Abstract
RNA silencing participates in several important functions: from the regulation of cell metabolism and organism development to sequence-specific antiviral defense. Most plant viruses have evolved proteins that suppress RNA silencing and that in many cases are multifunctional. Tobacco etch potyvirus (TEV) HC-Pro protein suppresses RNA silencing and participates in aphid-mediated transmission, polyprotein processing, and genome amplification. In this study, we have generated 28 HC-Pro amino acid substitution mutants and quantified their capacity as suppressors of RNA silencing in a transient expression assay. Most mutations either had no quantitative effect or completely abolished silencing suppression (10 in each class), 3 caused a significant decrease in the activity, and 5 significantly increased it, revealing an unexpected high frequency of mutations conferring hypersuppressor activity. A representative set of the mutant alleles, containing both hypo- and hypersuppressors, was further analyzed for their effect on TEV accumulation and the strength of induced symptoms. Whereas TEV variants with hyposuppressor mutants were far less virulent than wild-type TEV, those with hypersuppressor alleles induced symptoms that were not more severe than those characteristic of the wild-type virus, suggesting that there is not a perfect match between suppression and virulence.
SMALL RNAs, including micro RNAs (miRNAs) and short interfering RNAs (siRNAs), are key components of an evolutionarily conserved RNA-based gene regulation system documented in fungi, plants, and animals that is implicated in various biological processes from development to antiviral defenses (Ratcliff et al. 1997; Waterhouse et al. 2001; Baulcombe 2002; Voinnet 2002; Ding et al. 2004; Chen et al. 2005; Wilkins et al. 2005; Kim and Nam 2006; Ding and Voinnet 2007). The silencing pathway is triggered by the presence of double-stranded RNAs (dsRNA) or single-stranded RNAs with stem-loop structures that are processed by several Dicer proteins into ∼21- to 24-nt short RNAs, including siRNAs, miRNAs, and others, which are incorporated into an RNA-induced silencing complex (RISC) to promote a sequence-specific cleavage or translation arrest of transcripts of complementary sequence (Hammond et al. 2001; Waterhouse et al. 2001; Voinnet 2002; Carrington and Ambros 2003; Bartel 2004; Baulcombe 2004; Pfeffer et al. 2004; Brodersen and Voinnet 2006; Buchon and Vaury 2006; Chapman and Carrington 2007).
RNA silencing is responsible for important endogenous functions, including regulation of cellular transcripts, guiding heterochromatin formation and transcriptional repression of transposon, processing of noncoding RNA precursors that control developmental timing and leaf polarity, and regulation of stress (Hammond et al. 2001; Carrington and Ambros 2003; Bartel 2004; Xie et al. 2004; Voinnet 2005; Brodersen and Voinnet 2006). RNA silencing also represents a natural defense system against viruses because it is activated by the structured RNAs or the dsRNAs produced during the replication cycles of different classes of viruses and subviral pathogens (Ratcliff et al. 1997, 1999; Voinnet 2001; Waterhouse et al. 2001; Ding et al. 2004; Lecellier and Voinnet 2004; Chen et al. 2005; Wilkins et al. 2005; Buchon and Vaury 2006; Ding and Voinnet 2007).
Viruses are inducers and targets of RNA silencing, but they have also evolved strategies to counteract this defense mechanism (Kasschau and Carrington 1998; Voinnet et al. 1999; Roth et al. 2004; Li and Ding 2006). Silencing suppression is a common property of plant viruses and suppressor proteins are considered as pathogenicity determinants, needed for efficient accumulation. Found in most viruses, silencing suppressor proteins show a tremendous structural and sequence diversity that has been explained as an evolutionary convergence toward a common functional necessity (Li and Ding 2006). The sine qua non condition for the operation of natural selection is the existence of genetic variation affecting fitness. This condition is amply fulfilled by viral silencing suppressors. For example, focusing only on potyviruses, it has been shown that two amino acid replacements in the HC-Pro suppressor of Clover yellow vein virus are sufficient to attenuate the symptoms and reduce virus accumulation (Yambao et al. 2008), whereas a single mutation in Plum pox virus HC-Pro had a similar effect (González-Jara et al. 2005). As a final example, mutations in the conserved motifs of Zucchini yellow mosaic virus HC-Pro also produced attenuated viruses on the natural host squash and also abolished the ability to elicit hypersensitive responses in other local lesion hosts (Lin et al. 2007). All in all, viral suppressor proteins probably undergo strong selective pressures for optimal adaptation to the host since a successful infection will rely on the tight balance between the host silencing response and viral counterdefense mechanisms (Moissiard and Voinnet 2004).
As with many other viruses, Tobacco etch potyvirus (TEV) encodes in its genome a suppressor protein named HC-Pro (Kasschau et al. 1997; Anandalakshmi et al. 1998; Kasschau and Carrington 1998, 2001; Llave et al. 2000; Mallory et al. 2002). HC-Pro is a multifunctional protein involved in a wealth of functions (reviewed in Urcuqui-Inchima et al. 2001): it (i) acts as proteinase during the auto-proteolytic processing of the viral polyprotein, (ii) interacts with the stylet of aphids during transmission, (iii) displays RNA-binding activity and is involved with genome amplification, (iv) is required for entry into and exit from the vascular system, (v) interferes with the 20S proteasome (Ballut et al. 2005), (vi) limits methylation of viral-derived small RNAs (Ebhardt et al. 2005; Yu et al. 2006), and (vii) is essential for symptom development. Therefore, it participates in replication, systemic movement, and vector transmission. Mutagenesis analyses have allowed for defining several functional domains in potyvirus HC-Pro, although some functions are overlapping. Oversimplifying, the N-terminal region is essential for the transmission process but dispensable for infection; in the C-terminal region, the proteinase and movement domains overlap; and the central region is implicated in RNA silencing and genome amplification and overlaps with the movement domain (Kasschau et al. 1997; Plisson et al. 2003; Varrelmann et al. 2007).
In this work, we focused our attention on the RNA-silencing suppressor activity of HC-Pro. The HC-Pro suppression mechanism is complex, interfering with the RNA-silencing pathway at least two stages. First, HC-Pro reduces, although it does not completely eliminate, the processing of dsRNA by Dicer, since siRNAs can still be detected (Mallory et al. 2002; Dunoyer et al. 2004). Second, despite the presence of siRNAs, degradation of mRNA is prevented, suggesting that HC-Pro also likely interferes with the assembly or activity of RISC (Dunoyer et al. 2004; Chapman and Carrington 2007). This second point of action is also supported by the fact that the level of the labile intermediate in the miRNA biogenesis, miRNA*, increases in the presence of the protein (Mallory et al. 2002; Kasschau et al. 2003; Dunoyer et al. 2004; Chapman and Carrington 2007). Alterations of endogenous miRNAs by HC-Pro may have profound morphological effects, contributing to symptom severity (Kasschau et al. 2003; Dunoyer et al. 2004; Chapman and Carrington 2007).
In this study, a collection of HC-Pro amino acid substitution mutants was created. The suppressor activity of each mutant was quantified by determining the accumulation level of a reporter green fluorescent protein (GFP) mRNA in Nicotiana benthamiana leaves co-infiltrated with an Agrobacterium tumefaciens strain carrying the GFP reporter gene and HC-Pro (Voinnet et al. 2000; Johansen and Carrington 2001). Next, the in vivo effect of HC-Pro alleles with altered suppression activity on TEV accumulation and symptom development was explored. TEV infectious clones carrying a subset of the mutant HC-Pro proteins were assessed by inoculating N. benthamiana plants with RNA transcripts of each mutant genotype. Overall, mutants carrying hypersuppressor HC-Pro alleles accumulated at higher levels and were more virulent than mutants with hyposuppressor HC-Pro alleles, although hypersuppressors were not more virulent than wild-type TEV. The relationship between the viral ability to suppress host silencing and its virulence is discussed, including possible trade-offs between suppression and other HC-Pro functions.
MATERIALS AND METHODS
Plasmid constructs and infectious clones:
The plasmid pTEV7DA with an infectious TEV clone (GenBank accession DQ986288), kindly provided by James C. Carrington (Oregon State University), was used as a source of wild-type virus and template for site-directed mutagenesis. Mutants in the HC-Pro cistron were generated by site-directed mutagenesis using the Quickchange II XL kit (Stratagene) following the directions of the manufacturer. Eleven mutant genotypes, designated as CLA, corresponded to amino acid substitutions at sites that are conserved among all potyviral HC-Pro proteins so far characterized. The mutants T1 and T2 corresponded to amino acid substitutions at the homologous sites of those described by González-Jara et al. (2005) as essential for Plum pox virus HC-Pro suppression activity. The 11 PC mutants corresponded to a single random amino acid substitution and have been described before (Carrasco et al. 2007a,b). Finally, four previously described alanine-scanning mutants (Kasschau and Carrington 2001) were also generated (labeled as AS). All the mutants were sequenced with an Applied Biosystems PRISM DNA sequencer 3100, and the correctness of mutations was verified.
Vector pBIN61 (Bendahmane et al. 2000) and its derivative pBIN61-GFP, containing the mGFP5 cDNA (Haseloff et al. 1997), were the gift of David C. Baulcombe (Sainsbury Laboratory, John Innes Centre, Norwich, UK).
DNAs coding for mutant and wild-type HC-Pro proteins were amplified using the forward primer VP-680 [5′-ATGCGGGATCCATGAGCGACAAATCAATCTCTGAGG-3′; contains a BamHI site (underlined) upstream from the initiation codon (italics)] and the reverse primer VP-681 [5′-GATCGCCCCGGGTTATCCAACATTGTAAGTTTTCATTTCG-3′; contains a Cfr91 site (underlined) downstream from the termination codon (italics)]. PCR-amplified DNAs were digested with BamHI/Cfr91 and subcloned in pBIN61 vector using standard procedures (Sambrook et al. 1989).
Transient expression assays and quantification of suppression activity:
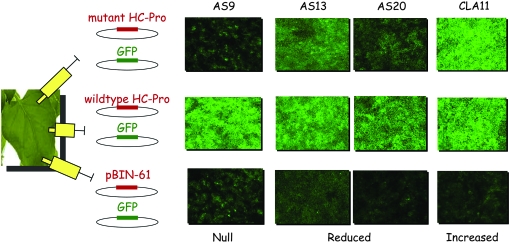
pBIN61-HC-Pro plasmids were electroporated into A. tumefaciens strain C58C1. Culture growth and induction was performed as described by Hamilton et al. (2002). Prior to co-infiltration, culture OD600 was adjusted to 0.1. Each N. bentamiana leaf was co-infiltrated with each of three different 1/1 culture mixes of transformed A. tumefaciens: (i) pBIN61-GFP and pBIN61, (ii) pBIN61-GFP and wild-type pBIN61-HC-Pro, and (iii) pBIN61-GFP and mutant pBIN61-HC-Pro (Figure 1). Each mutant was assayed following this scheme in four leaves of different plants for subsequent real-time quantitative RT–PCR (RT–qPCR) assays; independent cultures were infiltrated in 20 leaves from different plants for GFP fluorescence visualization.
Figure 1.—
Illustration of the co-infiltration experiment procedure. N. benthamiana leaves were infiltrated with three different mixtures: (i) pBIN61-GFP and mutant pBIN61-HC-Pro (first row), (ii) positive control pBIN61-GFP and wild-type pBIN61-HC-Pro (second row), and (iii) negative control pBIN61-GFP and pBIN61 (third row). GFP fluorescence was observed under the stereoscope 6 dpi. Four mutants are shown as examples.
Total RNA was extracted from agroinfiltrated areas 6 days post-inoculation (dpi). Up to 100 mg of plant tissue was processed with the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. To remove genomic and plasmid DNA, 600 ng of total RNA were treated using the TURBO DNA-free (Ambion) kit in a reaction volume of 20 μl. Each RNA sample was used as template for three independent reverse transcription reactions performed with TaqMan reverse transcription kit (Applied Biosystems). Volumes were set up to 20 μl, containing 120 ng of template DNA-free RNA and oligo(dT)16 primer. Primers for RT–qPCR were designed with the aid of Primer Express software (Applied Biosystems) using default parameters. Oligonuclotides GFP-F (5′-CGTGCAGGAGAGGACCATCT-3′) and GFP-R (5′-CGTGTCTTGTAGTTCCCGTCG-3′) amplify a 51-bp fragment from GFP mRNA; primers GADPH-F (5′-GGTGTCAAGCAAGCCTCTCAC-3′) and GAPDH-R (5′-GATGCCAAGGGTGGAGTCAT-3′) yield an equal-length fragment from glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA selected as internal control.
For relative quantitation, each cDNA was amplified in two separate reactions containing 2 μl cDNA in a 25-μl reaction, 1× Power SYBR PCR master mix (Applied Biosystems), and 300 nm GFP or 600 nm GAPDH primers. For standard curve quantitation, a single stock cDNA reaction was prepared as described above from 264 ng treated RNA/20 μl reaction, aliquoted, and stored at −20°. Twofold serial dilutions ranging from 264 to 16.5 ng RNA/20 μl RT reaction were prepared and amplified in triplicate. Amplification and quantification were done using Applied Biosystems Prism 7000 or 7005 sequence detection systems. The suppression activity was expressed as the average quantity of GFP mRNA in the areas co-infiltrated with each mutant HC-Pro relative to wild-type HC-Pro; both values were corrected by subtracting the GFP mRNA quantified in the absence of the suppressor. The expression of the GAPDH endogenous gene was used for mRNA normalization. To account for plant natural variability, each mutant was analyzed in four leaves from different plants, as variation within the plant was negligible (data not shown). To further reduce the effect of outliers, median values will be reported. Confidence intervals for the median were constructed by the Jackknife method.
GFP fluorescence was observed using a hand UV lamp UVGL-58 (UVP). GFP fluorescence images were obtained using a SMZ800 fluorescence stereoscope (Nikon) set with a EX 480/40 DM505 BA510 filter and acquired using the analySIS program (Soft Image System).
To validate the above RT–qPCR method, the relationship between the estimates obtained and the corresponding figures inferred following Kasschau and Carrington (2001) semiquantitative approach has been explored. Kasschau and Carrington (2001) used as a proxy for relative RNA-silencing suppression activity the number of GFP-positive reactions relative to the total number (n = 20) of infiltrated spots 6 dpi. Both estimates were highly associated (R2 = 0.896, F1,27 = 231.895, P < 0.001), indicating that the number of infiltration spots showing fluorescence strongly depended on the amount of GFP mRNA present on each spot.
In vitro transcription and infectivity assays:
Infectious plasmids were BglII digested and transcribed to 5′ capped RNAs using SP6 mmESSAGE mmACHINE kit (Ambion). Four-week-old N. benthamiana plants were inoculated by abrasion with ∼4 μg of transcript inoculum applied to the third true leaf. The whole procedure is detailed in Carrasco et al. (2007a).
Virions were partially purified from whole N. benthamiana plants as described before (Carrasco et al. 2007a). Briefly, 2 ml of 0.5 m borate buffer (pH 8.0) were added per gram of fresh tissue and partially homogenized and later clarified using CHCl3/CCl4; after centrifugation, virions were precipitated from the upper aqueous phase with PEG8000/NaCl, sedimented at 10,000 × g for 15 min, and resuspended in 20 μl/g 0.05 m borate buffer (pH 8.0, 5 mm EDTA).
For titering, 5 μl of partially purified virions and serial dilutions (0.5, 10−1, and 10−2) prepared in borate buffer were inoculated into four fully developed leaves of four Chenopodium quinoa (4-week-old) plants, using carborundum as an abrasive. To minimize plant effects, each plant was inoculated with every dilution (Kleczkowski 1949). Concentration of infectious viral particles could then be expressed as the number of lesion-forming units (LFU) per microliter of inoculum. Titer was estimated three times for each viral genotype on three independent full blocks. To control for block effect, wild-type TEV was titered on each block. The advantage of this method over techniques based on quantifying the amount of some viral molecule (e.g., RT–qPCR for detecting RNA molecules) is that it produces a biologically relevant quantity, namely the number of infectious particles produced, rather than properties that may be more or less correlated with infectious particles (e.g., RT–qPCR would count aberrant RNA molecules).
All plants were maintained in the greenhouse at 25° and 16 hr light.
Molecular confirmation of TEV infection:
To detect viral RNA in systemically infected leaves, total RNA was extracted from 100 mg of fresh tissue using a standard phenol/chloroform method and resuspended in 40 μl of DEPC-treated water. Primer sequences were HC-ProF (5′-CGGGATCCGATGCTCGTGCGAAGGTAAC-3′) and CTB2 (5′-GATCAACATCTCAATTGCACCTTGTG-3′). cDNA synthesis was performed using M-MuLV reverse transcriptase (Fermentas), according to instructions, from up to 200 μg of total RNA. PCR amplification reactions were performed using Taq DNA polymerase (Roche). Amplification mixture was prepared according to instructions provided for standard amplification. Cycling parameters were 95° 5 min for template denaturation, 40 cycles (95° 40 sec, 50° 30 sec, 72° 2 min) for DNA amplification, and a final elongation step of 72° 10 min.
To detect viral replication in inoculated leaves, total protein extraction and Western blot analysis were performed by standard protocols (Sambrook et al. 1989). Commercial-conjugated anti-TEV coat protein (Agdia) antibody was used for antigen detection and ECL substrate (Amersham) was used for peroxidase luminescent detection.
RESULTS
Distribution of mutational effects in HC-Pro RNA-silencing suppression activity:
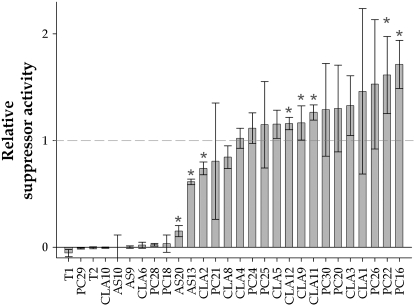
A collection of 28 TEV HC-Pro mutants was generated by site-directed mutagenesis in a pTEV7DA infectious clone. The location of each mutation, its nature, and the phenotype induced are summarized in Table 1. The suppressor activity of each mutant, relative to that of wild-type HC-Pro, was estimated by means of the co-infiltration assay depicted in Figure 1. As previously described, GFP mRNA was silenced after a maximum of 3 days in the absence of a suppressor activity, whereas in the presence of HC-Pro, fluorescence lasted longer (Johansen and Carrington 2001). Samples for quantification of GFP mRNA concentration by RT–qPCR were taken 6 days post-infiltration. The distribution of mutational effects on RNA-silencing suppression activity is shown in Figure 2. Each mutant HC-Pro was classified into one of four functional categories on the basis of the corresponding estimated 95% confidence interval of the median:
Null mutants that completely lack suppressor activity (= 0.0). Nine mutants belong to this category (Table 1 and Figure 2).
Mutants with a significant reduction in suppressor activity (>0.0 and <1.0). Three mutants are included in this category (Table 1 and Figure 2). Hereafter, these mutants will be defined as hyposuppressors since their median activity was 45.85% lower than that of wild type.
Mutants with no effect on the trait and thus performing as wild type (= 1.0). Eleven mutants belong to this category (Table 1 and Figure 2).
Mutants with increased activity (>1.0). Five mutants belong to this category (Table 1 and Figure 2). The median activity of these mutants was 22.0% larger than that of wild-type HC-Pro. Hereafter, these mutants will be referred to as hypersuppressors.
TABLE 1.
Description of TEV HC-Pro mutants and their phenotypes
| Mutant label | Codon(s) changeda | Amino acid(s) changed | Domainb | Suppressor activityc | Symptomsd |
|---|---|---|---|---|---|
| PC16 | 1086 AUA → UUA | I11L | Trans | Increased | Etch |
| PC18 | 1434 CGC → GGC | R127G | Sup | Null | None |
| T1 | 1458 GUA → GCA | V135A | Sup | Null | None |
| CLA1 | 1548 AGG → GGG | R165G | Sup | Wild type | Etch |
| CLA2 | 1629 GUG → GCG | V192A | Sup-Mov | Reduced | Mild etch |
| PC20 | 1632 AAU → UAU | N193Y | Sup-Mov | Wild type | None |
| PC21 | 1635 AAC → GAC | N194D | Sup-Mov | Wild type | None |
| PC22 | 1654 AAU → AGU | N200S | Sup-Mov | Increased | Etch |
| CLA3 | 1755 UAC → CAC | Y234H | Sup-Mov | Wild type | Etch |
| AS9 | 1773, 1774 AGG → GCG | R240A | Sup-Mov | Null | Nonee |
| 1776, 1777 AAA → GCA | K241A | ||||
| 1779, 1780 CAU → GCU | H242A | ||||
| AS10 | 1794, 1795 AGA → GCA | R247A | Sup-Mov | Null | Nonee |
| 1797, 1798 AAG → GCG | K248A | ||||
| PC24 | 1850 CAA → CAU | Q265H | Sup-Mov | Wild type | Etch |
| T2 | 1872 GAG → GCG | E273A | Sup-Mov | Null | None |
| AS13 | 1951 GAA → GCA | E299A | Sup-Mov-Pro | Reduced | Nonee |
| 1954 GAU → GCU | D300A | ||||
| PC25 | 1982 AAG → AAU | K309N | Mov-Pro | Wild type | Etch |
| CLA4 | 1986 CCA → CUA | P311L | Mov-Pro | Wild type | Etch |
| CLA5 | 2085 UAU → UCU | Y344S | Mov-Pro | Wild type | Mild etch |
| PC26 | 2118 GUG → UUG | V355L | Mov-Pro | Wild type | Etch |
| AS20 | 2134 GAG → GCG | E360A | Mov-Pro | Reduced | Mild etch |
| 2137 GAU → GCU | D361A | ||||
| PC28 | 2215 GCA → GAA | A387E | Mov-Pro | Null | None |
| CLA6 | 2223 UGC → UGG | C390W | Mov-Pro | Null | None |
| CLA8 | 2310 GUU → GCU | V419A | Mov-Pro | Wild type | Etch |
| PC29 | 2316 GAU → AAU | D421N | Mov-Pro | Null | None |
| CLA9 | 2322 UAU → CAU | Y423H | Mov-Pro | Increased | Etch |
| CLA10 | 2379 AUU → AUG | I442M | Mov-Pro | Null | Nonee |
| CLA11 | 2382 GAA → AAA | E443K | Mov-Pro | Increased | Etch |
| PC30 | 2411 GAA → GAU | E452D | Mov-Pro | Wild type | None |
| CLA12 | 2415 AAA → ACA | K454T | Mov-Pro | Increased | Etch |
Location of mutations on TEV nucleotide sequence (GenBank accession DQ986288) and on the deduced amino acid sequence of HC-Pro protein.
Corresponding functional domains: Trans, transmission; Sup, RNA-silencing suppression; Mov, viral movement; Pro, proteinase.
Relative to wild-type HC-Pro suppressor activity. Classification into functional classes was based on whether the confidence intervals in Figure 2 contained zero and/or one value.
Systemic symptoms displayed by N. benthamiana plants inoculated with infectious transcripts.
Asymptomatic infections in which TEV has been detected by RT–PCR in systemic and by Western blot in inoculated leaves.
Figure 2.—
Distribution of mutational effects on RNA-silencing suppressor activity for all HC-Pro mutant genotypes. Values are relative to the wild-type activity; median values are reported. Error bars represent ±1 standard error of the median and were computed by the Jackknife method. Asterisks indicate those cases that significantly differ both from wild type and from the negative control (background measurement).
Roughly speaking, the number of mutations positively affecting suppression activity was half the number of mutations negatively affecting suppression. The differences among these four groups were highly significant (Kruskal–Wallis test: H = 21.312, 3 d.f., P < 0.001) and, consequently, most of the observed variation (73.1%) was explained by true differences between categories rather than by measurement noise.
No association has been observed between the functional domain in which one mutation was generated and its inclusion in one of the above functional categories (χ2 = 3.400, 6 d.f., P = 0.757), even after collapsing the four categories into neutral and non-neutral (χ2 = 0.127, 1 d.f., P = 0.722). In a continuous trait scale, mutations in different domains do not differ in their effect on suppression activity (H = 2.829, 2 d.f., P = 0.243). These results are expected for overlapping functional domains.
Relationship between HC-Pro suppression activity and the severity of symptoms:
To assess mutant viability, transcripts from reconstituted TEV genomes containing the HC-Pro mutant alleles and wild-type TEV were mechanically inoculated on N. benthamiana plants and symptoms were recorded. Asymptomatic plants were analyzed in two different ways: (i) virus amplification and cell-to-cell local movement in inoculated leaves was detected by Western blot using anti-coat protein antibodies and (ii) systemic leaves were analyzed by RT–PCR to detect the presence of TEV (Table 1).
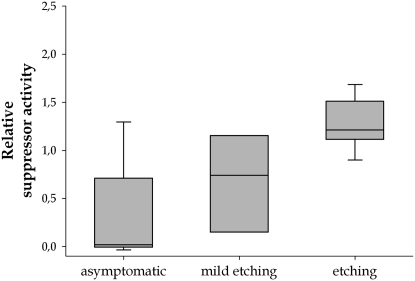
Figure 3 shows the relationship between relative suppressor activity and the symptoms described in Table 1. Activity significantly differed among symptom categories (H = 13.797, 2 d.f., P = 0.001), with asymptomatic infections associated, on average, with null or weak suppressors and stronger suppressors producing more severe etching. In other words, the stronger the suppressor, the more severe the symptoms that developed in plants (Pearson's correlation coefficient: r = 0.751, 27 d.f., P < 0.001). Plants infected with null mutants did not showed symptoms 15 dpi. In seven cases, mutant HC-Pro rendered nonviable TEV and the virus was not even detected in the inoculated leaves. However, in three cases (AS9, AS10, and CLA10), mutants were capable of systemic movement and were detected in upper asymptomatic leaves. The three TEV clones bearing HC-Pro mutants classified as hyposuppressors were viable but induced atypically mild etching (CLA2 and AS20) or no symptoms at all (AS13). The five TEV clones carrying HC-Pro mutants classified as hypersuppressors were all viable, inducing a similar etching pattern as wild type. In the case of CLA9, plants developed symptoms 1 day sooner than plants infected with wild-type TEV.
Figure 3.—
Relationship between symptom severity and relative suppressor activity. Boxes represent the 5 and 95% percentiles and error bars the 95% confidence interval. The horizontal line corresponds to the median activity.
In two cases, mutants performing wild-type suppressor activity induced altered symptoms, suggesting that mutations impaired another function. CLA5 produced mild etching in systemic leaves 15 dpi, suggesting that the mutation affected systemic movement, a suggestion compatible with the location of the mutation in the movement-proteinase domain. PC30 was not even detected in inoculated leaves, suggesting that the mutation affected the proteinase activity and thus rendered a nonfunctional protein. Concurrently, the mutation is located in the movement-proteinase domain.
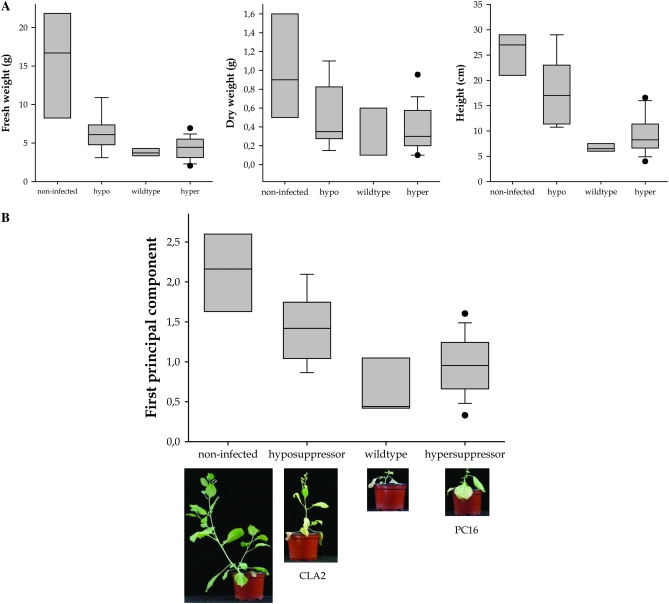
Symptoms described in Table 1 are, somehow, subjective. To avoid this drawback and to gain further insights into the relationship between suppression activity and virulence, the quantitative effect of viral infection on plant growth and vigor was studied for four hypersuppressor (CLA9, CLA11, CLA12, and PC16) and two symptom-producing hyposuppressor (CLA2 and AS20) alleles. Hypersuppressor PC22 was not incorporated in this study because we consistently failed to purify viral particles in large enough amounts to successfully infect new N. benthamiana plants. Fresh and dry (after 4 days in a desiccation oven at 100°) weights and the height of the canopy were measured 20 dpi (n = 7). Wild-type TEV and mock-inoculated plants served as controls. Figure 4A shows the effect of suppression categories on each morphological trait. Prior to analyses, variables were log transformed to achieve normality of data and homocedasticity of variances. Nested multivariate and univariate model II ANOVA tests were performed. Also, the three variables were collapsed into a first principal component, which explains up to 72.3% of observed variability. This principal component is positively correlated with the three morphological variables measured and its biological meaning is quite straightforward: it is large when plants are heavy and tall and small when plants are light and dwarfed (Figure 4). A nested model II ANOVA was also computed for the principal component values. All tests rendered the same results and thus only those from the analysis of the first principal component are reported. Significant differences exist between suppressor categories (Figure 4B; F4,4 = 170.196, P < 0.001) and among genotypes within categories (F4,48 = 3.818, P = 0.009). More interestingly, a Tukey's post-hoc test indicates that no apparent morphological differences exist between plants inoculated with wild-type TEV and hypersuppressor mutants (Figure 4B; P < 0.05) and that plants infected with TEV hyposuppressor mutants are, on average, bigger than plants infected with the wild type, although still significantly smaller than non-infected plants (Figure 4B).
Figure 4.—
(A) Morphological traits measured in mock-inoculated plants and plants infected with wild-type TEV and hyper- and hyposuppressor mutants. (B) First principal component computed from the three morphological traits: 0.927 × (log fresh weight) + 0.840 × (log height) + 0.779 × (log dry weight). Boxes represent the 5 and 95% percentiles and error bars the 95% confidence interval. The horizontal line corresponds to the median value. Circles represent outliers. Photos show representiative symptoms for each suppressor category.
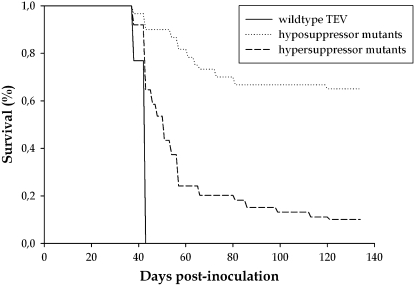
As an additional test of virulence, the effect of infection on plant survival was studied for wild-type TEV, three hyposuppressors (AS20, AS13, and CLA2), and four hypersupressors (CLA12, CLA9, CLA11, and PC16). Batches of N. benthamiana plants were infected with each genotype (median number of infected plants, 25; range 14–28). Infected plants were maintained in the greenhouse until systemic necrosis reached all leaves and roots or up to 134 dpi. Survival data were analyzed using a Kaplan–Meir regression in which genotypes were nested within their corresponding suppressor category. Figure 5 shows the average survival curves for each suppressor category; the mean survival time for each TEV genotype is reported in Table 2. Significant differences exist among suppressor categories (Figure 5; Mantel–Cox test: χ2 = 106.721, 2 d.f., P < 0.001). On average, a plant infected with wild-type TEV survived 41 dpi (95% C.I.: 40.416–41.738). Among hyposuppressors, AS20 and AS13 have no effect on plant survivorship and all plants survived until the end of the experiment even though they developed mild symptoms. By contrast, plants infected with CLA2 showed a significant reduction in survival time (Table 2). On average, plants infected with hyposuppressor mutants survived 108 dpi (95% CI: 98.518–117.449), 2.6 times longer than plants infected with wild-type TEV; this difference is statistically significant (non-overlapping 95% C.I.'s). The effect of hypersuppressor mutants was intermediated among wild-type TEV and hyposuppressors. Results ranged from CLA12, which shows the smallest impact on plant survival, and CLA9, which exerts the same impact as wild-type TEV on plant viability (Table 2). On average, a plant infected with a hypersuppressor mutant survived 1.5 times more than plants infected with wild-type TEV (61 dpi; 95% C.I.: 55.249–66.893), but 1.8 times less than plants infected with hyposuppressor mutants. As expected, symptom severity (Figure 4B) and plant survival time were negatively correlated (r = −0.819, 5 d.f., P = 0.024); that is, the severer the symptoms induced by the virus HC-Pro mutant, the shorter the plants that survived.
Figure 5.—
Survival curves of plants infected with wild-type TEV and hyper- and hyposuppressor mutants.
TABLE 2.
Survival time and virus accumulation in plants infected by TEV with HC-Pro mutants
| Suppression category | Mutant | Survival time (dpi) | Average survival time | Infectious titera (LFU/μl) | Average titer |
|---|---|---|---|---|---|
| Hyposuppressor | AS20 | 134.000 ± 0.000 | 107.983 ± 4.829 | 10.996 ± 5.153 | 16.461 ± 5.353 |
| AS13 | 134.000 ± 0.00 | ND | |||
| CLA2 | 56.650 ± 2.954 | 21.927 ± 9.320 | |||
| Wild type | 41.077 ± 0.337 | 50.446 ± 11.701 | |||
| Hypersuppressor | CLA12 | 99.250 ± 5.905 | 61.071 ± 2.970 | 60.406 ± 29.375 | 49.059 ± 12.930 |
| CLA9 | 41.167 ± 0.339 | 76.470 ± 33.703 | |||
| CLA11 | 47.429 ± 1.323 | 29.454 ± 18.599 | |||
| PC22 | ND | 4.306 ± 0.601 | |||
| PC16 | 49.346 ± 0.994 | 45.701 ± 13.127 |
ND, not determined.
Average number of local necrotic lesions per inoculum (LFU/μl) produced in C. quinoa leaves. In all cases, errors represent the standard error of the mean.
At face value, all these results indicate that, on average, hyposuppressor mutants are also less virulent than wild type but that viruses carrying hypersuppressor mutations are not more virulent than wild-type TEV. Phrased differently, hyposuppressors are hypovirulent whereas hypersuppressors are not hypervirulent. Therefore, other factors in addition to suppressor activity must be contributing to set the upper virulence value.
None of the mutants assayed in planta had reverted to the wild-type allele (C. Torres-Barceló and S. F. Elena, unpublished results).
Effect of HC-Pro mutations on virus accumulation:
To further investigate the consequences of suppression on viral accumulation, the number of infectious viral particles produced by wild-type TEV, two of three hyposuppressors (CLA2 and AS20), and five hypersuppressors (CLA9, CLA11, CLA12, PC22, and PC16) were estimated by counting the number of local necrotic lesions produced in C. quinoa leaves inoculated with serial dilutions of each genotype. AS13 hyposuppressor was not included in the analysis because it consistently failed to produce visible lesions in C. quinoa leaves. Table 2 shows the average titer for each of these genotypes. Under the culture's environmental conditions, the wild-type TEV produced visible lesions in C. quinoa leaves 10–11 dpi. The two hyposuppressor mutants developed lesions more slowly, being visible only after 15–18 dpi. Furthermore, on average, hyposuppressor mutations significantly reduced 67.37% of the number of infectious particles produced (Table 2; t-test: P = 0.018). Among the five hypersuppressors, CLA11 and CLA12 delayed the development of local lesions on C. quinoa leaves by 3 days (13–15 dpi), but the other three hypersuppressor genotypes did not differ from wild type. On average, hypersuppressor mutations have no significant effect on the accumulation of infectious viral particles (Table 2; t-test: P = 0.958).
Virus accumulation does not show a correlation with the intensity of symptoms induced in N. benthamiana plants. No significant differences in virus accumulation have been found between genotypes producing mild etching (AS20 and CLA2; average titer = 16.461 ± 5.465 LFU/μl) or wild-type-like etching (wild-type TEV, CLA9, CLA12, CLA11, PC16, and PC22; average titer = 44.464 ± 10.253 LFU/μl; Mann–Whitney U-test: P = 0.286).
DISCUSSION
Here, we have described the effect that amino acid substitutions exert on the suppressor activity of the multifunctional protein HC-Pro encoded by TEV. Mutational effects on suppressor activity, measured in a transient expression assay, ranged from no effect at all (neutral mutations) to complete elimination of activity (lethal mutations). Lying between these two extremes, three mutations induced a significant reduction in activity (from 1.4- to 7-fold reduction). We have qualified these HC-Pro mutants as hyposuppressor alleles. Interestingly, five mutations showed a significant increase in suppression activity (from 1.1- to 2.4-fold increase). We have qualified these HC-Pro mutants as hypersuppressor alleles. A set of HC-Pro alleles was reconstituted into the viral infectious cDNA and their effect on virus accumulation and symptom expression was explored. Suppression activity levels positively correlated with the intensity of symptoms induced, with hyposuppressors inducing milder symptoms and accumulating to lower levels than the wild-type TEV virus. By contrast, the hypersuppressor mutants induced symptoms and accumulated to levels that were not distinguishable from those characteristic of the wild-type virus. These results suggest an asymmetrical response of the virus to mutations affecting their suppressor activity; whereas hyposuppressor mutants clearly have impaired replication (lower accumulation) and mild symptoms, hypersuppressor alleles do not translate their stronger suppression to any quantitative effect on virus accumulation and symptom expression, at least to the resolution of our experimental procedures. Two cases are particularly interesting in this regard. The CLA12 hypersuppressor shows the second largest accumulation of infectious viral particles (Table 2) but affects the survival of plants in a way similar to hyposuppressors. By contrast, the CLA2 hyposuppressor affects plant survival almost similar to hypersuppressors, despite producing the second lowest amount of infectious particles (Table 2).
In a similar study, Stenger et al. (2006) created a collection of single-nucleotide substitution mutants of Wheat streak mosaic virus (WSMV) HC-Pro. The collection included both synonymous and nonsynonymous substitutions. Each mutant was evaluated for its ability to systemically move and the severity of the symptoms induced, although no quantitative information about the suppressor activity was provided. Briefly, they found that synonymous substitution did not have an effect on the virus's systemic movement nor on the strength of symptoms. Among the nonsynonymous mutants, most had no effect (57.69%), 15.38% resulted in attenuated systemic infection, and 26.92% abolished systemic infection. Interestingly, the latter mutants were not deficient in proteinase activity, thus suggesting that the mutation was affecting the silencing-suppression activity. These results are in excellent agreement with ours and support the conclusion that most mutations affecting HC-Pro negatively affect potyvirus systemic movement and accumulation.
These results provide further support to the well-established notion that HC-Pro is essential for symptom development and virus accumulation (Kasschau and Carrington 2001; Urcuqui-Inchima et al. 2001). In an extreme case of null mutants with no production of HC-Pro protein, it has been described that the potyvirus WSMV was still viable but infectivity was low and infection concurred with the development of very mild symptoms (Stenger et al. 2005). Experimental evolution of this deletion mutant by serial passages resulted in the recovery of infectivity and symptoms similar to wild type (Stenger et al. 2005), suggesting that hyposuppressor mutations do not represent an evolutionarily stable situation. Furthermore, it was also hypothesized that WSMV may encode a protein other than HC-Pro with RNA-silencing suppressor activity and thus HC-Pro may be functionally redundant and dispensable (Stenger et al. 2006). Supporting this possibility, Valli et al. (2006) found that a second copy of the P1 serine protease of Cucumber vein yellowing virus (a member of the genus Ipomovirus within the Potyviridae family) has RNA-silencing suppressor activity. Whether TEV may encode for a second suppressor protein is a tantalizing possibility that would explain why some suppression-defective mutants were still capable of producing a systemic infection.
Hypersuppressor alleles have not been described before, perhaps because only semiquantitative approaches have been applied to the characterization of HC-Pro mutants (e.g., Kasschau and Carrington 2001). The sensitivity of our RT–qPCR method for quantifying suppressor activity has allowed us to describe five HC-Pro alleles with increased suppression activity, representing 17.86% of the sample size analyzed here. This high frequency opens the question of whether hypersuppressor mutants may be common in nature or, alternatively, impose a fitness burden that precludes their spread in natural viral populations. In other words, has the intermediate suppression activity characteristic of wild type been optimized by natural selection? Although answering this question requires additional experiments, our observation that hypersuppressor mutants reach similar accumulation levels as wild type suggests that the benefits or costs of RNA-silencing hypersuppression, if any, are not associated with virus accumulation.
HC-Pro is a determinant of host range (Sáenz et al. 2002; Stenger and French 2004) and thus it may be argued that our estimates of virus accumulation may be affected by the fact that estimates were done by inoculating the mutants on a different host. However, we consider this possibility highly unlikely for the following reason: Stenger and French (2004) performed heterologous cistron replacement experiments in which WSMV HC-Pro was systematically replaced by homologous proteins from other members of the Potyviridae family. When the protein was replaced by that of a different isolate of WSMV (i.e., sequence identity ≥86%), host range was not altered. By contrast, when the replacing protein come from a virus belonging to a different genus (i.e., sequence identity ≤17%), then host range was largely affected (Stenger and French 2004). These results suggest that changes in host range are not easily achieved by a few mutations in HC-Pro, as they are in our case.
Here we have shown that the degree of RNA-silencing suppression shows a positive association both with virus accumulation and the strength of symptoms. However, we have failed to find such positive association between the severity of symptoms and the level of virus accumulation. That is, severer symptoms are not explained by a larger accumulation of viral particles. This lack of correlation is not an unexpected result since there are examples of uncorrelated changes in plant virus accumulation and virulence. For example, we have recently observed a lack of correlation between viral fitness and virulence for a collection of TEV random single-nucleotide substitution mutants (Carrasco et al. 2007b). Similarly, it was shown that when Barley stripe mosaic virus was evolved by serial horizontal transfers, its virulence increased with no concomitant increase in viral load (Stewart et al. 2005). As a final example, necrogenic and non-necrogenic variants of Cucumber mosaic virus did not differ in their accumulation in tomato plants (Escriu et al. 2000). Most theoretical models seeking to explain the evolution of virulence assume that it is a side effect of virus replication and accumulation (Ebert 1998; Brown et al. 2006). However, virulence would not depend on within-host replication if the extent of damage was not proportional to the amount of viral particles, as is the case with hypersensitive responses (Morel and Dangl 1997), or if expressing the systemic acquired resistance pathway is costly for the plant (Heidel et al. 2004), or if allocating resources to defense detracted plants from vegetative growth or reproductive effort (Heil 2001).
Expression of both Dicer proteins DCL4 and DCL2 is necessary for conferring antiviral immunity in Arabidopsis thaliana against RNA viruses (Deleris et al. 2006). DCL4 acts as the primary sensor and produces 21-nt siRNAs that guide RISC. In a second step, DCL2 forms 22-nt siRNAs with antiviral activity, but these siRNAs are manifested only when DCL4 is inactivated (Deleris et al. 2006). Plants deficient in DCL2 and DCL4 presented higher accumulation of Tobacco rattle virus RNA and stronger symptoms than wild-type plants, thus proving that virus accumulation depends on the strength of RNA silencing. This is in good agreement with our observation that the stronger the RNA-silencing suppression activity may be, the more viruses accumulate and the more severe the symptoms are. Indeed, this study also suggests a possible explanation for why TEV hypersuppressors do not significantly differ from wild type either in accumulation or in symptoms. If TEV HC-Pro kidnaps mainly the 21-nt siRNAs produced by DCL4 but has no significant activity in the 22-nt siRNAs produced by DCL2, a hypersuppressor mutant will more efficiently sequester 21-nt siRNA, but will be controlled by 22-nt siRNAs to the same extent as the wild-type virus.
In experimental evolution work currently in progress, we are exploring the pathways of compensatory evolution in restoring the wild-type level of suppressor activity and the molecular basis of such compensations.
Acknowledgments
We thank our lab mates for comments and fruitful discussion; A. Cuadrado and F. de la Iglesia for excellent technical assistance; and D. C. Baulcombe, O. Voinnet, and two anonymous referees for comments and suggestions. This work has been supported by grants from the Spanish Ministerio de Educación y Ciencia–Fondo Europeo de Desarrollo Regional (BFU2006-14819-C02-01/BMC), the Generalitat Valenciana (ACOMP07-263), and the European Molecular Biology Organization Young Investigator Program to S.F.E.
References
- Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory et al., 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut, L., M. Drucker, M. Pugnière, F. Cambon, S. Blanc et al., 2005. Hc-Pro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymic activities. J. Gen. Virol. 86 2595–2603. [DOI] [PubMed] [Google Scholar]
- Bartel, D. P., 2004. MicroRNA genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. C., 2002. RNA silencing. Curr. Biol. 12 R82–R84. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. C., 2004. RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A., M. Querci, K. Kanyuka and D. C. Baulcombe, 2000. Agrobacterium transient expression system as a tool for isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 21 73–81. [DOI] [PubMed] [Google Scholar]
- Brodersen, P., and O. Voinnet, 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280. [DOI] [PubMed] [Google Scholar]
- Brown, N. F., M. E. Wickham, B. K. Coombes and B. B. Finaly, 2006. Crossing the line: selection and evolution of virulence traits. PLoS Pathog. 2 e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon, N., and C. Vaury, 2006. RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity 96 195–202. [DOI] [PubMed] [Google Scholar]
- Carrasco, P., J. A. Daròs, P. Agudelo-Romero and S. F. Elena, 2007. a A real-time RT-PCR assay for quantifying the fitness of tobacco etch virus in competition experiments. J. Virol. Methods 139 181–188. [DOI] [PubMed] [Google Scholar]
- Carrasco, P., F. de la Iglesia and S. F. Elena, 2007. b Distribution of fitness and virulence effects caused by single-nucleotide substitution in Tobacco etch virus. J. Virol. 81 12979–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J. C., and V. Ambros, 2003. Role of microRNAs in plant and animal development. Science 301 336–338. [DOI] [PubMed] [Google Scholar]
- Chapman, E. J., and J. C. Carrington, 2007. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8 884–896. [DOI] [PubMed] [Google Scholar]
- Chen, W., M. Liu, G. Cheng, W. Yan, L. Fei et al., 2005. RNA silencing: A remarkable parallel to protein-based immune systems in vertebrates? FEBS Lett. 579 2267–2272. [DOI] [PubMed] [Google Scholar]
- Deleris, A., J. Gallego-Bartolomé, J. Bao, K. D. Kasschau, J. C. Carrington et al., 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313 68–71. [DOI] [PubMed] [Google Scholar]
- Ding, S. W., and O. Voinnet, 2007. Antiviral immunity directed by small RNAs. Cell 130 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S. W., H. Li, R. Lu, F. Li and W. X. Li, 2004. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 102 109–115. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., C. H. Lecellier, E. A. Parizotto, C. Himber and O. Voinnet, 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ebert, D., 1998. Experimental evolution of parasites. Science 282 1432–1435. [DOI] [PubMed] [Google Scholar]
- Ebhardt, H. A., E. P. Thi, M. B. Wang and P. J. Unrau, 2005. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl. Acad. Sci. USA 102 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escriu, F., A. Fraile and F. García-Arenal, 2000. Evolution of virulence in natural populations of the satellite RNA of Cucumber mosaic virus. Phytopathology 90 480–485. [DOI] [PubMed] [Google Scholar]
- González-Jara, P., F. A. Atencio, B. Martínez-García, D. Barajas, F. Tenllado et al., 2005. A single amino acid mutation in the Plum pox virus helper component-proteinase gene abolishes both synergistic and RNA silencing suppression activities. Phytopathology 95 894–901. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., O. Voinnet, L. Chappell and D. C. Baulcombe, 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S. M., A. A. Caudy and G. J. Hannon, 2001. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2 110–119. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., K. R. Siemering, D. C. Prasher and S. Hodge, 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel, A. J., J. D. Clarke, J. Antonovics and X. Dong, 2004. Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168 2197–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil, M., 2001. The ecological concept of costs of induced systemic resistance (ISR). Eur. J. Plant Pathol. 107 137–146. [Google Scholar]
- Johansen, L. K., and J. C. Carrington, 2001. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau, K. D., and J. C. Carrington, 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95 461–470. [DOI] [PubMed] [Google Scholar]
- Kasschau, K. D., and J. C. Carrington, 2001. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285 71–81. [DOI] [PubMed] [Google Scholar]
- Kasschau, K. D., S. Cronin and J. C. Carrington, 1997. Genome amplification and long-distance movement functions associated with the central domain of Tobacco etch potyvirus helper component-proteinase. Virology 228 251–262. [DOI] [PubMed] [Google Scholar]
- Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman et al., 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4 205–217. [DOI] [PubMed] [Google Scholar]
- Kim, V. N., and J. W. Nam, 2006. Genomics of microRNA. Trends Genet. 22 165–173. [DOI] [PubMed] [Google Scholar]
- Kleczkowski, A., 1949. The transformation of local lesion counts for statistical analysis. Am. Appl. Biol. 36 139–152. [DOI] [PubMed] [Google Scholar]
- Lecellier, C. H., and O. Voinnet, 2004. RNA silencing: No mercy for viruses? Immunol. Rev. 198 285–303. [DOI] [PubMed] [Google Scholar]
- Li, F., and S. W. Ding, 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60 503–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. S., H. W. Wu, F. J. Jan, R. F. Hou and S. D. Yeh, 2007. Modifications of the helper component-protease of Zucchini yellow mosaic virus for generation of attenuated mutants for cross protection against severe infection. Phytopathology 3 287–296. [DOI] [PubMed] [Google Scholar]
- Llave, C., K. D. Kasschau and J. C. Carrington, 2000. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A. C., B. J. Reinhart, D. Bartel, V. B. Vance and L. H. Bowman, 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99 15228–15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard, G., and O. Voinnet, 2004. Viral suppression of RNA silencing in plants. Mol. Plant Pathol. 5 71–82. [DOI] [PubMed] [Google Scholar]
- Morel, J. B., and J. L. Dangl, 1997. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 4 671–683. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S., M. Zavolan, F. A. Grässer, M. Chein, J. J. Russo et al., 2004. Identification of virus-encoded microRNAs. Science 304 734–736. [DOI] [PubMed] [Google Scholar]
- Plisson, C., M. Drucker, S. Blanc, S. German-Retana, O. Le Gall et al., 2003. Structural characterization of HC-Pro, a plant virus multifunctional protein. J. Biol. Chem. 278 23753–23761. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F. G., B. D. Harrison and D. C. Baulcombe, 1997. A similarity between viral defense and gene silencing in plants. Science 276 1558–1560. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F. G., S. A. MacFarlane and D. C. Baulcombe, 1999. Gene silencing without DNA: RNA-mediated cross-protection between viruses. Plant Cell 11 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, B. M., G. J. Pruss and V. B. Vance, 2004. Plant viral suppressors of RNA silencing. Virus Res. 102 97–108. [DOI] [PubMed] [Google Scholar]
- Sáenz, P., B. Salvador, C. Simón-Mateo, K. D. Kasschau, J. C. Carrington et al., 2002. Host-specific involvement of the HC protein in the long-distance movement of potyvirus. J. Virol. 76 1922–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Stenger, D. C., and R. French, 2004. Functional replacement of Wheat streak mosaic virus HC-Pro with the corresponding cistron from a diverse array of viruses in the family Potyviridae. Virology 323 257–267. [DOI] [PubMed] [Google Scholar]
- Stenger, D. C., R. French and F. E. Gildow, 2005. Complete deletion of Wheat streak mosaic virus HC-Pro: a null mutant is viable for systemic infection. J. Virol. 79 12077–12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger, D. C., B. A. Young and R. French, 2006. Random mutagenesis of Wheat streak mosaic virus HC-Pro: non-infectious interfering mutations in a gene dispensable for systemic infection of plants. J. Gen. Virol. 87 2741–2747. [DOI] [PubMed] [Google Scholar]
- Stewart, A. D., J. M. Logsdon, Jr. and S. E. Kelly, 2005. An empirical study of the evolution of virulence under both horizontal and vertical transmission. Evolution 59 730–739. [PubMed] [Google Scholar]
- Urcuqui-Inchima, S., A. L. Haenni and F. Bernardi, 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74 157–175. [DOI] [PubMed] [Google Scholar]
- Valli, A., A. M. Martín-Hernández, J. J. López-Moya and J. A. García, 2006. RNA silencing suppression by a second copy of P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HC-Pro. J. Virol. 80 10055–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrelmann, M., E. Maiss, R. Pilot and L. Palkovics, 2007. Use of pentapeptide-insertion scanning mutagenesis for functional mapping of the Plum pox virus helper component proteinase suppressor of gene silencing. J. Gen. Virol. 88 1005–1015. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., 2002. RNA silencing: small RNAs as ubiquitous regulators of gene expression. Curr. Opin. Plant Biol. 5 444–451. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6 206–221. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Y. M. Pinto and D. C. Baulcombe, 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., C. Lederer and D. C. Baulcombe, 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103 157–167. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P. M., M. B. Wang and T. Lough, 2001. Gene silencing as an adaptive defense against viruses. Nature 411 834–842. [DOI] [PubMed] [Google Scholar]
- Wilkins, C., R. Dishongh, S. Moore, M. A. Whitt, M. Chow et al., 2005. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 436 1044–1047. [DOI] [PubMed] [Google Scholar]
- Xie, Z., L. K. Johansen, A. M. Gustafson, K. D. Kasschau, A. D. Lellis et al., 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yambao, M. L. M., H. Yagihashi, T. Sekiguchi, T. Sasaki, M. Sato et al., 2008. Point mutations in helper component protease of Clover yellow vein virus are associated with the attenuation of RNA-silencing suppression activity and symptom expression in broad bean. Arch. Virol. 153 105–115. [DOI] [PubMed] [Google Scholar]
- Yu, B., E. J. Chapman, Z. Yang, J. C. Carrington and X. Chen, 2006. Transgenically expressed viral RNA silencing suppressors interfere with microRNA methylation in Arabidopsis. FEBS Lett. 580 3117–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]