Abstract
Recent molecular evidence of genetically distinct hantaviruses in shrews, captured in widely separated geographical regions, corroborates decades-old reports of hantavirus antigens in shrew tissues. Apart from challenging the conventional view that rodents are the principal reservoir hosts, the recently identified soricid-borne hantaviruses raise the possibility that other soricomorphs, notably talpids, similarly harbor hantaviruses. In analyzing RNA extracts from lung tissues of the Japanese shrew mole (Urotrichus talpoides), captured in Japan between February and April 2008, a hantavirus genome, designated Asama virus (ASAV), was detected by RT-PCR. Pairwise alignment and comparison of the S-, M-, and L-segment nucleotide and amino acid sequences indicated that ASAV was genetically more similar to hantaviruses harbored by shrews than by rodents. However, the predicted secondary structure of the ASAV nucleocapsid protein was similar to that of rodent- and shrew-borne hantaviruses, exhibiting the same coiled-coil helix at the amino terminus. Phylogenetic analyses, using the maximum-likelihood method and other algorithms, consistently placed ASAV with recently identified soricine shrew-borne hantaviruses, suggesting a possible host-switching event in the distant past. The discovery of a mole-borne hantavirus enlarges our concepts about the complex evolutionary history of hantaviruses.
Keywords: host switching, talpid, evolution, Japan
Dating from investigations conducted independently by Japanese and Russian medical scientists along opposite sides of the Amur River in the 1930s and 1940s, rodents have been suspected to harbor the etiological agent(s) of hemorrhagic fever with renal syndrome (HFRS) (1, 2). After a several decades-long impasse, the striped field mouse (Apodemus agrarius) was identified as the reservoir host of Hantaan virus (3), the prototype virus of HFRS (4). This seminal discovery made possible the identification of genetically distinct hantaviruses in other murinae and arvicolinae rodent species (5–12). Also, a previously unrecognized, frequently fatal respiratory disease, called hantavirus pulmonary syndrome (HPS) (13), is now known to be caused by hantaviruses harbored by neotominae and sigmodontinae rodents in the Americas, the prototype being Sin Nombre virus (SNV) in the deer mouse (Peromyscus maniculatus) (14). Remarkably, each of these hantaviruses appears to share a long coevolutionary history with a specific rodent host species. That is, based on phylogenetic analyses of full-length viral genomic and rodent mitochondrial DNA (mtDNA) sequences, these hantaviruses segregate into clades, which parallel the evolution of rodents in the murinae, arvicolinae, neotominae, and sigmodontinae subfamilies (15, 16).
Until recently, the single exception to the strict rodent association of hantaviruses was Thottapalayam virus (TPMV), a long-unclassified virus originally isolated from the Asian house shrew (Suncus murinus) (17, 18). Analysis of the recently acquired full genome of TPMV strongly supports an ancient non-rodent host origin and an early evolutionary divergence from rodent-borne hantaviruses (19, 20). Employing RT-PCR and oligonucleotide primers based on the TPMV genome, we have targeted the discovery of hantaviruses in shrew species from widely separated geographical regions, including the Chinese mole shrew (Anourosorex squamipes) from Vietnam (21), Eurasian common shrew (Sorex araneus) from Switzerland (22), northern short-tailed shrew (Blarina brevicauda), masked shrew (Sorex cinereus), and dusky shrew (Sorex monticolus) from the United States (23, 24) and Ussuri white-toothed shrew (Crocidura lasiura) from Korea (J.-W. Song and R. Yanagihara, unpublished observations). Many more shrew-hantavirus associations undoubtedly exist, as evidenced by preliminary studies of Sorex caecutiens and Sorex roboratus from Russia (H. J. Kang, S. Arai and R. Yanagihara, unpublished observations) and Sorex palustris, Sorex trowbridgii, and Sorex vagrans from North America (H. J. Kang and R. Yanagihara, unpublished observations).
In addition to challenging the view that rodents are the sole or principal reservoirs of hantaviruses, the discovery of soricid-borne hantaviruses predicts that other soricomorphs, notably talpids, might also harbor genetically distinct hantaviruses. In this regard, hantavirus antigens have been detected by enzyme immunoassay and fluorescence techniques in tissues of the European common mole (Talpa europea) captured in Russia (25) and Belgium (26), but no reports are available about hantavirus infection in shrew moles. Relying on oligonucleotide primers designed from our expanding sequence database of shrew-borne hantaviruses, we have identified a hantavirus genome, designated Asama virus (ASAV), in the Japanese shrew mole (Urotrichus talpoides). Genetic and phylogenetic analyses indicate that ASAV is distinct but related to hantaviruses harbored by Old World soricine shrews, suggesting a very ancient evolutionary history, probably involving multiple host-switching events in the distant past.
Results
RT-PCR Detection of Hantavirus Sequences.
In using RT-PCR to analyze RNA extracts, from lung tissues of three Laxmann's shrew (Sorex caecutiens), five slender shrew (Sorex gracillimus), six long-clawed shrew (Sorex unguiculatus), one dsinezumi shrew (Crocidura dsinezumi), and six Japanese shrew mole (Urotrichus talpoides), hantavirus sequences were not detected in shrew tissues, but were found in one of two and in two of three Japanese shrew moles (Fig. 1), captured in Ohtani (34 28′14.0° N; 136 45′46.2° E) and near Asama River (34 28′12.79° N; 136 45′45.81° E), respectively, located approximately 1 km apart at an elevation of 50 m in Mie Prefecture, during February and April 2008. After the initial detection of hantavirus sequences, amplification of the S-, M-, and L-genomic segments was accomplished by using oligonucleotide primers based on conserved regions.
Fig. 1.
Japanese shrew mole (Urotrichus talpoides) (family Talpidae, subfamily Talpinae), one of two endemic shrew mole species found only in Japan.
Nucleotide and Amino Acid Sequence Analysis.
The S, M, and L segments of ASAV, as amplified from tissues of three wild-caught Japanese shrew moles, indicated an overall genomic structure similar to that of other rodent- and soricid-borne hantaviruses. The nucleotide and deduced-amino acid sequences of each ASAV genomic segment were highly divergent from that of rodent-borne hantaviruses, differing by approximately 30–40% (Table 1).
Table 1.
Nucleotide and amino acid sequence similarity (%) between ASAV strain N10 and representative rodent- and shrew-borne hantaviruses
| Virus strain | S segment |
M segment |
L segment |
|||
|---|---|---|---|---|---|---|
| 1710 nt | 434 aa | 3604 nt | 1141 aa | 6126 nt | 2041aa | |
| HTNV 76–118 | 58.5 | 62.7 | 62.7 | 59.4 | 70.3 | 74.6 |
| SEOV 80–39 | 63.1 | 62.0 | 62.8 | 59.1 | 70.4 | 74.7 |
| SOOV SOO-1 | 62.8 | 62.9 | 63.3 | 59.7 | 70.2 | 74.3 |
| DOBV Greece | 62.2 | 62.2 | 63.0 | 59.3 | 70.2 | 75.7 |
| PUUV Sotkamo | 59.3 | 59.3 | 59.6 | 52.2 | 68.1 | 68.0 |
| TULV 5302v | 61.5 | 59.4 | 60.5 | 52.6 | 68.3 | 67.9 |
| PHV PH-1 | 60.7 | 59.3 | 59.3 | 51.9 | 66.4 | 67.1 |
| SNV NMH10 | 60.9 | 58.9 | 59.0 | 54.1 | 68.2 | 68.8 |
| RPLV MSB89866 | — | — | 68.8 | 63.5 | 75.2 | 83.2 |
| CBNV CBN-3 | 67.7 | 70.4 | 68.2 | 71.0 | 76.0 | 84.7 |
| ARRV MSB73418 | 65.7 | 66.6 | 70.9 | 77.0 | 73.8 | 83.5 |
| JMSV MSB89332 | 66.2 | 66.9 | — | — | 74.3 | 82.6 |
| SWSV mp70 | 63.8 | 69.9 | 75.2 | 79.5 | 75.0 | 83.2 |
| ASAV H4 | 98.9 | 100 | 99.3 | 99.6 | 98.2 | 99.6 |
| ASAV N9 | 100 | 100 | 99.9 | 100 | 100 | 100 |
| MJNV 05–11 | 57.2 | 46.0 | 56.1 | 44.4 | 65.8 | 61.5 |
| TPMV VRC | 58.0 | 45.8 | 57.7 | 43.0 | 64.3 | 62.0 |
Abbreviations: ARRV, Ash River virus; ASAV, Asama virus; CBNV, Cao Bang virus; DOBV, Dobrava virus; HTNV, Hantaan virus; JMSV, Jemez Spring virus; MJNV, Imjin virus; PHV, Prospect Hill virus; PUUV, Puumala virus; RPLV, Camp Ripley virus; SEOV, Seoul virus; SNV, Sin Nombre virus; SOOV, Soochong virus; SWSV, Seewis virus; TPMV, Thottapalayam virus; TULV, Tula virus. nt, nucleotides; aa, amino acids.
The S-genomic segment of ASAV (1,801 nucleotides for strains H4 and N9 and 1,756-nucleotides for strain N10) encoded a predicted nucleocapsid (N) protein of 434 amino acids, starting at nucleotide position 39, and a 3′-noncoding region (NCR) of approximately 465 nucleotides. The hypothetical NSs opening reading frame, typically found in the S segment of arvicolinae, neotominae, and sigmodontinae rodent-borne hantaviruses, was not found in ASAV. The interstrain variation of the S segment among the ASAV strains was negligible (1.1% at the nucleotide and 0% at the amino acid levels). In the hypervariable region of the N protein, between amino acid residues 244 and 269, ASAV differed by 18–20 and 20–22 amino acid from soricine shrew- and rodent-borne hantaviruses, respectively. Sequence similarity of the entire S-genomic segment of ASAV strains H4, N9, and N10 was higher with soricine shrew-borne hantaviruses than with hantaviruses harbored by rodents (Table 1).
The 3,646-nucleotide full-length M-genomic segment of ASAV encoded a predicted glycoprotein of 1,141 amino acids, starting at nucleotide position 41, and a 183-nucleotide 3′-NCR. Four potential N-linked glycosylation sites (three in Gn at amino acid positions 138, 352, 404, and one in Gc at position 933) were found in ASAV. In addition, the highly conserved WAASA amino acid motif, which in ASAV was WAVSA (amino acid positions 649–653), was present. An interstrain variation of 0.1–0.7% and 0–0.4% at the nucleotide and amino acid levels, respectively, was found among ASAV strains H4, N9, and N10. The full-length Gn/Gc amino acid sequence of ASAV exhibited the highest similarity with Seewis virus (79.5%) from the Eurasian common shrew (Table 1).
Analysis of the nearly full-length 6,126-nucleotide (2,041-amino acid) L segment of ASAV revealed the five conserved motifs (A–E), identified among all hantavirus RNA polymerases. The overall high sequence similarity of the L segment among ASAV and rodent- and soricid-borne hantaviruses was consistent with the functional constraints on the RNA-dependent RNA polymerase (Table 1).
Secondary Structure of N Protein.
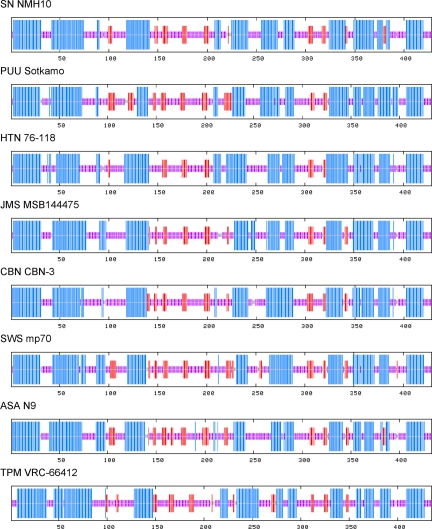
Secondary structure analysis revealed striking similarities, as well as marked differences, among the N protein sequences of ASAV and 13 representative rodent- and soricid-hantaviruses. Each sequence appeared to adopt a two-domain, predominantly α-helical structure joined by a central β-pleated sheet. Whereas the length of the N-terminal domain was mostly invariant, the length of the central β-pleated sheet and of the adjoining C-terminal α-helical domain showed systematic reciprocal structural changes according to the genetic relationship and evolutionary descent of the individual sequences.
The N-terminal α-helical domain, from residues 1 to approximately 140, was composed of four helices connected by large loops (representative viruses shown in Fig. 2). The C-terminal α-helical domain, from residues 210/230 to 430, contained seven to nine helices that were connected by tighter loops (Fig. 2). And the central β-pleated region, from residues 140 to 210/230, was composed of three to five possible anti-parallel strands. Interestingly, an increasing number of strands in this section were observed when the hantaviral sequences were arranged according to their positions in the phylogenetic tree. This resulted in a widening of the central β-pleated region with a concomitant shortening of the C-terminal α-helical domain while preserving the total length of the protein. The helix adjoining the central β-sheet progressively shortened in this architectural change. These structural alterations were reversed in TPMV, which was evolutionarily more distant from the other sequences (Fig. 2).
Fig. 2.
Consensus secondary structure of N protein of ASAV and representative rodent- and soricid-borne hantaviruses, predicted using a high-performance method implemented on the NPS@ structure server (47). As shown, the ASAV N protein was very similar to that of other hantaviruses, characterized by the same coiled-coil helix at the amino terminal end and similar secondary structure motifs at their carboxyl terminals. The predicted structures were represented by colored bars to visualize the schematic architecture: α-helix, blue; β-sheet, red; coil, magenta; unclassified, gray. For simplicity, turns and other less frequently occurring secondary structural elements were omitted. All sequences are numbered from Met-1.
Phylogenetic Analysis.
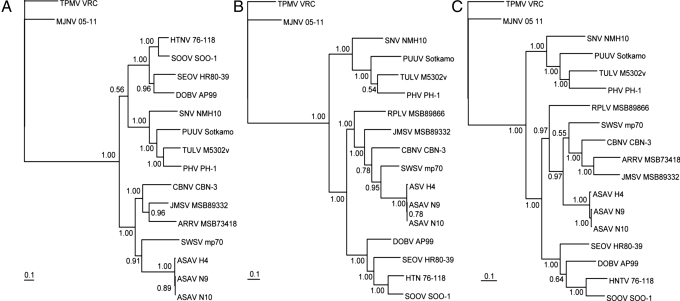
Exhaustive phylogenetic analyses based on nucleotide and deduced amino acid sequences of the S-, M-, and L-genomic segments, generated by the maximum-likelihood (ML) method, indicated that ASAV was distinct from rodent-borne hantaviruses (with high posterior node probabilities based on 30,000 trees) (Fig. 3). Nearly identical topologies were consistently derived, by using various algorithms and different taxa and combinations of taxa, suggesting an ancient evolutionary origin. The most strikingly consistent feature was the phylogenetic position of ASAV with soricine shrew-borne hantaviruses, rather than being placed as an outgroup beyond TPMV, the prototype crocidurine shrew-borne hantavirus. That is, the prediction that a shrew mole-associated hantavirus would be phylogenetically distant from hantaviruses harbored by shrews was not validated.
Fig. 3.
Phylogenetic trees generated by the ML method, using the GTR+I+G model of evolution as estimated from the data, based on the alignment of the coding regions of the full-length (A) 1,302-nucleotide S and (B) 3,423-nucleotide M segments, and partial (C) 6,126-nucleotide L-genomic segment of ASAV. The phylogenetic positions of ASAV strains H4, N9, and N10 are shown in relationship to representative murinae rodent-borne hantaviruses, including Hantaan virus (HTNV 76–118, NC_005218, NC_005219, NC_005222), Soochong virus (SOOV SOO-1, AY675349, AY675353, DQ056292), Dobrava virus (DOBV AP99, NC_005233, NC_005234, NC_005235), and Seoul virus (SEOV HR80–39, NC_005236, NC_005237, NC_005238); arvicolinae rodent-borne hantaviruses, including Tula virus (TULV M5302v, NC_005227, NC_005228, NC_005226), Puumala virus (PUUV Sotkamo, NC_005224, NC_005223, NC_005225), and Prospect Hill virus (PHV PH-1, Z49098, X55129, EF646763); and a neotominae rodent-borne hantavirus, Sin Nombre virus (SNV NMH10, NC_005216, NC_005215, NC_005217). Also shown are Thottapalayam virus (TPMV VRC, AY526097, EU001329, EU001330) from the Asian house shrew (Suncus murinus); Imjin virus (MJNV 05–11, EF641804, EF641798, EF641806) from the Ussuri white-toothed shrew (Crocidura lasiura); Cao Bang virus (CBNV CBN-3, EF543524, EF543526, EF543525) from the Chinese mole shrew (Anourosorex squamipes); Ash River virus (ARRV MSB 73418, EF650086, EF619961) from the masked shrew (Sorex cinereus); Jemez Springs virus (JMSV MSB89332, EF619962, EF619960) from the dusky shrew (Sorex monticolus); and Seewis virus (SWSV mp70, EF636024, EF636025, EF636026) from the Eurasian common shrew (Sorex araneus). The numbers at each node are posterior node probabilities based on 30,000 trees: two replicate MCMC runs consisting of six chains of 3 million generations each sampled every 1,000 generations with a burn-in of 7,500 (25%). The scale bar indicates nucleotide substitutions per site. GenBank accession numbers: ASAV S segment (H4, EU929070; N9, EU929071; N10, EU929072); ASAV M segment (H4, EU929073; N9, EU929074; N10, EU929075); and ASAV L segment (H4, EU929076; N9, EU929077; N10, EU929078).
Sequence and Phylogenetic Analysis of Mole mtDNA.
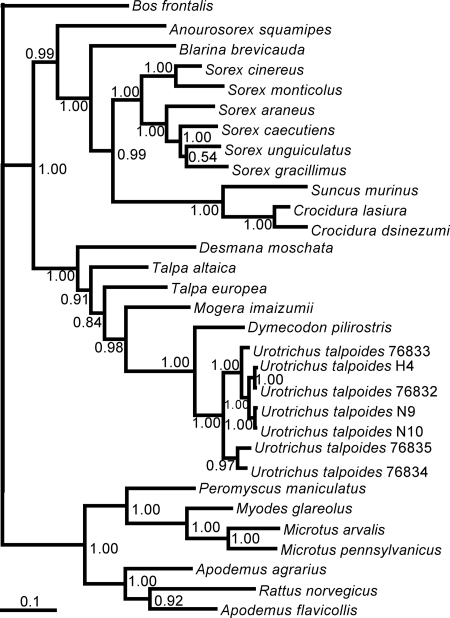
Molecular confirmation of the taxonomic identification of the hantavirus-infected Japanese shrew moles based on morphological features was achieved by amplification and sequencing of the 1,140-nucleotide mtDNA cytochrome b gene. Phylogenetic analysis showed distinct grouping of hantavirus-infected U. talpoides from this study with other U. talpoides mtDNA sequences available in GenBank, rather than with soricids or rodents (Fig. 4).
Fig. 4.
Confirmation of host identification of ASAV-infected Urotrichus talpoides by mtDNA sequencing. Phylogenetic tree, based on the 1,140-nucleotide cytochrome b (cyt b) gene, was generated by the ML method. The phylogenetic positions of Urotrichus talpoides H4 (EU918369), N9 (EU918370), and N10 (EU918371) are shown in relationship to other Urotrichus talpoides cyt b sequences from GenBank (Ut76835: AB076835; Ut76834: AB076834; Ut76833: AB076833; Ut76832: AB076832), as well as other talpids, including Desmana moschata (AB076836), Talpa altaica (AB037602), Talpa europea (AB076829), Mogera imaizumii (AB037616), Dymecodon pilirostris (AB076830), and Bos frontalis (EF061237). Also shown are representative murinae rodents, including Apodemus agrarius (AB303226), Apodemus flavicollis (AB032853), and Rattus norvegicus (DQ439844); arvicolinae rodents, including Microtus arvalis (EU439459), Myodes glareolus (DQ090761), and Microtus pennsylvanicus (AF119279); and a neotominae rodent, Peromyscus maniculatus (AF119261), as well as crocidurinae shrews, including Suncus murinus (DQ630386), Crocidura lasiura (AB077071), and Crocidura dsinezumi (AB076837); and soricinae shrews, including Anourosorex squamipes (AB175091), Blarina brevicauda (DQ630416), Sorex cinereus (EU088305), Sorex monticolus (AB100273), Sorex araneus (DQ417719), Sorex caecutiens (AB028563), Sorex unguiculatus (AB028525), and Sorex gracillimus (AB175131). The numbers at each node are posterior node probabilities based on 30,000 trees: two replicate MCMC runs consisting of six chains of 3 million generations each sampled every 1,000 generations with a burn-in of 7,500 (25%). The scale bar indicates nucleotide substitutions per site.
Discussion
Newfound Shrew Mole-Borne Hantavirus.
Despite reports of hantavirus antigens in tissues of the Eurasian common shrew (Sorex araneus), alpine shrew (Sorex alpinus), Eurasian water shrew (Neomys fodiens), and common mole (Talpa europea) (25–28), shrews and moles have been generally dismissed as being unimportant in the transmission dynamics of hantaviruses. With the recent demonstration that TPMV and other newly identified soricid-borne hantaviruses are genetically distinct and phylogenetically distant from rodent-borne hantaviruses (19–24), the conventional view that rodents are the principal or primordial reservoir hosts of hantaviruses is being challenged. In its wake, a compelling conceptual framework, or paradigm shift, is emerging that supports an ancient origin of hantaviruses in soricomorphs (or insectivores). To this emerging concept must now be added the first molecular evidence of a newfound hantavirus, designated ASAV, in the Japanese shrew mole (family Talpidae, subfamily Talpinae). The demonstration of ASAV sequences in this endemic shrew mole species captured at different times and in two separate locations in Mie Prefecture argues strongly against this being an isolated or coincidental event. Instead, these data suggest a well established coexistence of this newfound hantavirus in the Japanese shrew mole and further solidifies the notion of a long-standing evolutionary association between soricomorphs and hantaviruses.
Shrew moles differ from typical or true moles in that they look like shrews and are much less specialized for burrowing. The greater Japanese shrew mole, which morphologically resembles semifossorial shrew moles in China (Scaptonyx) and North America (Neurotrichus), is widely distributed in the lowlands and peripheral islands of Japan, except Hokkaido, and is not found on mainland Asia (29, 30). Also endemic in Japan, the lesser Japanese shrew mole (Dymecodon pilirostris) is largely restricted to mountainous regions on Honshu, Shikoku, and Kyushu and is considered the more ancestral species. As determined by cytochrome b mtDNA and nuclear recombination activating gene-1 (RAG1) sequence analyses, the greater and lesser Japanese shrew moles are closely related, but their evolutionary origins and biogeography remain unresolved (31, 32). The existence of two distinct chromosomal races of U. talpoides, geographically separated by the Fuji and Kurobe rivers in central Honshu (33, 34), provides an opportunity to further clarify the evolutionary origins of shrew mole-borne hantaviruses in Japan. Studies, now underway, will examine whether ASAV is harbored by U. talpoides in locations east of Mie Prefecture, as well as ascertain whether D. pilirostris also serves as a reservoir of ASAV-related hantaviruses.
Although our RT-PCR attempts have failed to detect hantavirus sequences in other talpid species, including the long-nosed mole (Euroscaptor longirostris) (21) and eastern mole (Scalopus aquaticus) (H. J. Kang, J.-W. Song, and R. Yanagihara, unpublished observations), it may be because appropriate primers were not used. That is, based on the vast genetic diversity of soricid-borne hantaviruses, talpid-associated hantaviruses may be even more highly divergent and would require designing very different primers for amplification.
Finally, as for shrew-borne hantaviruses, the importance of this newfound shrew mole-associated hantavirus to human health warrants careful inquiry. Virus isolation attempts have been unsuccessful to date. In the meantime, an ASAV recombinant N protein is being prepared for use in enzyme immunoassays. In this regard, as evidenced by the corresponding sequence of YIEVNGIRKP in the ASAV N protein, the monoclonal antibody E5/G6, which recognizes the epitope YEDVNGIRKP (with variations) in rodent-borne hantaviruses (35), might be useful as a capturing antibody. In addition, other sensitive technologies, including nucleic acid and protein microarrays, are being developed to establish whether ASAV is pathogenic for humans.
Secondary Structure of Hantavirus N Protein.
The overall N protein secondary structure of ASAV and other hantaviruses was compatible with a putative bilobed, three-dimensional protein architecture, which would allow the protein to clamp around the RNA as often observed in a variety of RNA-binding proteins. Whereas the core elements of the central β-pleated sheet appeared also to be conserved, more evolutionary variability was seen in the number of constituent strands and in the adjoining connecting elements and helices. This variability may reflect the function of this region as a flexible spacer element that can determine the relative orientation and separation of the two main α-helical domains and can accommodate the conformational changes upon RNA binding. The connecting regions could act as hinges of variable size leading to opening of the nucleocapsid. The flexible domain linkage would allow the interaction with the differently sized virus-specific RNA structures may modulate the oligomerization or assembly of the N protein in an evolutionarily and systematically changing fashion.
Phylogeny of Hantaviruses.
Just as the identification of novel hantaviruses in the Therese shrew (Crocidura theresae) (36) and the northern short-tailed shrew (Blarina brevicauda) (23) heralded the discovery of other soricid-borne hantaviruses (21, 22, 24), the detection of ASAV in the Japanese shrew mole forecasts the existence of other hantaviruses in talpids. Perhaps more importantly, these findings emphasize that the evolutionary history and transmission dynamics of hantaviruses are far more rich and complex than originally imagined. That is, instead of a single progenitor virus being introduced into the rodent lineage more than 50 million years ago, mounting evidence supports a more ancient virus lineage with parallel coevolution of hantaviruses in crocidurine and soricine shrews. And given the sympatric and synchronistic coexistence of moles, shrews, and rodents, through a long continuum dating from the distant past to the present time, it seems plausible that ongoing exchanges of hantaviruses continues to drive their evolution.
In this regard, several rodent species may occasionally serve as reservoir hosts for the same hantavirus. For example, Vladivostok virus (VLAV) may be found in its natural host, the reed vole (Microtus fortis) (37–39), as well as an ancillary host, the tundra or root vole (Microtus oeconomus) (40). Similarly, the Maximowiczi vole (Microtus maximowiczii) is the natural reservoir of Khabarovsk virus (KHAV), which may also be harbored by Microtus fortis (10, 39, 40). Moreover, a KHAV-related hantavirus, named Topografov virus (TOPV), has also been found in the Siberian lemming (Lemmus sibiricus) (41). This is a far more extreme situation in which a hantavirus has switched from its natural rodent reservoir host and become well established in a rodent host of a different genus. Such host-switching or species-jumping events may account for the extraordinarily close phylogenetic relationship between TOPV and KHAV (41). That is, whereas Lemmus and Microtus are very distantly related, TOPV and KBRV are monophyletic.
In much the same way, as evidenced by the polyphylogenetic relationship between ASAV and other soricid-associated hantaviruses, the progenitor of ASAV may have ‘jumped’ from its natural soricine shrew host to establish itself in the Japanese shrew mole, or vice versa. That is, burrows and shallow tunnel systems excavated by Japanese shrew moles may be occasionally shared with sympatric species, including shrews, allowing opportunities for virus transmission through interspecies wounding or contaminated nesting materials. Such a host-switching event may have occurred in the distant past, possibly before the present-day Japanese shrew mole became endemic in Japan. Accordingly, intensive investigations of shrews in Japan and elsewhere in Far East Asia may provide further insights into the evolutionary origins of hantaviruses.
Materials and Methods
Trapping.
Sherman traps (H.B. Sherman) and pit-hole traps were used to capture shrews and shrew moles in Japan between October 2006 and April 2008. Traps were set at intervals of 4 to 5 m during the evening hours of each day, over a four-day period, at sites in Hokkaido (Hamatonbetsu, Saruhutsu, and Nopporo) and Honshu (Nara and Mie), where soricomorphs had been captured. Species, gender, weight, reproductive maturity, and global positioning system (GPS) coordinates of each captured animal were recorded.
Specimen Processing.
Lung tissues, dissected using separate instruments, were frozen on dry ice, and then stored at −80°C until used for testing. In some instances, portions of tissues were also placed in RNAlater RNA Stabilization Reagent (QIAGEN, Inc.) and processed for RT-PCR within 4 weeks of tissue collection.
RNA Extraction and cDNA Synthesis.
Total RNA was extracted from tissues, by using the PureLink Microto-Midi total RNA purification kit (Invitrogen), in a laboratory in which hantaviruses had never been handled. cDNA was then prepared by using the SuperScript™ III RNase H- reverse transcriptase kit (Invitrogen) with a primer based on the conserved 5′-terminus of the S, M and L segments of hantaviruses (5′-TAGTAGTAGACTCC-3′).
RT-PCR.
Touchdown-PCR was performed by using oligonucleotide primers designed from TPMV and other hantaviruses: S (outer: 5′-TAGTAGTAGACTCCTTRAARAGC-3′ and 5′-AGCTCIGGATCCATITCATC-3′; inner: 5′-AGYCCIGTIATGRGWGTIRTYGG-3′ and 5′-AIGAYTGRTARAAIGAIGAYTTYT T-3′); M (outer: 5′-GGACCAGGTGCADCTTGTGAAGC-3′ and 5′-GAACCCCADGCCCCITCYAT-3′; inner: 5′-TGTGTICCWGGITTYCATGGIT-3′ and 5′-CATGAYATCTCCAGGGTCHCC-3′); and L (outer: 5′-ATGTAYGTBAGTGCWGATGC-3′ and 5′-AACCADTCWGTYCCRTCATC-3′; inner: 5′-TGCWGATGCHACIAARTGGTC-3′ and 5′-GCRTCRTCWGARTGRTGDGCAA-3′).
First- and second-round PCR were performed in 20-μL reaction mixtures, containing 250 μM dNTP, 2.5 mM MgCl2, 1 U of LA Taq polymerase (Takara) and 0.25 μM of each primer (24). Initial denaturation at 94°C for 2 min was followed by two cycles each of denaturation at 94°C for 30 sec, two-degree step-down annealing from 46°C to 38°C for 40 sec, and elongation at 72°C for 1 min, then 30 cycles of denaturation at 94°C for 30 sec, annealing at 42°C for 40 sec, and elongation at 72°C for 1 min, in a GeneAmp PCR 9700 thermal cycler (Perkin–Elmer). PCR products were separated by agarose gel electrophoresis and purified by using the Qiaex Gel Extraction Kit (Qiagen). Amplified DNA was sequenced directly by using an ABI Prism 3130 Avant Genetic Analyzer (Applied Biosystems).
Genetic and Phylogenetic Analyses.
Sequences were processed by using the Genetyx version 9 software (Genetyx Corporation) and aligned using Clustal W and W2 (42). For phylogenetic analysis, ML consensus trees were generated by the Bayesian Metropolis–Hastings Markov Chain Monte Carlo (MCMC) tree-sampling methods as implemented by Mr. Bayes (43) using a GTR+I+G model of evolution, as selected by hierarchical likelihood-ratio test (hLRT) in MrModeltest2.3 (http://www.abc.se/∼nylander/mrmodeltest2/mrmodeltest2.html) (44), partitioned by codon position.
An initial ML estimate of the model of evolutionary change among aligned viruses was generated by MrModeltest2.3. ML tree estimation in PAUP (45) was conducted starting with a neighbor-joining (NJ) tree based on this initial ML model of evolution, and proceeding with successive rounds of heuristic tree-searches to select the single most likely ML tree. Support for topologies was generated by bootstrapping for 1,000 NJ replicates (under the ML model of evolution, implemented in PAUP) and for 100 ML replicates (data not shown). Phylogenetic relationships were further confirmed using amino acid sequences analyzed by Bayesian tree sampling, using the WAG model (46) implemented by Bayes (43).
Secondary Structure Prediction.
Secondary structure prediction of the N protein was performed using the NPS@ structure server (47). To achieve 70–80% accuracy and to validate the prediction, five different methods were used jointly: DSC (48), HNN (49), PHD (50), PREDATOR (51), and MLRC (49), which in turn were based on GOR4 (52), SIMPA96 (53), and SOPMA (54). The minimum number of conformational states was set to four (helix, sheet, turn, and coil) for each analysis, and the results were combined into a consensus structure where the most prevalent predicted conformational state was reported for each residue. For convenience in visualization of the predicted structures, the NPS@ server also provided graphic outputs for the individual sequences which were subsequently combined into a multipart joint image.
PCR Amplification of Shrew Mole mtDNA.
Total DNA, extracted from liver tissues using the QIAamp Tissue Kit (QIAGEN), was used to verify the identity of the hantavirus-infected shrew moles. The 1,140-nucleotide mtDNA cytochrome b gene was amplified by PCR, using described universal primers (5′- CGAAGCTTGATATGAAAAACCATCGTTG-3′; 5′- AACTGCAGTCATCTCCGGTTTACAAGAC-3′) (55). PCR was performed in 50-μl reaction mixtures, containing 200 μM dNTP and 1.25 U of rTaq polymerase (Takara). Cycling conditions consisted of an initial denaturation at 95°C for 4 min followed by 40 cycles with denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and elongation at 72°C for 1 min in a GeneAmp PCR9700 thermal cycler.
Acknowledgments.
We thank Dr. Akio Shinohara (Frontier Science Research Center, University of Miyazaki) for kindly providing the photo of the Japanese shrew mole (Fig. 1). This work was supported by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Science, and Culture of Japan (18300136), Gakujyutsu-Frontier Cooperative Research at Rakuno Gakuen University, and National Institute of Allergy and Infectious Diseases Grant R01AI075057, Centers of Biomedical Research Excellence Grant P20RR018727, and Research Centers in Minority Institutions Grant G12RR003061 from the National Center for Research Resources, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
References
- 1.Yanagihara R. Hantavirus infection in the United States: Epizootiology and epidemiology. Rev Infect Dis. 1990;12:449–457. doi: 10.1093/clinids/12.3.449. [DOI] [PubMed] [Google Scholar]
- 2.Yanagihara R, Gajdusek DC. In: CRC Handbook of Viral and Rickettsial Hemorrhagic Fevers. Gear JHS, editor. Boca Raton: CRC Press; 1988. pp. 151–188. [Google Scholar]
- 3.Lee HW, Lee P-W, Johnson KM. Isolation of the etiologic agent of Korean hemorrhagic fever. J Infect Dis. 1978;137:298–308. doi: 10.1093/infdis/137.3.298. [DOI] [PubMed] [Google Scholar]
- 4.Schmaljohn CS, Hasty SE, Harrison SA, Dalrymple JM. Characterization of Hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J Infect Dis. 1983;148:1005–1012. doi: 10.1093/infdis/148.6.1005. [DOI] [PubMed] [Google Scholar]
- 5.Brummer-Korvenkontio M, et al. Nephropathia epidemica: Detection of antigen in bank voles and serologic diagnosis of human infection. J Infect Dis. 1980;141:131–134. doi: 10.1093/infdis/141.2.131. [DOI] [PubMed] [Google Scholar]
- 6.Lee HW, Baek LJ, Johnson KM. Isolation of Hantaan virus, the etiologic agent of Korean hemorrhagic fever from wild urban rats. J Infect Dis. 1982;146:638–644. doi: 10.1093/infdis/146.5.638. [DOI] [PubMed] [Google Scholar]
- 7.Lee P-W, et al. Partial characterization of Prospect Hill virus isolated from meadow voles in the United States. J Infect Dis. 1985;152:826–829. doi: 10.1093/infdis/152.4.826. [DOI] [PubMed] [Google Scholar]
- 8.Avsic-Zupanc T, et al. Characterization of Dobrava virus: A hantavirus from Slovenia, Yugoslavia. J Med Virol. 1992;38:132–137. doi: 10.1002/jmv.1890380211. [DOI] [PubMed] [Google Scholar]
- 9.Plyusnin A, et al. Tula virus: A newly detected hantavirus carried by European common voles. J Virol. 1994;68:7833–7839. doi: 10.1128/jvi.68.12.7833-7839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hörling J, et al. Khabarovsk virus: A phylogenetically and serologically distinct hantavirus isolated from Microtus fortis trapped in far-east Russia. J Gen Virol. 1996;77:687–694. doi: 10.1099/0022-1317-77-4-687. [DOI] [PubMed] [Google Scholar]
- 11.Nemirov K, et al. Isolation and characterization of Dobrava hantavirus carried by the striped field mouse (Apodemus agrarius) in Estonia. J Gen Virol. 1999;80:371–379. doi: 10.1099/0022-1317-80-2-371. [DOI] [PubMed] [Google Scholar]
- 12.Baek LJ, et al. Soochong virus: A genetically distinct hantavirus isolated from Apodemus peninsulae in Korea. J Med Virol. 2006;78:290–297. doi: 10.1002/jmv.20538. [DOI] [PubMed] [Google Scholar]
- 13.Duchin JS, et al. Hantavirus pulmonary syndrome: A clinical description of 17 patients with a newly recognized disease. N Engl J Med. 1994;330:949–955. doi: 10.1056/NEJM199404073301401. [DOI] [PubMed] [Google Scholar]
- 14.Nichol ST, et al. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 15.Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: Genome structure, expression and evolution. J Gen Virol. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- 16.Hughes AL, Friedman R. Evolutionary diversification of protein-coding genes of hantaviruses. Mol Biol Evol. 2000;17:1558–1568. doi: 10.1093/oxfordjournals.molbev.a026254. [DOI] [PubMed] [Google Scholar]
- 17.Carey DE, Reuben R, Panicker KN, Shope RE, Myers RM. Thottapalayam virus: A presumptive arbovirus isolated from a shrew in India. Indian J Med Res. 1971;59:1758–1760. [PubMed] [Google Scholar]
- 18.Zeller HG, et al. Electron microscopic and antigenic studies of uncharacterized viruses. II. Evidence suggesting the placement of viruses in the family Bunyaviridae. Arch Virol. 1989;108:211–227. doi: 10.1007/BF01310935. [DOI] [PubMed] [Google Scholar]
- 19.Song J-W, Baek LJ, Schmaljohn CS, Yanagihara R. Thottapalayam virus: A prototype shrewborne hantavirus. Emerg Infect Dis. 2007;13:980–985. doi: 10.3201/eid1307.070031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav PD, Vincent MJ, Nichol ST. Thottapalayam virus is genetically distant to the rodent-borne hantaviruses, consistent with its isolation from the Asian house shrew (Suncus murinus) Virol J. 2007;4:80. doi: 10.1186/1743-422X-4-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song J-W, et al. Newfound hantavirus in Chinese mole shrew, Vietnam. Emerg Infect Dis. 2007;13:1784–1787. doi: 10.3201/eid1311.070492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song J-W, et al. Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus) Virol J. 2007;4:114. doi: 10.1186/1743-422X-4-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arai S, et al. Hantavirus in northern short-tailed shrew, United States. Emerg Infect Dis. 2007;13:1420–1423. doi: 10.3201/eid1309.070484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arai S, et al. Phylogenetically distinct hantaviruses in the masked shrew (Sorex cinereus) and dusky shrew (Sorex monticolus) in the United States. Am J Trop Med Hyg. 2008;78:348–351. [PMC free article] [PubMed] [Google Scholar]
- 25.Tkachenko EA, et al. Potential reservoir and vectors of haemorrhagic fever with renal syndrome (HFRS) in the U.S.S.R. Ann Soc Belg Med Trop. 1983;63:267–269. [PubMed] [Google Scholar]
- 26.Clement J, et al. In: Virus Infections of Rodents and Lagomorphs. Horzinek MC, editor. Amsterdam: Elsevier Science BV; 1994. pp. 295–316. [Google Scholar]
- 27.Gavrilovskaya IN, et al. Features of circulation of hemorrhagic fever with renal syndrome (HFRS) virus among small mammals in the European U.S.S.R. Arch Virol. 1983;75:313–316. doi: 10.1007/BF01314898. [DOI] [PubMed] [Google Scholar]
- 28.Gligic A, et al. Hemorrhagic fever with renal syndrome in Yugoslavia: Epidemiologic and epizootiologic features of a nationwide outbreak in 1989. Eur J Epidemiol. 1992;8:816–825. doi: 10.1007/BF00145326. [DOI] [PubMed] [Google Scholar]
- 29.Ishii N. Size and distribution of home ranges of the Japanese shrew-mole Urotrichus talpoides. J Mamm Soc Jpn. 1993;18:87–98. [Google Scholar]
- 30.Yokohata Y. A brief review of the biology on moles in Japan. Mammal Study. 2005;30:S25–S30. [Google Scholar]
- 31.Shinohara A, Campbell KL, Suzuki H. Molecular phylogenetic relationships of moles, shrew moles, and desmans from the new and old worlds. Mol Phylogenet Evol. 2003;27:247–258. doi: 10.1016/s1055-7903(02)00416-5. [DOI] [PubMed] [Google Scholar]
- 32.Shinohara A, Campbell KL, Suzuki H. An evolutionary view on the Japanese talpids based on nucleotide sequences. Mammal Study. 2005;30:S19–S24. [Google Scholar]
- 33.Kawada S, Obara Y. Reconsideration of the karyological relationship between two Japanese species of shrew-moles, Dymecodon pilirostris and Urotrichus talpoides. Zoolog Sci. 1999;16:167–174. [Google Scholar]
- 34.Harada M, Ando A, Tsuchiya K, Koyasu K. Geographic variation in chromosomes of the greater Japanese shrew-mole, Urotrichus talpoides (Mammalia: Insectivora) Zoolog Sci. 2001;18:433–442. [Google Scholar]
- 35.Okumura M, et al. Development of serological assays for Thottapalayam virus, an insectivore-borne hantavirus. Clin Vaccine Immunol. 2007;14:173–181. doi: 10.1128/CVI.00347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klempa B, et al. Novel hantavirus sequences in shrew, Guinea. Emerg Infect Dis. 2007;13:520–552. doi: 10.3201/eid1303.061198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kariwa H, et al. Genetic diversities of hantaviruses among rodents in Hokkaido, Japan and Far East Russia. Virus Res. 1999;59:219–228. doi: 10.1016/s0168-1702(98)00141-5. [DOI] [PubMed] [Google Scholar]
- 38.Zou Y, et al. Isolation and genetic characterization of hantaviruses carried by Microtus voles in China. J Med Virol. 2008;80:680–688. doi: 10.1002/jmv.21119. [DOI] [PubMed] [Google Scholar]
- 39.Zou Y, et al. Genetic analysis of hantaviruses carried by reed voles Microtus fortis in China. Virus Res. 2008;137:122–128. doi: 10.1016/j.virusres.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 40.Plyusnina A, et al. Genetic analysis of hantaviruses carried by Myodes and Microtus rodents in Buryatia. Virol J. 2008;5:4. doi: 10.1186/1743-422X-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vapalahti O, et al. Isolation and characterization of a hantavirus from Lemmus sibiricus: Evidence for host switch during hantavirus evolution. J Virol. 1999;73:5586–5592. doi: 10.1128/jvi.73.7.5586-5592.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 44.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 45.Swofford DL. Sunderland, Massachusetts: Sinauer Associates; 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Version 4. [Google Scholar]
- 46.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 47.Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: Network Protein Sequence Analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 48.King RD, Sternberg MJ. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 1996;5:2298–2310. doi: 10.1002/pro.5560051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guermeur Y, Geourjon C, Gallinari P, Deléage G. Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics. 1999;15:413–421. doi: 10.1093/bioinformatics/15.5.413. [DOI] [PubMed] [Google Scholar]
- 50.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 51.Frishman D, Argos P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 52.Garnier J, Gibrat J-F, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 53.Levin JM, B., Robson B, Garnier J. SIMPA96: An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett. 1986;205:303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- 54.Geourjon C, Deléage G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 55.Irwin DM, Kocher TD, Wilson AC. Evolution of the cytochrome b gene of mammals. J Mol Evol. 1991;32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]