obtained within few weeks.
Currently, screening of antibody libraries is typically done in a number of different cellular and noncellular environments, such as bacteria (11, 12, 30), yeast (16), or in vitro in a test tube (14, 15). The key advantage of the mammalian antibody display system chosen here is that the antibodies are expressed in their natural environment and due to the presence of the cellular components normally involved in antibody synthesis will be properly folded and glycosylated. Therefore, it is likely that the repertoire of antibodies isolated by using this expression system is less biased by properties other than binding to the antigen. In support of this assumption, it was shown that eukaryotic expression (i.e., yeast display) captures the antibody repertoire more completely than prokaryotic expression (i.e., phage display) (31). Mammalian cells may even be better suited than yeast for the representative expression of antibody repertoires.
One screening system based on cell surface expression of antibodies in mammalian cells has been described recently (32). In this method, a scFv expression library is introduced into HEK 293T cells by means of transient transfection of plasmid DNA, leading to pools of cells expressing scFv antibodies on their surface. FACS-based screening can then be used to identify cells expressing antibodies specific for a particular antigen. A major limitation of this technology is that all transfection methods lead to the delivery of multiple plasmid molecules to each cell. Therefore, each transfected cell expresses several different antibodies, significantly complicating the screening and the chances to identify the right antibody. This method not only increases the selective disadvantage of poorly expressed or otherwise problematic antibodies, but also requires multiple rounds of selection to isolate an antibody of interest. In contrast, the use of the Sindbis expression system allows to precisely dose infection rates, ensuring expression of no more than one antibody per cell, and to directly isolate cells expressing an antibody of interest after a single round of selection.
One major drawback of antibody screening in mammalian cells is the relatively small number of cells that can be handled at a time. Thus, whereas phage display routinely allows for the screening of 1012 to even 1013 clones in a single panning round (33), the throughput of a mammalian screening procedure in a one antibody per cell format is in the range of ≈106 to 107 clones that can be analyzed concomitantly, severely limiting the complexity of libraries amenable to screening. We solved this problem by generating libraries from B cells preselected for antigen-specificity rather than from total B cells. Because libraries are typically constructed from pools of <500 antigen-specific B cells, the resulting random combinatorial libraries have only small theoretical diversities and can, therefore, readily be screened in mammalian cells.
Several methods for the isolation of mAbs from human peripheral B cells have been described, including their cloning from single or amplified B cell clones (9, 34) and from EBV-immortalized memory B cells (8). Whereas efficiency of these methods has been shown, the mammalian display method described here is advantageous, because it does not involve cumbersome and technically challenging PCR amplification of VRs from single B cells, nor does it require manipulation and propagation of viable B cells in vitro. Whereas the efficiency of single cell PCR and thus cloning of human antibodies can be improved by using plasma cells rather than B cells (24), also this approach is limited because plasma cells are only found in blood within a very narrow time frame (i.e., few days) after encounter with the antigen. In contrast, the method described here isolates mAbs from memory B cells, which are present for years in immune individuals.
In the present study, we have isolated mAbs from immunized individuals. However, immunization may often not be required, in particular not for the isolation of pathogen-specific antibodies due to natural immunity in previously infected individuals. Also, even for hormone- or cytokine-specific antibodies, immunization may not be necessary, because individuals with spontaneous high levels of such antibodies are relatively frequent. Indeed, humans with high levels of anti-IL6 or anti-IL1α antibodies are found at frequencies of 1:100 to 1:4,000 (35, 36). Also, by screening as little as 50 blood samples, we found one blood donor exhibiting high levels of antibodies specific for ghrelin, a hormone involved in appetite regulation (data not shown).
All of the antibodies we isolated showed signs of hypermutation, indicating that they went through the germinal center machinery yielding high-affinity antibodies. This may offer a clear advantage over humanized mice, from which it is often difficult to isolate high-affinity antibodies that have undergone isotype-switching as well as hypermutation.
In conclusion, the technology described here allows for the rapid isolation of highly expressed, clinically useful fully human antibodies.
Materials and Methods
Analysis of Antibody Sequences.
HCVR and LCVR sequences were analyzed by using the International ImMunoGeneTics Information System (http://imgt.cines.fr) (37).
Antibodies.
Unless stated otherwise, all antibodies were purchased from BD Biosciences PharMingen and Jackson ImmunoResearch Laboratories.
Isolation of Qβ-Specific B-Cells.
After informed consent was obtained, PBMC were isolated from 20 ml of heparinized blood of a Qβ-vaccinated volunteer by a standard Ficoll-HypaqueTM Plus (Amersham Biosciences) gradient method. PBMC were stained with: Alexa 647 nm-labeled Qβ (4 μg/ml); FITC-labeled mouse anti-human IgM, mouse anti-human IgD, mouse anti-human CD4, mouse anti-human CD8, rat anti-mouse/human CD11b, and mouse anti-human CD11c (eBioscience); and PE-labeled mouse anti-human CD19. After 30 min, cells were washed, filtered, and stained with propidium iodide (PI) to exclude dead cells. Qβ-specific B cells (Qβ-/CD19-positive, IgM-/IgD-/CD4-/CD8-/CD11b-/Gr1-/PI-negative) were sorted and used for library construction. All sorts were done on a FACS Vantage SE flow cytometer (Becton Dickinson).
Isolation of Nicotine-Specific B-Cells.
After informed consent was obtained, PBMC were isolated from 32 ml of heparinized blood of a Qβ-nicotine vaccinated volunteer, by using the Vacutainer CPT Tube method (BD Bioscience). PBMC were preincubated with Alexa 647 nm-labeled Qβ (3 μg/ml) and mouse gamma globulin (10 μg/ml) and then stained with: Qβ-Nicotine (1 μg/ml) in combination with a Alexa 488 nm-labeled Qβ-specific mouse mAb and a Alexa 488 nm-labeled Nicotine-specific mouse mAb (1 μg/ml); PE-labeled mouse anti-human IgM, mouse anti-human IgD, mouse anti-human CD14, and mouse anti-human CD3 antibodies; and PE-TexasRed-labeled mouse anti-human CD19 antibody (Caltag Laboratories). After staining, cells were washed and filtered, and Nicotine-specific B cells (Qβ-Nicotine-/CD19-positive, Qβ-/IgM-/IgD-/CD3-/CD14-negative) were sorted and used for library construction.
Construction of scFv Libraries.
Total RNA was isolated from antigen-specific human B cells by using TRI reagent (Molecular Research, Inc.). Double-stranded cDNA was produced by using the SMART PCR cDNA synthesis kit (Clontech). HCVRs and LCVRs were amplified and assembled to scFv coding regions by PCR by using primers and protocols described previously (33). The resulting PCR products encoded a 5′ LCVR (either κ or λ) and a 3′ HCVR, linked by an 18-aa flexible linker, and were flanked by two SfiI restriction sites. For the Qβ-specific scFv library, the κ- and λ-containing scFv fragments were pooled in equimolar ratio. In the case of the nicotine-specific library, the κ- and λ-containing scFv fragments were processed separately. The scFv fragments were digested with the restriction endonuclease SfiI and cloned into the SfiI-digested vector pDel-SP-TM, a derivative of pSinRep5 (21, 38); pDel-SP-TM allows for expression of scFv fused to an N-terminal mouse Igκ-signal sequence (SP) and a C-terminal TM domain of the human PDGFR beta chain, to ascertain cell surface localization. DNA was isolated from pooled colonies by using the HiSpeed Plasmid Maxi Kit (Qiagen).
Isolation of BHK Cells Displaying Antigen-Specific scFv by FACS.
Sixty million subconfluent (80%) BHK cells were infected with each scFv library or an empty viral vector as a negative control at a MOI of 0.2. After 5 h, cells were detached with cell dissociation buffer (Sigma), washed, and stained. To isolate cells displaying Qβ-specific scFv antibodies, half of the cells were stained with Alexa 647 nm-labeled Qβ. The remaining cells were stained with Alexa 546 nm-labeled Qβ and a rabbit anti-sindbis serum, followed by staining with Cy5-labeled donkey anti-rabbit IgG. All cells were then washed, filtered, and stained with PI to exclude dead cells. Single cell sorting was performed for, respectively, Alexa 647 nm-positive and PI-negative and Alexa 546 nm-positive, Sindbis-positive, and PI-negative cells. Cells displaying nicotine-specific scFv antibodies were isolated by staining with RNase-nicotine in combination with an RNase-specific rabbit polyclonal antibody (Abcam) and a FITC-labeled donkey anti-rabbit IgG antibody, followed by sorting of FITC-positive, PI-negative cells. Each positive cell was sorted into a well of a 24-well plate containing 50% confluent BHK feeder cells. On days 2 and 3 after sorting, wells showing typical signs of viral infection were tested by FACS analysis for, respectively, Qβ or nicotine binding to identify virus clones encoding antigen-specific scFv antibodies.
Expression and Purification of Antibodies.
Fusion proteins were generated carrying an N-terminal human scFv fused to a C-terminal human Fc-γ1 domain. Thus, scFv coding regions were digested with the restriction endonuclease SfiI and cloned into the expression vector pCEP-SP-Sfi-Fc. This vector is a derivative of the episomal mammalian expression vector pCEP4 (Invitrogen), carrying the EBV replication origin and encoding the EBV nuclear antigen (EBNA-1) to permit extrachromosomal replication, and contains a puromycin selection marker in place of the original hygromycin B resistance gene. The resulting plasmids drive expression of secreted scFv-Fc domain fusion proteins under the control of a CMV promoter.
For expression as IgG2 or Fab, the HCVR and LCVR coding segments of Qβ- or nicotine-specific scFv antibodies were amplified by PCR by using VR-specific transfer primers, essentially as described in ref. 9. Fully human κ LC, λ LC, γ2 HC, and γ2 Fd chain expression constructs were assembled in pCMV-Script (Stratagene). HC and LC coding regions were then combined into a single, EBV-based episomal expression vector, pCB15. This vector contains two CMV promoters for concomitant expression of HC and LC and carries a puromycin selection marker. Thus, for expression of a whole IgG2, a κ or λ LC coding region was combined with a γ2 HC coding region. To generate a Fab expression vector, a LC coding region was combined with a Fd coding region.
Expression of antibodies was done by transfecting the expression vectors into HEK-293T cells using Lipofectamin Plus (Invitrogen). For large scale production and purification, stable protein-expressing cells were enriched by selection in the presence of puromycin (Sigma). Pools of resistant cells were maintained in serum-free medium on Poly-L-Lysine coated dishes or in roller bottles for up to three weeks. Supernatants containing the respective antibodies were collected twice a week. All antibodies were purified by affinity chromatography over protein A-Sepharose or protein G-Sepharose columns (GE healthcare), except Fab, which were purified by affinity chromatography over a goat anti-human F(ab′)2 column.
Evaluation of Nicotine Distribution in Plasma and Brain in Mice.
All animal experiments were carried out in accordance with Swiss guidelines. Groups of 5 to 12 female BALB/c mice were injected i.p. with 0.1 to 3 mg of each of the recombinant human IgG2 mAbs. One day later, a small sample of blood was collected to determine the serum concentration of nicotine-specific antibody by ELISA. On the same day, inhibition of nicotine entry into the brain was analyzed by i.v. injection of 750 ng of tritium-labeled (−)-nicotine (Amersham) into the tail vein (5 μCi in 150-μl PBS per mouse). Mice were killed 1, 5, or 30 min later by CO2 asphyxiation and blood and brains were collected. Nicotine concentrations in serum and brain were calculated from the radioactivity present. Brain nicotine concentration was corrected for the blood content of brain (3 μl/100 mg). The percentage reduction of nicotine uptake into the brain was calculated relative to the nicotine concentrations found in brains of mice injected with control IgG. Brain nicotine concentrations were compared by using a two-tailed Student's t test assuming equal variance. The following criteria were used in all experiments to exclude mice from the analysis: inefficient relocation of antibody to serum on i.p. injection (at least five times lower titer than average of other mice of same group) and significant loss of nicotine solution during i.v. injection procedure.
Footnotes
Conflict of interest statement: All authors are employees and owners of stock options of Cytos Biotechnology AG, and therefore declare potential competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805942105/DCSupplemental.
References
- 1.Reichert JM, Rosensweig CJ, Faden LB, Dewitz MC. Monoclonal antibody successes in the clinic. Nat Biotechnol. 2005;23:1073–1078. doi: 10.1038/nbt0905-1073. [DOI] [PubMed] [Google Scholar]
- 2.Miller RA, Oseroff AR, Stratte PT, Levy R. Monoclonal antibody therapeutic trials in seven patients with T-cell lymphoma. Blood. 1983;62:988–995. [PubMed] [Google Scholar]
- 3.Ratner B. Allergy, Anaphylaxis and Immunotherapy, Basic Principles and Practice. Baltimore: Williams and Wilkins; 1943. [Google Scholar]
- 4.Foote J, Winter G. Antibody framework residues affecting the conformation of the hypervariable loops. J Mol Biol. 1992;224:487–499. doi: 10.1016/0022-2836(92)91010-m. [DOI] [PubMed] [Google Scholar]
- 5.Riechmann L, Clark M, Waldmann H, Winter G. Reshaping human antibodies for therapy. Nature. 1988;332:323–327. doi: 10.1038/332323a0. [DOI] [PubMed] [Google Scholar]
- 6.Lonberg N. Human antibodies from transgenic animals. Nat Biotechnol. 2005;23:1117–1125. doi: 10.1038/nbt1135. [DOI] [PubMed] [Google Scholar]
- 7.Mendez MJ, et al. Functional transplant of megabase human immunoglobulin loci recapitulates human antibody response in mice. Nat Genet. 1997;15:146–156. doi: 10.1038/ng0297-146. [DOI] [PubMed] [Google Scholar]
- 8.Traggiai E, et al. An efficient method to make human monoclonal antibodies from memory B cells: Potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weitkamp JH, et al. Generation of recombinant human monoclonal antibodies to rotavirus from single antigen-specific B cells selected with fluorescent virus-like particles. J Immunol Methods. 2003;275:223–237. doi: 10.1016/s0022-1759(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 10.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 11.Barbas CF, 3rd, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: The gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 13.Krebs B, et al. High-throughput generation and engineering of recombinant human antibodies. J Immunol Methods. 2001;254:67–84. doi: 10.1016/s0022-1759(01)00398-2. [DOI] [PubMed] [Google Scholar]
- 14.Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He M, Taussig MJ. Antibody-ribosome-mRNA (ARM) complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res. 1997;25:5132–5134. doi: 10.1093/nar/25.24.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 17.Malin DH, et al. Passive immunization against nicotine prevents nicotine alleviation of nicotine abstinence syndrome. Pharmacol Biochem Behav. 2001;68:87–92. doi: 10.1016/s0091-3057(00)00436-6. [DOI] [PubMed] [Google Scholar]
- 18.Maurer P, et al. A therapeutic vaccine for nicotine dependence: Preclinical efficacy, and Phase I safety and immunogenicity. Eur J Immunol. 2005;35:2031–2040. doi: 10.1002/eji.200526285. [DOI] [PubMed] [Google Scholar]
- 19.Tissot AC, et al. Effect of immunisation against angiotensin II with CYT006-AngQb on ambulatory blood pressure: A double-blind, randomised, placebo-controlled phase IIa study. Lancet. 2008;371:821–827. doi: 10.1016/S0140-6736(08)60381-5. [DOI] [PubMed] [Google Scholar]
- 20.Gatto D, Ruedl C, Odermatt B, Bachmann MF. Rapid response of marginal zone B cells to viral particles. J Immunol. 2004;173:4308–4316. doi: 10.4049/jimmunol.173.7.4308. [DOI] [PubMed] [Google Scholar]
- 21.Koller D, et al. A high-throughput alphavirus-based expression cloning system for mammalian cells. Nat Biotechnol. 2001;19:851–855. doi: 10.1038/nbt0901-851. [DOI] [PubMed] [Google Scholar]
- 22.Sirena D, et al. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J Virol. 2004;78:4454–4462. doi: 10.1128/JVI.78.9.4454-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friguet B, Chaffotte AF, Djavadi-Ohaniance L, Goldberg ME. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 24.Meijer PJ, et al. Isolation of human antibody repertoires with preservation of the natural heavy and light chain pairing. J Mol Biol. 2006;358:764–772. doi: 10.1016/j.jmb.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 25.Keyler DE, et al. Monoclonal nicotine-specific antibodies reduce nicotine distribution to brain in rats: Dose- and affinity-response relationships. Drug Metab Dispos. 2005;33:1056–1061. doi: 10.1124/dmd.105.004234. [DOI] [PubMed] [Google Scholar]
- 26.Maurer P, Bachmann MF. Vaccination against nicotine: An emerging therapy for tobacco dependence. Expert Opin Investig Drugs. 2007;16:1775–1783. doi: 10.1517/13543784.16.11.1775. [DOI] [PubMed] [Google Scholar]
- 27.Lindblom N, et al. Active immunization against nicotine prevents reinstatement of nicotine-seeking behavior in rats. Respiration. 2002;69:254–260. doi: 10.1159/000063629. [DOI] [PubMed] [Google Scholar]
- 28.Celie PH, et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 29.Benowitz NL, Zevin S, Jacob P., 3rd Sources of variability in nicotine and cotinine levels with use of nicotine nasal spray, transdermal nicotine, and cigarette smoking. Br J Clin Pharmacol. 1997;43:259–267. doi: 10.1111/j.1365-2125.1997.00566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazor Y, Van Blarcom T, Mabry R, Iverson BL, Georgiou G. Isolation of engineered, full-length antibodies from libraries expressed in Escherichia coli. Nat Biotechnol. 2007;25:563–565. doi: 10.1038/nbt1296. [DOI] [PubMed] [Google Scholar]
- 31.Bowley DR, Labrijn AF, Zwick MB, Burton DR. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng Des Sel. 2007;20:81–90. doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]
- 32.Ho M, Nagata S, Pastan I. Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc Natl Acad Sci USA. 2006;103:9637–9642. doi: 10.1073/pnas.0603653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbas CF, 3rd, Burton DR, Scott JK, Silverman GJ. Phage Display: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 34.Lagerkvist AC, Furebring C, Borrebaeck CA. Single, antigen-specific B cells used to generate Fab fragments using CD40-mediated amplification or direct PCR cloning. BioTechniques. 1995;18:862–869. [PubMed] [Google Scholar]
- 35.Galle P, Svenson M, Bendtzen K, Hansen MB. High levels of neutralizing IL-6 autoantibodies in 0.1% of apparently healthy blood donors. Eur J Immunol. 2004;34:3267–3275. doi: 10.1002/eji.200425268. [DOI] [PubMed] [Google Scholar]
- 36.Jouvenne P, Fossiez F, Banchereau J, Miossec P. High levels of neutralizing autoantibodies against IL-1 alpha are associated with a better prognosis in chronic polyarthritis: A follow-up study. Scand J Immunol. 1997;46:413–418. doi: 10.1046/j.1365-3083.1997.d01-139.x. [DOI] [PubMed] [Google Scholar]
- 37.Scaviner D, Barbie V, Ruiz M, Lefranc MP. Protein displays of the human immunoglobulin heavy, kappa and lambda variable and joining regions. Exp Clin Immunogenet. 1999;16:234–240. doi: 10.1159/000019115. [DOI] [PubMed] [Google Scholar]
- 38.Bredenbeek PJ, Frolov I, Rice CM, Schlesinger S. Sindbis virus expression vectors: Packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]