Abstract
Transposable elements (TEs) constitute >80% of the wheat genome but their dynamics and contribution to size variation and evolution of wheat genomes (Triticum and Aegilops species) remain unexplored. In this study, 10 genomic regions have been sequenced from wheat chromosome 3B and used to constitute, along with all publicly available genomic sequences of wheat, 1.98 Mb of sequence (from 13 BAC clones) of the wheat B genome and 3.63 Mb of sequence (from 19 BAC clones) of the wheat A genome. Analysis of TE sequence proportions (as percentages), ratios of complete to truncated copies, and estimation of insertion dates of class I retrotransposons showed that specific types of TEs have undergone waves of differential proliferation in the B and A genomes of wheat. While both genomes show similar rates and relatively ancient proliferation periods for the Athila retrotransposons, the Copia retrotransposons proliferated more recently in the A genome whereas Gypsy retrotransposon proliferation is more recent in the B genome. It was possible to estimate for the first time the proliferation periods of the abundant CACTA class II DNA transposons, relative to that of the three main retrotransposon superfamilies. Proliferation of these TEs started prior to and overlapped with that of the Athila retrotransposons in both genomes. However, they also proliferated during the same periods as Gypsy and Copia retrotransposons in the A genome, but not in the B genome. As estimated from their insertion dates and confirmed by PCR-based tracing analysis, the majority of differential proliferation of TEs in B and A genomes of wheat (87 and 83%, respectively), leading to rapid sequence divergence, occurred prior to the allotetraploidization event that brought them together in Triticum turgidum and Triticum aestivum, <0.5 million years ago. More importantly, the allotetraploidization event appears to have neither enhanced nor repressed retrotranspositions. We discuss the apparent proliferation of TEs as resulting from their insertion, removal, and/or combinations of both evolutionary forces.
GENOMES of higher eukaryotes, and particularly those of plants, vary extensively in size (Bennett and Smith 1976, 1991; Bennett and Leitch 1997, 2005). This is observed not only among distantly related organisms, but also between species belonging to the same family or genus (Chooi 1971; Jones and Brown 1976). More than 90% of genes are conserved in sequenced plant genomes (Bennetzen 2000a; Sasaki et al. 2005; Jaillon et al. 2007) and thus differences in gene content explain only a small fraction of the genome size variation. It is widely accepted that whole-genome duplication by polyploidization (Blanc et al. 2000; Paterson et al. 2004; Adams and Wendel 2005) and differential proliferation of transposable elements (TEs) are the main driving forces of genome size variation. The differential proliferation of TEs results from their transposition (SanMiguel et al. 1996; Bennetzen 2000b, 2002a,b; Kidwell 2002; Bennetzen et al. 2005; Hawkins et al. 2006; Piegu et al. 2006; Zuccolo et al. 2007) as well as the differential efficiency of their removal (Petrov et al. 2000; Petrov 2002a,b; Wendel et al. 2002).
Polyploidization and differential proliferation of TEs are particularly obvious in the case of wheat species belonging to the closely related Triticum and Aegilops genera. Rice (Oryza sativa), Brachypodium, and diploid Triticum or Aegilops species underwent the same whole-genome duplications (Adams and Wendel 2005; Salse et al. 2008), but Triticum or Aegilops genomes are >10 times larger (Bennett and Smith 1991), mainly due to proliferation of repetitive DNA, which represents >80% of the genome size (Smith and Flavell 1975; Vedel and Delseny 1987). Diploid wheat species can differ in their genome sizes by hundreds or even thousands of megabases (Bennett and Smith 1976, 1991; http://data.kew.org/cvalues/homepage.html). For example, the genome size of Triticum monococcum (6.23 pg) is 1.3 pg greater than that of Triticum urartu (4.93 pg) (Bennett and Smith 1976, 1991), although these species diverged <1.5 million years ago (MYA) (Dvorak et al. 1993; Huang et al. 2002; Wicker et al. 2003b). Similarly, the calculated size of the B genome of polyploid wheat species (7 pg) is higher than that of any diploid wheat species (http://data.kew.org/cvalues/homepage.html).
The genome size variation within wheat is also accentuated by frequent allopolyploidization events, among which two successive events have led to the formation of the allohexaploid bread wheat Triticum aestivum (2n = 6x = 42, AABBDD). The first event led to the formation of the allotetraploid Triticum turgidum (2n = 4x = 28, AABB) and occurred <0.5–0.6 MYA between the diploid species T. urartu (2n = 2x = 14, AA), donor of the A genome, and an unidentified diploid species of the Sitopsis section, donor of the B genome (Feldman et al. 1995; Blake et al. 1999; Huang et al. 2002; Dvorak et al. 2006). The second allopolyploidization event occurred 7000–12,000 years ago, between the early domesticated tetraploid T. turgidum ssp. dicoccum and the diploid species Aegilops tauschii (2n = 14), donor of the D genome, resulting in hexaploid wheat (Feldman et al. 1995).
The amount of available wheat genomic sequences is very limited, compared to other organisms (reviewed by Sabot et al. 2005; Stein 2007; http://genome.jouy.inra.fr/triannot/index.php and http://www.ncbi.nlm.nih.gov/). Individual bacterial artificial chromosome (BAC) clones, selected primarily because they contained genes of agronomic interest, have been sequenced. Analyses of randomly chosen BAC clones from wheat have been also performed (Devos et al. 2005), and 2.9 Mb of sequences from a whole-genome shotgun library of Ae. tauschii were analyzed by Li et al. (2004). More recently, a detailed analysis of 19,400 BAC-end sequences of chromosome 3B, representing a cumulative sequence length of nearly 11 Mb (1.1% of the estimated chromosome length) was reported (Paux et al. 2006). Altogether, these sequencing efforts have confirmed previous estimates of the amount of repetitive DNA in the wheat genome (∼80%) (Smith and Flavell 1975; Vedel and Delseny 1987) and have identified the major types of TEs (Wicker et al. 2002; Sabot et al. 2005).
Because of the limited genomic sequence information, the extent to which various TEs contribute to the wheat genome and affect its size variation, or how they are distributed among different genomes, remains unexplored. Little is known about the dynamics of TEs, their proliferation processes, and whether they proliferated gradually or in waves of sudden bursts of insertions. In this study, 10 genomic regions from wheat chromosome 3B were sequenced and used to constitute, along with three other genomic sequences, 1.98 Mb of sequence from the wheat B genome. Transposable element dynamics and proliferation in these B-genome sequences were analyzed and compared to those in 3.63 Mb of sequence from 19 genomic regions of the wheat A genome. Our study provides novel insights into the dynamics and differential proliferation of TEs as well as their important role in the evolution and divergence of the wheat B and A genomes.
MATERIALS AND METHODS
Plant material and genomic DNA isolation:
Hexaploid wheat deletion lines used to map the 10 BAC clones on different deletion bins of chromosome 3B (see results) were originally described by Qi et al. (2003) and kindly provided by Catherine Feuillet (INRA, Clermont-Ferrand, France). Hexaploid wheat genotypes were kindly provided by Joseph Jahier (INRA, Rennes, France). Tetraploid wheat genotypes were kindly provided by Moshe Feldman (Weizemann Institute). Genomic DNA was extracted from leaves as described by Graner et al. (1990).
Primer design and PCR-based tracing of retrotransposon insertions:
The program Primer3 (Rozen and Skaletsky 2000) was used to design oligonucleotide primers on the basis of TE–TE or TE-unassigned DNA junctions. We often designed and used several couples (including nested) of PCR primers. Internal controls (PCR primers designed within the TE) were also used. Primer sequences are given in supplemental Table 1. PCR reactions were carried out in a final volume of 10 μl with 200 μm of each dNTP, 500 nm each of forward and reverse primers, 0.2 units Taq polymerase (Perkin Elmer). PCR amplification was conducted using the following “touchdown” procedure: 14 cycles (30 sec 95°, 30 sec 72° minus 1° for each cycle, 30 sec 72°), 30 cycles (30 sec 95°, 30 sec 55°, 30 sec 72°), and one additional cycle of 10 min 72°. Amplification products were visualized using standard 2% agarose gels.
BAC sequencing, sequence assembly, and annotation:
BAC shotgun sequencing was performed at the Centre National de Sequencage (Evry, France) essentially as described by Chantret et al. (2005). Genes, TEs, and other repeats were identified by computing and integrating results on the basis of BLAST algorithms (Altschul et al. 1990, 1997), predictor programs, and different software and procedures, detailed below. Cross-analysis of the information obtained for genes and TEs as well as for repeats and unassigned DNA was integrated into ARTEMIS (Rutherford et al. 2000). Sequence annotation and analysis were performed as described in supplemental Method 1. The 10 BAC clone sequences were submitted to EMBL and under the following accession nos.: TA3B54F7, AM932680; TA3B63B13, AM932681; TA3B63B7, AM932682; TA3B81B7, AM932683; TA3B95C9, AM932684; TA3B95F5, AM932685; TA3B95G2, AM932686; TA3B63C11, AM932687; TA3B63E4, AM932688; TA3B63N2, AM932689. Accession numbers for the three publicly available genomic sequences from the wheat B genome (Sabot et al. 2005; Gu et al. 2006; Dvorak et al. 2006) are CT009588, AY368673, DQ267103.
Publicly available genomic sequences from the wheat A genome:
The retained publicly available A-genome sequences consist of 19 sequenced and well annotated BAC clones or contigs (SanMiguel et al. 2002; Yan et al. 2002, 2003; Wicker et al. 2003b; Chantret et al. 2005; Isidore et al. 2005; Dvorak et al. 2006; Gu et al. 2006; Miller et al. 2006), representing >3.5 Mb. Accession numbers for the analyzed BAC sequences are the following: diploid A genome—AF326781, AF488415, AY146588, AY188331, AY188332, AY188333, AY491681, AY951944, AY951945, DQ267106, AF459639; tetraploid A genome—AY146587, AY485644, AY663391, CT009587, DQ267105; hexaploid A genome—AY663392, CT009586, DQ537335.
Chromosome 3B BAC clones and fluorescent in situ hybridization:
The 10 BAC clones and/or their subclones were originally mapped by fluorescence in situ hybridization (FISH) on flow-sorted 3B chromosomes using the Cot-1 fraction as blocking DNA to suppress hybridization of repeated sequences (Dolezel et al. 2004; Safar et al. 2004; M. Kubalakova and J. Dolezel, personal communication). Further FISH hybridization experiments were conducted, without Cot-1 DNA, on mitotic metaphase chromosomes of hexaploid wheat (T. aestivum) cv. Chinese Spring. The FISH hybridization protocol is presented in supplemental Method 2.
Estimation of Long Terminal Repeat-retrotransposon insertion dates:
For all genomic sequences of the B and A genomes of wheat, retrotransposon copies with both 5′ and 3′ long terminal repeats (LTRs), and target-site duplications (TSD) were considered as corresponding to original insertions and analyzed by comparing their 5′ and 3′ LTR sequences. The two LTRs were aligned and the number of transition and transversion mutations was calculated using MEGA3 software (Kumar et al. 2004). A mutation rate of 1.3 × 10−8 substitutions/site/year (SanMiguel et al. 1998; Ma et al. 2004; Ma and Bennetzen 2004; Wicker et al. 2005; Gu et al. 2006) was used. The insertion dates and their standard errors (SE) were estimated using the formula T = K2P/2r (Kimura 1980).
Statistical analysis:
All statistical analyses and the different tests (Kolmogorov–Smirnov, Bootstrap, and probability density functions) were done with the R-package (http://www.r-project.org). Kolmogorov–Smirnov tests (Férignac 1962) were applied to check whether the distribution of insertion dates of retrotransposons deviates from uniformity, and whether they are different when comparing different TE families or superfamilies within and between the B and A genomes. Probability density of TE insertion dates was estimated using Gaussian kernel density estimation (Silverman 1986), taking into account measured standard deviation for each individual insertion date (Kimura 1980).
RESULTS
Constitution of a genomic sequence data set representative of the wheat B genome—analysis of 10 BAC sequences from the wheat chromosome 3B:
Only three large well-annotated genomic sequences (BAC clones), representing 0.55 Mb of sequence, were available for the wheat B genome (Sabot et al. 2005; Dvorak et al. 2006; Gu et al. 2006). To obtain more representative genomic sequences, we sequenced and annotated 10 BAC clones of wheat chromosome 3B, representing 0.15% of the chromosome length (1.43 Mb) (Figure 1). Detailed annotation files are deposited at EMBL/GenBank Data Libraries.
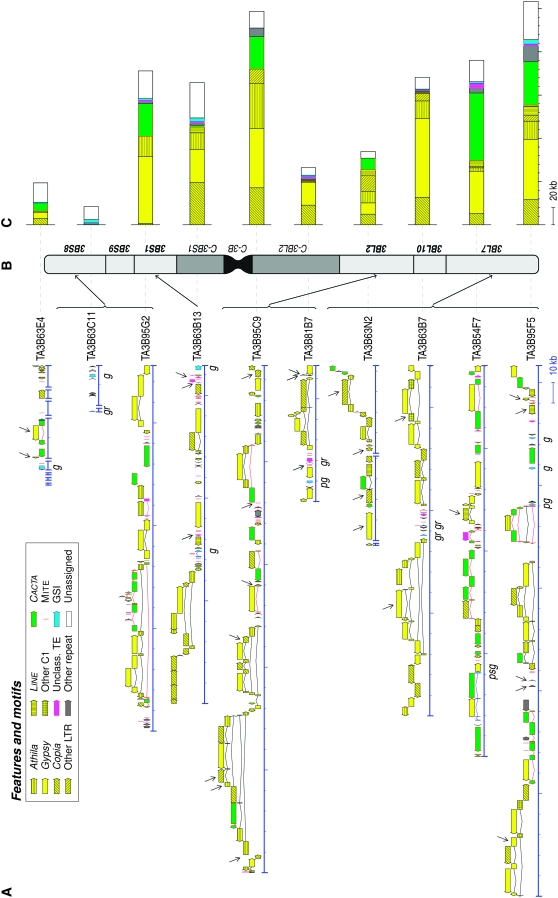
Figure 1.—
Detailed annotation, BIN map positions, and sequence composition of 10 sequenced BAC clones of wheat chromosome 3B. (A) Detailed annotations of the 10 sequenced BAC clones. Main TEs, other repeats, and gene sequence information (GSI) are represented with distinct features and motifs (detailed in the “features and motifs” key). g, genes; pg, putative genes; gr, gene relics; and psg, pseudogenes. For nested insertions of TEs, the newly inserted TE is presented above the split one. Complete reconstruction of split TEs was done and the different parts are linked with a line to visualize the entire element. Some BAC clones are represented by several unordered contigs (TA3B63E4, TA3B63C11, TA3B63N2). EMBL BAC clone references and annotation files are given in materials and methods. Detailed coding sequence and TE descriptions are supplied in supplemental Text 1 and supplemental Text 2. Arrows indicate novel TEs identified in this study and described in supplemental Text 2 and supplemental Table 5. (B) BIN map position of nine of the BAC clones. The wheat chromosome 3B bins are according to Qi et al. (2003). Details of the genotyping results are given in supplemental Table 6. (C) Proportions of the main sequence classes and types. See “features and motifs” in A for an explanation of colors. Details are given in supplemental Table 2.
These sequenced genomic regions show a high proportion of TEs, which represent 79.1% of the cumulative sequence length (Figure 1, supplemental Table 2). Other repeated DNA sequences represent 2.4% and unassigned DNA sequences account for 17.5% of the cumulative sequence length.
We conducted gene prediction analysis for the remaining 18.5% non-TEs and nonrepeated DNA, using different search programs (see supplemental Method 1 and supplemental Text 1 for detailed description). Genes of known and unknown functions or putative genes were defined on the basis of predictions and the existence of rice or other Triticeae homologs. Hypothetical genes were identified on the basis of prediction programs only. Pseudogenes were not well predicted and frameshifts need to be introduced within the coding sequences (CDS) structure to better fit a putative function on the basis of BLASTX (mainly with rice). Truncated pseudogenes (genes disrupted by large insertion or deletion) and highly degenerated CDS sequences were considered as gene-relics. Combined together, all these types of gene sequence information (GSI) account for only 1.0% of the sequence and are present in seven BAC clones (one or two genes per clone) while the remaining three BAC clones (TA3B95C9, TA3B95G2, TA3B63N2) contain no genes (indicated in Figure 1A and detailed in supplemental Text 1, supplemental Table 3, and supplemental Table 4).
Six genes (of known or unknown function) and two putative genes were identified using the FGENESH prediction software (http://www.softberry.com) and by identification of homologs in rice (Figure 1A, supplemental Table 3). Six additional “gene-relics” or “pseudogenes” were also identified on the basis of colinearity with rice (Figure 1A, supplemental Table 3). Finally, 10 CDS, designated as “hypothetical genes,” were identified according to the FGENESH prediction program only (Figure 1A, supplemental Table 4).
TE prediction, annotation, classification, and nomenclature were performed essentially as suggested by the unified classification system for eukaryotic TEs (Wicker et al. 2007) with two modifications. The Athila retrotransposons were analyzed separately from the other Gypsy retrotransposons (see also supplemental Methods 1). The Sukkula retrotransposons were considered as belonging to the Gypsy superfamily because of similarities with the Erika (Gypsy) elements. The 79.1% of TEs were shown to be composed of a wide variety of TEs, distributed as follows: 61.9% class I (171 TEs from 48 families), 16.2% class II (113 TEs from 28 families), and 1.0% unclassified TEs (18 TEs from 9 families) (Figure 1). The CACTA TEs represent the majority (96%) of class II TEs. More details about the TE composition in the 10 different BAC clones of wheat chromosome 3B are provided in supplemental Text 2.
Twenty-one transposable element families, some of which are present in several copies, were identified for the first time in this study (Figure 1A, indicated by arrows). They account for 9.8% by number and 7.9% by length of the overall sequences. Class I retrotransposons are the category for which we found the majority of novel TE families (17). Description of these novel TEs, their features, and the suggested nomenclature are presented in supplemental Text 2 and supplemental Table 5.
The 10 sequenced BAC clones or their subclones were originally mapped by FISH on flow-sorted 3B chromosomes, using the Cot − 1 fraction as blocking DNA to suppress hybridization of repeated sequences (Dolezel et al. 2004; Safar et al. 2004; M. Kubalakova and J. Dolezel, personal communication). As described by Devos et al. (2005) and Paux et al. (2006), specific PCR markers, based on TE–TE or TE-unassigned DNA junctions, were used to confirm the different BAC clone map positions on the deletion bins (Qi et al. 2003) of chromosome 3B (except TA3B63E4) (Figure 1B). Details of PCR markers and genotyping results are given in supplemental Table 6.
Representation of transposable elements and the wheat B genome:
Five BAC clone sequences were publicly available from the B genome of wheat (Sabot et al. 2005; Dvorak et al. 2006; Gu et al. 2006). Four of these were sequenced for two orthologous regions in tetraploid and hexaploid wheat species (one BAC clone per region and per species) (Sabot et al. 2005; Gu et al. 2006). As they share nearly identical sequences (99%) with common TE insertions, they were considered as redundant in our study and only the longest BAC clone sequences (three in total) were counted in calculation and appreciation of TE proliferation. These, added to the above-described 10 genomic region sequences of wheat chromosome 3B, constitute 1.98 Mb of sequence from the wheat B genome. Four main TE superfamilies occupy 66.5% of the analyzed B-genome loci: the Athila superfamily (54 elements), the Copia superfamily (57 elements), the Gypsy superfamily (79 elements), and the CACTA superfamily (70 elements) (Table 1). Interestingly, proportions of the Athila, Copia, and Gypsy retrotransposons (respectively, 10.8, 14.2, and 28.1%) (Table 1) are very similar to estimates based on 11 Mb of the chromosome 3B sequence BAC end (Paux et al. 2006). The major deviation concerns the proportion of CACTA class II TEs, which is higher in the 13 genomic regions (13.4%) than in the overall BAC-end sequences (4.9%), probably due to their clustering in some BAC clones that we have sequenced, such as TA3B54F7 (40.5% of CACTA TEs) (Figure 1).
TABLE 1.
Details of TEs from the four most represented superfamilies in 13 genomic regions of the wheat B genome, compared to publicly available sequences from 19 genomic regions of the wheat A genome
| 13 genomic regions of the wheat B genome (1.98 Mb)a
|
19 publicly available genomic regions of the wheat A genome (3.63 Mb)b
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Athila | Copia | Gypsy | CACTA | Athila | Copia | Gypsy | CACTA | |
| Observed number of TEs | 54 | 57 | 79 | 70 | 72 | 149 | 123 | 53 |
| Sequence proportion (means ±SE)%c | 10.8 ± 1.6 | 14.2 ± 2.5 | 28.1 ± 3.8 | 13.4 ± 3.3 | 10.4 ± 1.8 | 21.8 ± 1.8 | 19.7 ± 2.9 | 9.4 ± 1.9 |
| Bootstrap means deviationd | −0.07 | +0.02 | +0.02 | −0.05 | +0.01 | −0.02 | −0.03 | −0.09 |
| Complete TEs with TSD (%) | 13 | 18 | 39 | 19 | 19 | 60 | 38 | 32 |
| Incomplete (truncated) TEs | 41 | 39 | 40 | 51 | 53 | 89 | 85 | 21 |
| LTR-mediated homologous recombination | ||||||||
| Entire TE without TSD | 3 | 7 | 0 | — | 0 | 4 | 0 | — |
| Solo LTR | 4 | 2 | 2 | — | 5 | 15 | 9 | — |
| Illegitimate recombination | 34 | 30 | 38 | 51 | 48 | 70 | 76 | 21 |
| Complete TEs/incomplete (truncated) TEs | 0.32 | 0.46 | 0.98 | 0.37 | 0.36 | 0.67 | 0.45 | 1.52 |
This corresponds to 1.43 Mb from the 10 genomic regions sequenced in this study and 0.55 Mb from three other publicly available genomic regions from Sabot et al. (2005), Gu et al. (2006), and Dvorak et al. (2006). See materials and methods for BAC clone sequence references.
Nineteen genomic regions available for the A genome (SanMiguel et al. 2002; Yan et al. 2002, 2003; Wicker et al. 2003b; Chantret et al. 2005; Isidore et al. 2005; Dvorak et al. 2006; Gu et al. 2006; Miller et al. 2006). See materials and methods for BAC clone sequence references.
Relative to cumulative sequence length. SE, standard errors for estimated means.
Differences between arithmetic means (line above) and bootstrap analysis (Efron 1979) with 10,000 resamplings.
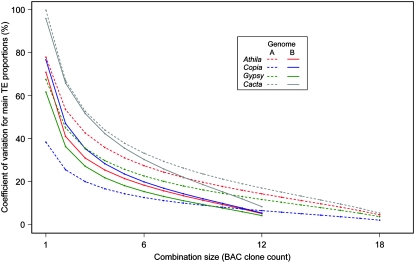
The 13 sequences represent only ∼0.03% of the B genome. However, statistical tests, using SE as well as a bootstrap analysis with 10,000 resamplings, confirm the robustness of estimations of sequence proportions of the Gypsy, Copia, Athila, and CACTA TE superfamilies (Table 1). We also evaluated the variation of mean sequence proportions estimated for the four TE superfamilies by comparing all possible clone number representations and combinations (from 1 to 12 BAC clones) (Figure 2). Results show that representing the wheat B genome with a low number of BAC clones results in very variable proportions of the TE sequences (Figure 2). These variations decrease significantly by increasing the number of considered BAC clones (Figure 2). This confirms the usefulness of our effort in sequencing more BAC clones for better representation of the wheat B genome.
Figure 2.—
Changes of the coefficient of variation of proportions (in percentages) of the main transposable element superfamilies calculated over all possible BAC clone combinations and simulated over a size varying from 1 to 12 BAC clones for the wheat B genome and 1 to 18 for the wheat A genome (combination size). For each number of considered BAC clones (x-axis), sequence proportions (in percentages) were calculated for all possible BAC clone combinations, and the coefficient of variation between these proportions was calculated (y-axis).
It is also interesting to note that direct FISH hybridization, using the whole BAC clone as a probe, resulted in dispersed and mostly homogenous signals across all wheat chromosomes for 8 of all 10 BAC clones of wheat chromosome 3B (except TA3B63C11 and TA3B54F7) (Safar et al. 2004 and supplemental Figure 1), thus confirming sequencing results that show high TE composition.
Constitution of a genomic sequence data set representative of the wheat A genome:
The publicly available A-genome sequences that we were able to use are more abundant and consist of 20 sequenced and well-annotated BAC clones or contigs. Ten of these were comparatively sequenced for five orthologous regions of the wheat A genome at the diploid, tetraploid, and/or hexaploid levels and were partially overlapping (Wicker et al. 2003b; Chantret et al. 2005; Isidore et al. 2005; Dvorak et al. 2006; Gu et al. 2006), while others were determined at only one ploidy level (mostly diploid) (SanMiguel et al. 2002; Yan et al. 2002, 2003; Miller et al. 2006). Comparisons show that no shared TE insertions were observed between orthologous regions (from two ploidy levels), except in the region of the high-molecular-weight (HMW) glutenin gene, the sequences of which were nearly identical at the tetraploid and hexaploid levels (Gu et al. 2006). Thus, we used only the sequence from hexaploid wheat to represent the HMW glutenin gene region and considered all the other different orthologous regions (from different ploidy levels) separately. This led to 19 BAC clones, representing 3.63 Mb of sequence, that were analyzed for the wheat A genome.
The Gypsy TEs were found to occupy 19.7%, the Athila TEs 10.4%, the Copia TEs 21.8%, and the CACTA TEs 9.4% of the cumulative sequence length (Table 1). Similarly, for the B-genome sequences, we also analyzed and validated the robustness of the estimation of sequence proportions of the main TE superfamilies and their representation of the A genome (Figure 2). Similar proportions of the Gypsy, Copia, Athila, and CACTA TEs were found whether the 11 genomic sequences from the diploid A genome or those determined from A genomes of tetraploid (six regions) and hexaploid (three regions) wheat species were considered separately or combined (data not shown).
Comparison of TE sequence proportions and ratios of complete to truncated copies:
Our analysis showed a significantly higher number of Gypsy retrotransposons in the wheat B-genome sequences than in the A genome (Table 1). Conversely, a higher proportion of Copia retrotransposons is observed in genomic sequences of the wheat A genome than in the B genome (Table 1). Proportions of the Athila and CACTA TEs were not statistically different between the two genomes (Table 1).
Major differences were found between the three main retrotransposon superfamilies in the ratio of complete (intact) copies, defined as having both LTRs and target-site TSD, as compared to degenerated and truncated copies that resulted from LTR-mediated unequal homologous recombinations or illegitimate DNA recombination (Devos et al. 2002; Ma et al. 2004; Ma and Bennetzen 2004; Vitte and Bennetzen 2006) (Table 1). In the B-genome sequences, the Athila and Copia retrotransposons show low ratios of complete to incomplete retrotransposons (respectively, 0.32 and 0.46), whereas the Gypsy retrotransposons show the highest ratio (0.98) (Table 1). In comparison, the 3.63 Mb of genomic sequence of the wheat A genome shows a lower ratio (0.45) of complete to incomplete Gypsy retrotransposons whereas proportions of intact Copia retrotransposons are relatively higher than those observed in the B genome (0.67) (Table 1). The Athila retrotransposon ratio in the A genome is comparable to the ratio in the B genome (0.36 and 0.32, respectively).
CACTA TE original insertions are characterized by the “CACTA” sequence and 3-bp TSD sequence motifs surrounding terminal inverted repeats (TIR) at both ends. We used these signatures to define complete CACTA copies, where the “CACTA,” TIR, and TSD sequence motifs are observed at both ends, and truncated copies, where the “CACTA” and TSD motifs are absent from one or both ends. The ratio of complete to incomplete copies of the CACTA class II TEs was about five times lower in the wheat B genome (ratio of 0.37) than in the A genome (ratio of 1.52) (Table 1).
Insertion dates and proliferation of LTR retrotransposons:
To understand differences in sequence proportions and the ratios of complete to truncated copies between retrotransposon superfamilies, as well as between the B and A genomes, we compared TE proliferation periods and rates.
The two LTRs are identical at the time of retrotransposon insertion and their sequence divergence reflects time lapsed since the insertion (SanMiguel et al. 1998). Several studies have shown that LTRs evolve at approximately twice the rate of genes and UTR regions, and we used a rate of 1.3 × 10−8 substitutions/site/year (Ma et al. 2004; Ma and Bennetzen 2004; Wicker et al. 2005; Gu et al. 2006).
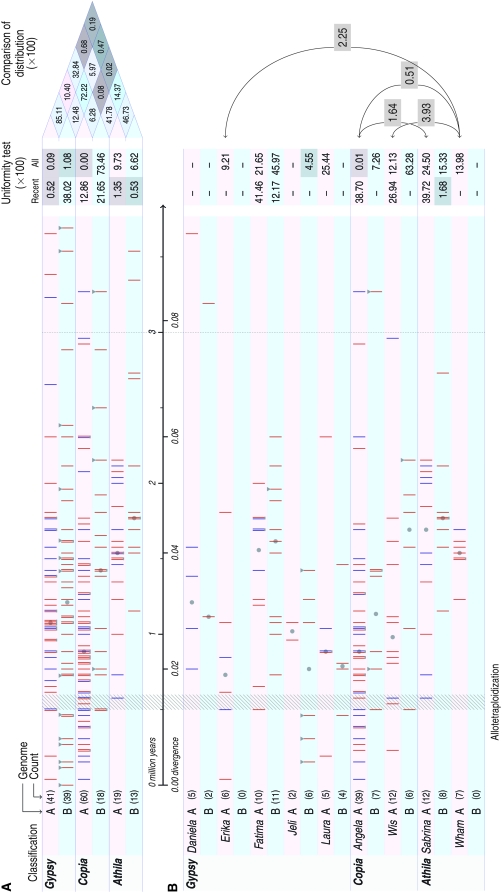
We calculated the LTR divergence and dates of insertion of the Athila, Copia, and Gypsy retrotransposon (complete copies with both LTRs and TSD) found in the wheat B and A genomes (Figure 3). Such TE insertion dates offer a very important insight into the relative timing of various events, regardless of the approaches used to estimate nucleotide substitution rates or the molecular clock calibration points used in these calculations.
Figure 3.—
Distribution of insertion dates estimated for LTR retrotransposons in the B and A-genome sequences of wheat (divergence and MYA). (A) All dated LTR retrotransposons combined at the three main superfamily levels (Athila, Gypsy, and Copia). (B) The most abundant retrotransposon families, showing five or more dated copies in at least one of the A or B wheat genomes. Mean insertion dates calculated for retrotransposons are represented by vertical bars. For the A-genome sequences, blue indicates retrotransposons detected from the diploid and red from the polyploid genomic sequences. The genomic sequences of the B genome (red) were obtained from the polyploid wheat. Copies of a given retrotransposon superfamily or family showing identical mean insertion dates are presented by adjacent vertical bars that are joined with a lower horizontal gray bar. The number within parentheses corresponds to the total number of considered retrotransposon copies. Gray triangles indicate retrotransposon insertions that have been traced using PCR in a collection of genotypes of T. aestivum and T. turgidum. The interval period of the allotetraploidization event (0.5–0.6 MYA, divergence 0.013–0.016) is highlighted in gray. “Uniformity test” refers to Kolmogorov–Smirnov (Férignac 1962) tests determining probabilities (P-value) that the distribution of insertion dates of retrotransposons deviates from uniformity (thus confirming a burst of higher proliferation): “All” refers to the last 3 million years (0.078 divergence); “Recent” refers to the most recent periods, estimated when dividing the LTR–retrotransposon insertions by the median (indicated by gray circles). Tests were done on families that show five copies or more. “Comparison of distribution” indicates the same Kolmogorov–Smirnov tests determining probabilities that distributions of insertion dates for the last 3 million years (0.078 divergence) are different in the retrotransposon superfamilies and families as well as the in B and A genomes of wheat.
The vast majority of complete retrotransposons in the B and A genomes of wheat (86 and 92%, respectively) were estimated to be <3 million years old (Figure 2) in agreement with several previous studies of grasses and other plants species (SanMiguel et al. 1998, 2002; Wicker et al. 2003b, 2005; Gao et al. 2004; Ma et al. 2004; Du et al. 2006; Piegu et al. 2006; Wicker and Keller 2007). This is explained by the fact that LTR retrotransposons are continuously removed by unequal homologous recombination and illegitimate DNA recombination as new ones are inserted (Vicient et al. 1999; Devos et al. 2002; Ma et al. 2004; Pereira 2004). Insertion of the Egug element (RLGa_Egug_TA3B95C9-1 ∼5 MYA; divergence of 0.131) is the oldest such event found in our study and the most recent one is the Sukkula insertion (RLG_Sukkula_TA3B63B7-2) for which only a 1-base indel differentiates the two LTRs of 4192/4193 bp.
Comparison of LTR divergence dates revealed that different LTR–retrotransposon superfamilies and families proliferated at different periods and rates during evolution of the wheat B and A genomes (Figure 3). We applied Kolmogorov–Smirnov tests to check whether within the last 3 million years (0.078 divergence) the distribution of insertion dates of retrotransposons deviates from uniformity (thus confirming a burst of higher proliferation), and whether these dates are different when comparing different retrotransposon families or superfamilies within and between the wheat B and A genomes (thus illustrating differential proliferation). This was done for all complete copies of the three main retrotransposon superfamilies as well as for the most abundant retrotransposon families (nine) that have five or more complete copies in the B and/or A genomes (Figure 3).
Superfamily level comparison:
The combination of all complete retrotransposon copies at the superfamily level (Figure 3A) indicated that the distribution of the Gypsy retrotransposon insertion dates in both B and A genomes and that of Copia retrotransposons in the A genome were significantly different from uniform (P-value <0.01) because of their higher proliferation during the last 2 million years (Figure 3A). Proliferation of the Copia retrotransposons in the B genome was uniform and low all across the 3-million-year period, whereas proliferation of the Athila retrotransposons was different from a uniform distribution in both genomes at P-value <0.1.
One possible reason for the non-uniform distributions of retrotransposon insertion dates within the 3-million-year period is because older insertions are more likely to be removed (completely or partially) from the genome (see above). Therefore, we checked whether distributions of insertions are significantly different from a uniform distribution for the most recent period of evolution during which the impact of DNA removal should be lower. To carry out this analysis, we divided the LTR–retrotransposon insertions according to the median (of their distribution) that varies depending on the retrotransposon superfamily and family (Figure 3A, gray circle). Kolmogorov–Smirnov (Férignac 1962) tests were then conducted on half of the complete copies, which show the most recent insertion dates. Distribution of insertion dates of the Gypsy retrotransposons in the wheat B genome and that of the Copia retrotransposons in the B and A genomes can be considered as uniform (P-value >0.05, Figure 3A), indicating that they have constantly proliferated during this most recent period. In contrast, the distribution of Athila retrotransposons in the wheat B and A genomes and that of Gypsy retrotransposons in the A genome are not uniform (P-value <0.05, Figure 3A), consistent with a decreasing proliferation during the most recent period.
Comparison of the proliferation of the three retrotransposon superfamilies shows that distribution of the Athila retrotransposons is statistically different from that of the Gypsy retrotransposons (Figure 3A, P-value <0.05) in the B genome. The Athila distribution is significantly different from that of the Gypsy and Copia retrotransposons (Figure 3A, P-value <0.05) in the A genome.
Comparison of the distributions of the three retrotransposon superfamilies between the B and A genomes shows that Copia distributions are significantly different (Figure 3A, P-value = 0.628) due to their higher proliferation and more recent insertions in the A genome. Both genomes show similar old distribution of the Athila retrotransposons (Figure 3A). Distributions of the Gypsy retrotransposons were not statistically different between the two genomes for the entire 3-million-year period (Figure 3A, P-value >0.05). However, separate Kolmogorov–Smirnov tests for the most recent period show that these have proliferated less in the wheat A genome (P-value = 0.052, Figure 3A), unlike in the wheat B genome (P-value = 0.38, Figure 3A).
Distribution of the most abundant retrotransposon families:
Some specific retrotransposon families were abundant in the B and/or A genomes. This is the case of the Angela and Wis families, together representing 72 and 85% of the Copia superfamily in the B and A genomes, respectively (Figure 3B). This is also the case of the Sabrina family representing 62 and 63% of the Athila superfamily in the B and A genomes, respectively (Figure 3B). There are more families that compose the Gypsy retrotransposon superfamily, the most abundant being Fatima, representing 25% in both genomes (Figure 3B).
Kolmogorov–Smirnov tests show nonsignificant deviations (P-value >0.05) from uniform distributions for all nine retrotransposon families (with five or more observed complete copies in at least one genome), with the exception of the Jeli (Gypsy) elements in the B genome and the Angela (Copia) elements in the A genome, which have more recently proliferated (Figure 3B). Separate analysis for the most recent period, corresponding to half of the complete copies, shows that, as expected from the superfamily-level analysis, the Wham family in the A genome and the Sabrina family in the B genome have not recently proliferated (P-value <0.05, Figure 3B).
Distribution of insertion dates of the Wham and Sabrina families is different from almost all the other seven families within and between the B and A genomes (P-value <0.05). Distribution of insertion dates of the Angela family in the wheat A genome is statistically different (P-value <0.05) from that of the Fatima family in both genomes. Distributions of insertion dates of the remaining families do not show statistical differences (P-value >0.05) within and between the wheat B and A genomes (Figure 3B).
Moreover, some retrotransposon families were abundant and present in several complete copies in only one genome (Romani, Daniela, Erika, and Wham for the A genome; Egug and Jeli for the B genome) but absent or presenting few copies in the other (Figure 3B). It is likely that this corresponds to differential proliferation of the considered retrotransposons, as different copies were detected in different genomic regions of wheat B or A genomes.
LTR–retrotransposon proliferation was neither enhanced nor repressed by the allotetraploidization event:
The allotetraploidization event that brought the B and A genomes of wheat together in one nucleus was estimated to occur no more than 0.5–0.06 MYA (Huang et al. 2002; Dvorak et al. 2006; Chalupska et al. 2008). This corresponds to a divergence interval of 0.013–0.016, using the corrected rate of 1.3 × 10−8 substitutions/site/year for more rapid divergence of LTRs (Ma et al. 2004; Ma and Bennetzen 2004; Dvorak et al. 2006).
Comparisons show that retrotransposon insertions continued in wheat B and A genomes during the last 0.5–0.6 million years, apparently without being enhanced nor repressed by the allotetraploidization event (Figure 3). For example, analysis of genomic sequences available from the three ploidy levels of the A genome does not show differences in proliferation periods and rates of retrotransposons (Figure 3).
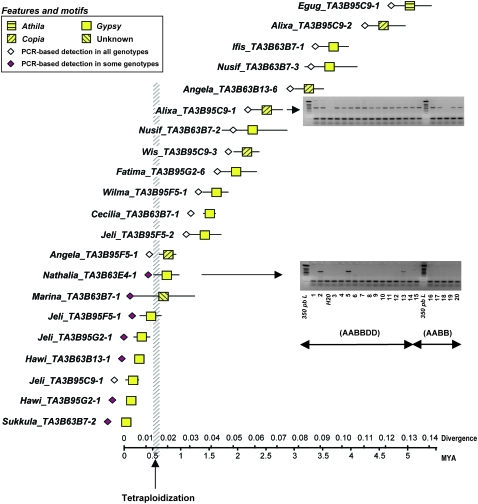
To check the accuracy of these observations and to calibrate the divergence rate used for coding sequences, on one hand, and that used for LTRs of retrotransposons, on the other hand, we traced several retrotransposons for their insertion prior or posterior to the allopolyploidization event. A PCR-based tracing strategy, derived from the retrotransposon-based insertion polymorphism method (Flavell et al. 1998; Devos et al. 2005; Paux et al. 2006), was developed for 21 retrotransposon insertions from the B genome, sampled as having different estimated insertion dates (Figure 3, indicated by gray triangles). It simply relies on primers designed in both the retrotransposon and its flanking sequences (either unassigned DNA or an older preinserted TE sequence) so that PCR amplification will be specific to the retrotransposon insertion. As the diploid wheat species donor of the B genome is unknown (Feldman et al. 1995; Blake et al. 1999; Huang et al. 2002), we analyzed the occurrence (i.e., presence or absence) of the 21 retrotransposon insertions in hexaploid (T. aestivum) and tetraploid (T. turgidum) wheat genotypes, which carry the wheat B genome. Examples of PCR-based tracing of the 21 original retrotransposon insertions in the wheat genotypes compared with their estimated insertion dates (±SE) are presented in Figure 4. Full tracing results are supplied in supplemental Table 7 and sequences of the PCR primers in supplemental Table 1. With the exception of Jeli_TA3B95C9-1, all the other 7 most recently inserted retrotransposons, which have calculated insertion date intervals (means ±SE) equal to or less than the 0.5–0.6 MYA interval (divergence 0.013–0.016), were detected in some but not all genotypes carrying the B genome, suggesting their occurrence after the tetraploidization event (Figure 4 and supplemental Table 7). In contrast, all 13 retrotransposon insertions, which have calculated insertion intervals (means ±SE) >0.7 MYA, were detected in all tested genotypes carrying the B genome, suggesting their occurrence prior to the allotetraploidization event (Figure 4 and supplemental Table 7). Given the uncertainty in calculating intervals of insertion dates, the PCR-based tracing method confirms the calibration of LTR divergence on that of gene divergence. More importantly, it also confirms that retrotranspositions (insertions) were not enhanced or repressed by the alloteraploidization event.
Figure 4.—
PCR-based tracing of series of retrotransposons, inserted at different dates in wheat chromosome 3B across a collection of wheat tetraploid (T. turgidum) and hexaploid (T. aestivum) genotypes. Averages and intervals (means ±SE) of retrotransposon insertion dates are presented. The interval of the allotetraploidization event (0.5–0.6 MYA), calculated according to gene sequence divergence (Huang et al. 2002; Dvorak et al. 2006; Chalupska et al. 2008), is in gray. Retrotransposons for which insertion dates were estimated on the basis of divergence of their LTR prior to the tetraploidization event were generally detected in almost all genotypes, whereas those posterior to the tetraploidization event were detected in only some genotypes. Gels show PCR-based detection of insertion of RLC_Alixa_TA3B95C9-1 (into DTC_Caspar_TA3B95C9-1) in all tested genotypes, except one and insertion of RLG_Nathalia_TA3B63E4-1 (into DTC_Vincent_TA3B63E4-2) in some genotypes. Primer sequences, details of insertion dates (averages and intervals), and PCR-based detection in different wheat genotypes are given in supplemental Table 1 and supplemental Table 7. AABBDD (hexaploid wheat accessions): -1—T. aestivum cv. Renan; -2—T. aestivum cv. Chinese Spring; -3—T. aestivum spelta, Erge 27216; -4—T. aestivum spelta, Erge 2776; -5—T. aestivum spelta, Erge 2771; -6—T. aestivum spelta Rouquin, Erge 6329; -7—T. aestivum macha 1793, Erge 27240; -8—T. aestivum compactum rufulum 71V, Erge 26786; -9—T. aestivum compactum crebicum 72V, Erge 26787; -10—T. aestivum compactum clavatum 73V, Erge 26788; -11—T. aestivum compactum icterinum 74V, Erge 26789; -12—T. aestivum compactum erinaceum 75V, Erge 26790; -13—T. aestivum sphaerococcum tumidum perciv globosum, Erge 27016; -14—T. aestivum cv. Soisson. AABB (T. turgidum, tetraploid wheat accessions): -15—T. turgidum durum cv. Langdon; -16—T. turgidum durum; -17—T. turgidum dicoccum, -18—T. turgidum dicoccoides; -19—T. turgidum polinicum; -20—T. turgidum turgidum.
Relative proliferation periods of the CACTA class II transposable elements:
The CACTA class II DNA TEs represent an important proportion of the B- and A-genome sequences (13.4 and 9.4%, respectively). As for the main LTR–retrotransposon superfamilies, ratios of complete to truncated copies are very different for B (0.37) and A (1.52) genomes (Table 1). In contrast to LTR retrotransposons, the CACTA TEs do not have long repeats or other features, which would allow determination of their insertion dates on the basis of sequence divergence. Therefore, their proliferation periods and rates were evaluated indirectly, relative to their level of insertions into or by other CACTA TEs and, more importantly, by elements of the three main LTR–retrotransposon superfamilies for which proliferation periods and rates were evaluated on the basis of the dates of insertions (described above). This was calculated for all CACTA TE copies as well as for complete and truncated copies separately (Table 2).
TABLE 2.
Associations of CACTA transposable elements with the four most represented TE superfamilies and other DNA sequence classes in 13 genomic regions of the wheat B genome and 19 publicly available genomic sequences of the wheat A genome
| 13 genomic regions of the wheat B genome (1.98 Mb)a
|
19 publicly available genomic regions of the wheat A genome (3.63 Mb)b
|
|||
|---|---|---|---|---|
| DNA sequence classes | CACTA TEs inserted into other DNA sequencesc | Other DNA sequences inserted into CACTA TEsc | CACTA TEs inserted into other DNA sequences | Other DNA sequences inserted into CACTA TEs |
| Athila TEs | 12: 4/8 | 6: 0/6 | 7: 7/0 | 0: 0/0 |
| Copia TEs | 1: 0/1 | 5: 0/5 | 10: 7/3 | 8: 6/2 |
| Gypsy TEs | 1: 1/0 | 6: 0/6 | 5: 4/1 | 6: 5/1 |
| CACTA TEs | 7: 4/3 | 6: 1/5 | 4: 4/0 | 3: 3/0 |
| Other TEs | 3: 3/0 | 0: 0/0 | 2: 1/1 | 0: 0/0 |
| Unclear TE associationsd | 2: 0/2 | 2: 0/2 | 4: 0/4 | 4: 0/0 |
| Unassigned DNA | 44: 7/37 | — | 21: 9/12 | — |
| Total | 70: 19/51 | 25: 1/24 | 53: 32/21 | 21: 14/7 |
This corresponds to 1.43 Mb from the 10 genomic regions sequenced in this study and 0.55 Mb from three other publicly available genomic regions from Sabot et al. (2005), Gu et al. (2006), and Dvorak et al. (2006). See materials and methods for BAC clone sequence references.
Nineteen genomic regions available for the A genome (SanMiguel et al. 2002; Yan et al. 2002, 2003; Wicker et al. 2003b; Chantret et al. 2005; Isidore et al. 2005; Dvorak et al. 2006; Gu et al. 2006; Miller et al. 2006). See materials and methods for BAC clone sequence references.
Results are as follows: total CACTA TE copies: complete CACTA TE copies/truncated CACTA TE copies.
From cases where we cannot be certain that a CACTA TE is inserted into or by another TE element.
In the wheat B genome, the majority of CACTA TE insertions (mainly those detected as truncated copies) occurred in DNA annotated as unassigned (Table 2). For the rest, significantly higher insertions of CACTA TEs into Athila and other CACTA TEs than into Copia and Gypsy retrotransposons were observed. The two latter retrotransposon superfamilies were significantly more inserted into, rather than by, CACTA TEs (Table 2). These observations indicate that proliferation of the CACTA TEs in the B genome of wheat started before, and continued during and after Athila retrotransposon proliferation, whereas very few insertions occurred during the last waves of high proliferation of Copia and Gypsy.
Similarly, a high level of insertions into unassigned DNA was observed for the CACTA TEs in the A genome. However, for the remaining insertions, no clear period of proliferation could be determined as these show similar levels of insertions into or by all other TE superfamilies (Table 2). These observations, combined with the observed higher level of complete copies (Table 1), suggest that the CACTA TE proliferation continued in the wheat A genome during the last waves of proliferation of Copia and Gypsy, unlike those in the B genome.
DISCUSSION
To constitute representative genomic sequences of the wheat B genome, in this study we have sequenced 10 BAC clones of the chromosome 3B, representing the most important number of genomic regions sequenced for a single wheat chromosome and a cumulative sequence length of 1.429 Mb (0.15% of the chromosome length). As expected, TE proliferation was pronounced (representing 79.1%). Five of these were revealed as gene-containing BAC clones at a density of one or two genes per clone; two other BAC clones contain gene relics or pseudogenes, whereas the three remaining BAC clones were missing genes. This confirms the previous conclusion about the more random distribution of genes on the wheat genome (Devos et al. 2005). Interestingly and in comparison with rice, a high level of “truncated genes” was revealed [six gene relic or pseudogenes, several of which because of TE insertions (three confirmed cases)]. If the confirmed gene number (excluding hypothetical genes) identified in the 1.43-Mb sequences (eight) is extrapolated to the whole wheat chromosome 3B of 1 Gb estimated size, then 5594 genes might be present. A slightly higher number (6000) was calculated from BAC-end sequence analysis (Paux et al. 2006).
Representation of transposable elements:
In this study, TE dynamics, proliferation, and evolutionary pathways were analyzed and compared in 1.98 Mb of sequence from 13 BAC clones of the wheat B genome and 3.63 Mb of sequence from 19 BAC clones of the wheat A genome. These genomic sequences represent very small fractions (<0.03%) of their respective genomes. Nevertheless, it has been argued that, for studying abundant repeats, sequencing and annotation of a small proportion of the genome can be representative (Brenner et al. 1993; Vitte and Bennetzen 2006; Liu et al. 2007). We have been able to confirm the adequate representation where less variation in the proportion of the main TE superfamilies was observed when analyzing a large number of BAC clones (Figure 2). Interestingly, TE proportions observed in the 13 genomic regions of the B genome of wheat are similar to those obtained from 11 Mb of BAC-end sequences of wheat chromosome 3B (Paux et al. 2006). Similarly, TE proportions were not significantly different for the wheat A genome when they were compared with the different ploidy levels (see results).
Although they are representative of abundant wheat TEs available in the TREP database (Wicker et al. 2002; http://wheat.pw.usda.gov.ITMI/Repeats), the class I and class II TEs observed in the genomic sequences of the wheat B and A genomes may not cover all wheat TEs. It is expected that more wheat TEs will be identified, as more wheat genomic sequences will become available. This is particularly supported by the identification in this study of >21 different novel TE families, most of which (17) are retrotransposons. We also believe that low-copy TEs and those that tend to “compartmentalize” in specific regions, such as pericentromeric heterochromatin regions (which is not the case in our regions), would be missed, over-, or underrepresented in this study (Ma and Bennetzen 2006; Liu et al. 2007). This could be the case for the CACTA TEs, which show the highest variation in sequence proportion between regions because they tend to be clustered in the Triticeae genomes (our unpublished results and Wicker et al. 2003a, 2005).
Transposable elements proliferated differentially in the B and A genomes of wheat:
Abundance of TEs varies widely across different organisms. Human (Homo sapiens) DNA is composed of 45% (Lander et al. 2001) repetitive sequences, Drosophila melanogaster of 3.9% (Kaminker et al. 2002), and maize of 67% (Haberer et al. 2005; Liu et al. 2007) whereas TE content in the wheat genomic sequences analyzed in this study or in other studies (Li et al. 2004; Gu et al. 2006; Paux et al. 2006) is ∼80%. Proportions of different classes of TEs also vary among organisms. Class II TEs are almost >10 times less abundant than class I TEs and constitute a small fraction (<2%) of the human, rice (Piegu et al. 2006), maize (Kronmiller and Wise 2008), Arabidopsis, and cotton (Hawkins et al. 2006) genomes. In comparison, class II TE abundance is relatively high in the wheat B and A genomes (14.1 and 9.9%, respectively), the majority of which (95%) are CACTA TEs, which are particularly abundant in the Triticeae genomes (Wicker et al. 2003a, 2005). Class I retrotransposon abundance is relatively high in several plant genomes, 58.7 and 56.6% estimated in this study for the wheat B and A genomes, respectively; 40–50% in cotton species (Hawkins et al. 2006); 35–60% in rice species (Piegu et al. 2006); and 64% in maize (Liu et al. 2007; Kronmiller and Wise 2008).
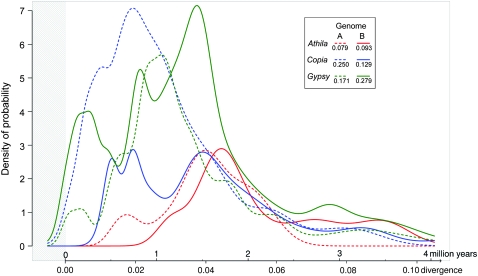
In this study, combination of TE sequence analysis and classification, comparison of proportions of complete to incomplete copies, TE insertion date estimations, and PCR-based tracing of insertions allow us to compare TE proliferation periods and rates in the wheat B and A genomes (Figure 5). It is evident that TEs appear to proliferate differentially in waves of high activity followed by periods of low activity (Figure 5). Both genomes show similar rates and relatively old proliferation periods for the Athila retrotransposons (Figure 5). However, the Copia retrotransposons have proliferated relatively more recently in the A genome whereas a more recent Gypsy proliferation is observed in the B genome. Due to their biology and replication mechanism, it was not possible to directly estimate the CACTA class II TE insertion dates. We have estimated their proliferation periods and rates relative to that of the three main LTR retrotransposon superfamilies. In the wheat B genome, the CACTA TE high proliferation period started before and overlaps with that of the Athila retrotransposons. In the wheat A genome, in addition to the relatively old proliferation similar to that in the B genome, CACTA TEs continued to proliferate during the same period as Gypsy and Copia retrotransposons. Determining the ancient proliferation periods of CACTA TEs partially explains why CACTA TEs often tend to be clustered together (see results and Wicker et al. 2003a, 2005), although they were detected in almost all analyzed BAC clones. Differential proliferation of TEs provides a valid explanation for the size variation of closely related wheat genomes (Bennett and Smith 1976, 1991; http://data.kew.org/cvalues/homepage.html).
Figure 5.—
Proliferation periods and rates of the main retrotransposon superfamilies in the wheat B and A genomes. Expressed as probability density functions, where the area under each curve was calculated on the basis of the estimated insertion dates of retrotransposons (in Figure 3) and their corresponding standard errors, using Gaussian kernel density estimation (Silverman 1986). The curves have been scaled with respect to the number of observations, so that the sum of their areas (given for each retrotransposon superfamily in the key) equals the probability of 1 and comparisons between genomes and retrotransposon superfamilies can be performed. When calculated standard errors were very low, a minimum value of 80,000 years (corresponding to 0.002 divergence) was used. The shaded field is due to uncertainty in very recent insertion date estimations.
Four families (Angela, Wis, Sabrina, Fatima) were abundant, representing the majority of LTR retrotransposons in the B and A genomes of wheat, some of which proliferated differentially (see results). Proliferation of specific types of TEs in specific genomes (or species), leading to rapid genome size variation and sequence divergence, has also been observed in other plant species. Analysis of maize (Zea mays) genomic sequences suggests that the high percentage of LTR retrotransposons is due to proliferation of only a few families of TEs (Meyers et al. 2001; Liu et al. 2007; Kronmiller and Wise 2008). Similarly, comparison of TE proportions between various cotton species (Gossypium species) revealed differential lineage-specific expansion of various LTR–retrotransposon superfamilies and families, leading to threefold genome size differences (Hawkins et al. 2006). Species-specific differential retrotransposon expansions are also the cause of the size doubling of the Oryza australiensis genome as compared to cultivated rice (O. sativa) (Piegu et al. 2006).
This is the first time that dynamics as well as proliferation periods and rates of TEs have been compared between two closely related wheat genomes. This was possible only because in this study we sequenced 10 different genomic regions that constituted a genomic sequence data set representative of the wheat B genome. For the wheat A genome, more representative genomic sequence data were rendered publicly available. There have been initial attempts to evaluate TE proliferation in the wheat genomes. Li et al. (2004) analyzed the D genome of the diploid Ae. tauschii and showed that the copy number of most TEs have increased gradually following polyploidization. However, they used dot blots, which are not very accurate. Sabot et al. (2005) have updated TE annotation in wheat genomic sequences and reported their composition and distribution in relation to genes. They suggested that Copia TEs have been most active in the wheat A, B, and D genomes, combined together (Sabot et al. 2005). Accurate comparison of dynamics as well as proliferation periods and rates between individual genomes of wheat could not be conducted in the study of Sabot et al. (2005) as, in the genomic sequences available at that time, the A genome was overrepresented whereas the B genome was underrepresented. By using more representative genomic sequences in this study, we showed the more recent activation of the Copia and CACTA TEs in the wheat A genome but not in the B genome in which a more recent Gypsy proliferation is observed. Overrepresentation of the A-genome sequences in the study of Sabot et al. (2005) may explain the reason why they found that Copia TEs have been most active in the wheat A, B, and D genomes combined together. Thus our analysis, using representative sequence data sets, for the first time shows differential proliferation of TEs between the wheat A and B genomes and illustrates the inadequacy of combining sequence data sets from different genomes as was previously done.
Neither enhancement nor repression of transposable element proliferation following allotetraploidization:
As estimated from their insertion dates and confirmed by PCR-based tracing analysis, the majority of the differential proliferation of TEs in B and A genomes of wheat (87% and 83, respectively) occurred prior to the allotetraploidization event that brought them together in T. turgidum and T. aestivum <0.5 MYA (Huang et al. 2002; Dvorak et al. 2006; Chalupska et al. 2008). More importantly, the allotetraploidization event appears to have neither enhanced nor repressed retrotranspositions. We suggest that, in addition to the Ph1 gene preventing homeologous recombination (Griffiths et al. 2006), differential proliferation of TEs has also contributed to the rapid divergence of the B and A genomes of the wheat diploid progenitors and the relative stability of the natural wheat allopolyploids that occurs thereafter.
Different levels of stability, estimated as elimination of DNA sequences, were observed in newly synthesized wheat allopolyploids, depending on wheat genome combinations (Feldman and Levy 2005 and our unpublished results). The natural wheat allopolyploids combining the B and A genomes are relatively stable and cannot be exactly resynthesized because the diploid progenitor of the B genome is unidentified (Feldman et al. 1995; Blake et al. 1999; Huang et al. 2002; Dvorak et al. 2006). Nevertheless, by studying a synthetic wheat allotetraploid combining the A and S genomes (the closest identified diploid relatives to the progenitors of the A and of the B genomes of natural wheat polyploids), Kashkush et al. (2003) reported on transcriptional activation of the Wis LTR retrotransposon but not its transposition following allotetraploidization. This is in agreement with the lack of enhancement of transpositions observed in this study in wheat natural allopolyploids combining the A and B genomes. Comparatively, less TE proliferation, estimated as the increased rate of deletions and the decreased rate of insertions, was recently observed in the cotton polyploid species Gossypium hirsutum as compared to its diploid progenitors Gossypium arboretum and Gossypium raimondii (Grover et al. 2008).
Apparent transposable element proliferation as a balance between two evolutionary forces: TEs “transposition” and also their removal:
As in this study, the vast majority of complete retrotransposons studied so far were also estimated to be <3 million years old (SanMiguel et al. 1998, 2002; Wicker et al. 2003b, 2005; Gao et al. 2004; Ma et al. 2004; Du et al. 2006; Piegu et al. 2006; Wicker and Keller 2007). These findings imply that there are mechanisms of active deletion of LTR retrotransposons from the genome, such as unequal homologous recombination and illegitimate recombination (Vicient et al. 1999; Devos et al. 2002; Ma et al. 2004; Pereira 2004). Proliferation periods and rates estimated for TEs at a given evolutionary period are the result of both antagonist evolutionary forces: TE insertion activity (transpositions) (Bennetzen and Kellogg 1997) and the removal of TEs (Petrov et al. 2000; Petrov 2002a). Thus, it is not clear whether the insertions and/or truncation (removal) rates of TEs are constant or vary during genome evolution. The “burst of insertions” described for TEs could correspond to periods of (i) high insertion activity, (ii) low rates of TE removal, and/or (iii) combinations of both evolutionary forces.
The fact that Copia retrotransposons have been active until recently in the Arabidopsis thaliana genome allowed Pereira (2004) to calculate the rate of their elimination (or half-life) as 472,000 years, outside of centromeric regions. Using this method and assuming that repetitive sequences are removed from the genome at a constant rate, a higher half-life (79,000 years) was calculated for Copia removal in rice (Wicker and Keller 2007). As the insertion-date distribution of Copia retrotransposons in Triticeae (wheat and barley) is not exponential, Wicker and Keller (2007) suggested that their half-life is much longer than in rice, thus representing a major difference between small and large genomes of plants. Similar distributions are observed in our study for all three retrotransposon superfamilies in both B and A genomes of wheat. Our analysis suggests that lower proliferation of the LTR retrotransposons during the most recent period could account for these apparent nonexponential distributions of insertion dates (including Copia retrotransposons) (Figure 5).
Our study clearly shows that, during their evolution, specific types of TEs have undergone differential proliferation in specific wheat genomes (or species) but not in others, leading to rapid sequence divergence. Little is known about the mechanistic causes that lead to differential proliferation of a single or related group of TEs across the genome of a specific species. These rapid TE expansions could correspond to periods of relaxed selection pressure such as genome duplication, interspecific hybridizations (although this was not revealed in our study), or stress conditions. It is also possible that TE proliferation could be caused by advantageous mutations in the TE sequence. A third alternative is differential deregulation of epigenetic silencing that allows specific TE families to proliferate in specific genomes.
Acknowledgments
We sincerely thank J. Dolezel and M. Kubalakova (Institute of Experimental Botany, Olomouc, Czech Republic) for providing FISH mapping information for BAC clones B95G2, B95C9, B63B7, and B54F7; Joseph Jahier [Institut National de la Recherche Agronomique (INRA), Rennes, France] and Moshe Feldman (Weizmann Institute of Science) for valuable discussions and for providing wheat genotypes; Catherine Feuillet (INRA, Clermont-Ferrand, France) for providing the wheat deletion lines; Thomas Wicker (Zurich University) for valuable advice on novel transposable element classifications and CACTA TE evolution; Piotr Gornicki (University of Chicago) and anonymous reviewers for valuable discussion and constructive criticisms; and Heather McKhann (Centre National de Génotypage, Etude du Polymorphisme Génomique Vegetal-INRA, Evry, France) for valuable discussion and revision of the manuscript. This project was supported by the National Center for Sequencing (Centre National de Séquençage-Génoscope)/APCNS2003-Project: Triticum species comparative genome sequencing in wheat (http://www.genoscope.cns.fr/externe/English/). PCR-based tracing of retrotransposons insertions was funded by the Agence Nationale pour la Recherche Biodiversité Project (ANR-05-BDIV-015) and the ANR-05-Blanc project-ITEGE.
References
- Adams, K. L., and J. F. Wendel, 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8 135–141. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M. D., and I. J. Leitch, 1997. Nuclear DNA amounts in angiosperms: 583 new estimates. Ann. Bot. 80 169–196. [Google Scholar]
- Bennett, M. D., and I. J. Leitch, 2005. Plant genome size research: a field in focus. Ann. Bot. 95 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M. D., and J. B. Smith, 1976. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 274 227–274. [DOI] [PubMed] [Google Scholar]
- Bennett, M. D., and J. B. Smith, 1991. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 334 309–345. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., 2000. a Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell 12 1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J. L., 2000. b Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42 251–269. [PubMed] [Google Scholar]
- Bennetzen, J. L., 2002. a Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 115 29–36. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., 2002. b The rice genome: opening the door to comparative plant biology. Science 296 60–63. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and E. A. Kellogg, 1997. Do plants have a one-way ticket to genomic obesity? Plant Cell 9 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J. L., J. Ma and K. M. Devos, 2005. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 95 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, N. K., B. R. Lehfeldt, M. Lavin and L. E. Talbert, 1999. Phylogenetic reconstruction based on low copy DNA sequence data in an allopolyploid: the B genome of wheat. Genome 42 351–360. [PubMed] [Google Scholar]
- Blanc, G., A. Barakat, R. Guyot, R. Cooke and M. Delseny, 2000. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., G. Elgar, R. Sandford, A. Macrae, B. Venkatesh et al., 1993. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature 366 265–268. [DOI] [PubMed] [Google Scholar]
- Chalupska, D., H. Y. Lee, J. D. Faris, A. Evrard, B. Chalhoub et al., 2008. Acc homoeoloci and the evolution of wheat genomes. Proc. Natl. Acad. Sci. USA 105 9691–9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantret, N., J. Salse, F. Sabot, S. Rahman, A. Bellec et al., 2005. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chooi, W. Y., 1971. Variation in nuclear DNA content in the genus Vicia. Genetics 68 195–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K. M., J. K. Brown and J. L. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K. M., J. Ma, A. C. Pontaroli, L. H. Pratt and J. L. Bennetzen, 2005. Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc. Natl. Acad. Sci. USA 102 19243–19248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel, J., M. Kubalakova, J. Bartos and J. Macas, 2004. Flow cytogenetics and plant genome mapping. Chromosome Res. 12 77–91. [DOI] [PubMed] [Google Scholar]
- Du, C., Z. Swigonova and J. Messing, 2006. Retrotranspositions in orthologous regions of closely related grass species. BMC Evol. Biol. 6 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, J., P. Diterlizzi, H.-B. Zhang and P. Resta, 1993. The evolution of polyploid wheats: identification of the A genome donor species. Genome 36 21–31. [DOI] [PubMed] [Google Scholar]
- Dvorak, J., E. D. Akhunov, A. R. Akhunov, K. R. Deal and M. C. Luo, 2006. Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol. Biol. Evol. 23 1386–1396. [DOI] [PubMed] [Google Scholar]
- Efron, B., 1979. Bootstrap methods: another look at the jackknife. Ann. Stat. 7 1–26. [Google Scholar]
- Feldman, M., and A. A. Levy, 2005. Allopolyploidy: a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109 250–258. [DOI] [PubMed] [Google Scholar]
- Feldman, M., F. G. H. Lupton and T. E. Miller, 1995. Wheats, pp.184–192 in Evolution of Crops, Ed. 2, edited by J. Smartt and N. W. Simmonds. Longman Scientific, London.
- Férignac, P., 1962. Test de Kolmogorov-Smirnov sur la validité d'une fonction de distribution. Rev. Stat. Appl. 10 13–32. [Google Scholar]
- Flavell, A. J., M. R. Knox, S. R. Pearce and T. H. Ellis, 1998. Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J. 16 643–650. [DOI] [PubMed] [Google Scholar]
- Gao, L., E. M. McCarthy, E. W. Ganko and J. F. McDonald, 2004. Evolutionary history of Oryza sativa LTR retrotransposons: a preliminary survey of the rice genome sequences. BMC Genomics 5: 18. [DOI] [PMC free article] [PubMed]
- Graner, A., H. Siedler, A. Jahoor, R. G. Herrman and G. Wenzal, 1990. Assessment of the degree and the type of restriction fragment length polymorphism in barley (Hordeum vulgare). Theor. Appl. Genet. 80 826–832. [DOI] [PubMed] [Google Scholar]
- Griffiths, S., R. Sharp, T. N. Foote, I. Bertin, M. Wanous et al., 2006. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439 749–752. [DOI] [PubMed] [Google Scholar]
- Grover, C. E., Y. Yu, R. A. Wing, A. H. Paterson and J. F. Wendel, 2008. A phylogenetic analysis of indel dynamics in the cotton genus. Mol. Biol. Evol. 25 1415–1428. [DOI] [PubMed] [Google Scholar]
- Gu, Y. Q., J. Salse, D. Coleman-Derr, A. Dupin, C. Crossman et al., 2006. Types and rates of sequence evolution at the high-molecular-weight glutenin locus in hexaploid wheat and its ancestral genomes. Genetics 174 1493–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer, G., S. Young, A. K. Bharti, H. Gundlach, C. Raymond et al., 2005. Structure and architecture of the maize genome. Plant Physiol. 139 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, J. S., H. Kim, J. D. Nason, R. A. Wing and J. F. Wendel, 2006. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 16 1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., A. Sirikhachornkit, X. Su, J. Faris, B. Gill et al., 2002. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidore, E., B. Scherrer, B. Chalhoub, C. Feuillet and B. Keller, 2005. Ancient haplotypes resulting from extensive molecular rearrangements in the wheat A genome have been maintained in species of three different ploidy levels. Genome Res. 15 526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon, O., J. M. Aury, B. Noel, A. Policriti, C. Clepet et al., 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–467. [DOI] [PubMed] [Google Scholar]
- Jones, R. N., and L. M. Brown, 1976. Chromosome evolution and DNA variation in Crepis. Heredity 36 91–104. [Google Scholar]
- Kaminker, J. S., C. M. Bergman, B. Kronmiller, J. Carlson, R. Svirskas et al., 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3 RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., M. Feldman and A. A. Levy, 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33 102–106. [DOI] [PubMed] [Google Scholar]
- Kidwell, M. G., 2002. Transposable elements and the evolution of genome size in eukaryotes. Genetica 115 49–63. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111–120. [DOI] [PubMed] [Google Scholar]
- Kronmiller, B. A., and R. P. Wise, 2008. TE nest: automated chronological annotation and visualization of nested plant transposable elements. Plant Physiol. 146 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- Li, W., P. Zhang, J. P. Fellers, B. Friebe and B. S. Gill, 2004. Sequence composition, organization, and evolution of the core Triticeae genome. Plant J. 40 500–511. [DOI] [PubMed] [Google Scholar]
- Liu, R., C. Vitte, J. Ma, A. A. Mahama, T. Dhliwayo et al., 2007. A GeneTrek analysis of the maize genome. Proc. Natl. Acad. Sci. USA 104 11844–11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and J. L. Bennetzen, 2004. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and J. L. Bennetzen, 2006. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc. Natl. Acad. Sci. USA 103 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., K. M. Devos and J. L. Bennetzen, 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B. C., S. V. Tingey and M. Morgante, 2001. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A. K., G. Galiba and J. Dubcovsky, 2006. A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum. Mol. Genet. Genomics 275 193–203. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., J. E. Bowers and B. A. Chapman, 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paux, E., D. Roger, E. Badaeva, G. Gay, M. Bernard et al., 2006. Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J. 48 463–474. [DOI] [PubMed] [Google Scholar]
- Pereira, V., 2004. Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol. 5 R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D. A., 2002. a Mutational equilibrium model of genome size evolution. Theor. Popul. Biol. 61 531–544. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., 2002. b DNA loss and evolution of genome size in Drosophila. Genetica 115 81–91. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., T. A. Sangster, J. S. Johnston, D. L. Hartl and K. L. Shaw, 2000. Evidence for DNA loss as a determinant of genome size. Science 287 1060–1062. [DOI] [PubMed] [Google Scholar]
- Piegu, B., R. Guyot, N. Picault, A. Roulin, A. Saniyal et al., 2006. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, L., B. Echalier, B. Friebe and B. S. Gill, 2003. Molecular characterization of a set of wheat deletion stocks for use in chromosome bin mapping of ESTs. Funct. Integr. Genomics 3 39–55. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. [DOI] [PubMed] [Google Scholar]
- Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice et al., 2000. Artemis: sequence visualization and annotation. Bioinformatics 16 944–945. [DOI] [PubMed] [Google Scholar]
- Sabot, F., R. Guyot, T. Wicker, N. Chantret, B. Laubin et al., 2005. Updating of transposable element annotations from large wheat genomic sequences reveals diverse activities and gene associations. Mol. Genet. Genomics 274 119–130. [DOI] [PubMed] [Google Scholar]
- Safar, J., J. Bartos, J. Janda, A. Bellec, M. Kubalakova et al., 2004. Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J. 39 960–968. [DOI] [PubMed] [Google Scholar]
- Salse, J., S. Bolot, M. Throude, V. Jouffe, B. Piegu et al., 2008. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., A. Tikhonov, Y. K. Jin, N. Motchoulskaia, D. Zakharov et al., 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274 765–768. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., B. S. Gaut, A. Tikhonov, Y. Nakajima and J. L. Bennetzen, 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20 43–45. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P. J., W. Ramakrishna, J. L. Bennetzen, C. S. Busso and J. Dubcovsky, 2002. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A(m). Funct. Integr. Genomics 2 70–80. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., W. Jianzhong, T. Itoh and T. Matsumoto, 2005. [Complete rice genome sequence information: the key for elucidation of Rosetta stones of other cereal genome] Tanpakushitsu Kakusan Koso 50 2167–2173. [PubMed] [Google Scholar]
- Silverman, B. W., 1986. Density Estimation for Statistics and Data Analysis. Chapman & Hall, London/New York.
- Smith, D., and R. Flavell, 1975. Characterization of the wheat genome by renaturation kinetics. Chromosoma 50 223–242. [Google Scholar]
- Stein, N., 2007. Triticeae genomics: advances in sequence analysis of large genome cereal crops. Chromosome Res. 15 21–31. [DOI] [PubMed] [Google Scholar]
- Vedel, F., and M. Delseny, 1987. Repetivity and variability of higher plant genomes. Plant Physiol. Biochem. 25 191–210. [Google Scholar]
- Vicient, C. M., A. Suoniemi, K. Anamthawat-Jonsson, J. Tanskanen, A. Beharav et al., 1999. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11 1769–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte, C., and J. L. Bennetzen, 2006. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 103 17638–17643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F., R. C. Cronn, J. S. Johnston and H. J. Price, 2002. Feast and famine in plant genomes. Genetica 115 37–47. [DOI] [PubMed] [Google Scholar]
- Wicker, T., and B. Keller, 2007. Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res. 17 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., D. Matthews and B. Keller, 2002. TREP: a database for Triticeae repetitive elements. Trends Plant. Sci. 7 561–562. [Google Scholar]
- Wicker, T., R. Guyot, N. Yahiaoui and B. Keller, 2003. a CACTA transposons in Triticeae: a diverse family of high-copy repetitive elements. Plant Physiol. 132 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., N. Yahiaoui, R. Guyot, E. Schlagenhauf, Z. D. Liu et al., 2003. b Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., W. Zimmermann, D. Perovic, A. H. Paterson, M. Ganal et al., 2005. A detailed look at 7 million years of genome evolution in a 439 kb contiguous sequence at the barley Hv-eIF4E locus: recombination, rearrangements and repeats. Plant J. 41 184–194. [DOI] [PubMed] [Google Scholar]
- Wicker, T., F. Sabot, A. Hua-Van, J. L. Bennetzen, P. Capy et al., 2007. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8 973–982. [DOI] [PubMed] [Google Scholar]
- Yan, L., V. Echenique, C. Busso, P. SanMiguel, W. Ramakrishna et al., 2002. Cereal genes similar to Snf2 define a new subfamily that includes human and mouse genes. Mol. Genet. Genomics 268 488–499. [DOI] [PubMed] [Google Scholar]
- Yan, L., A. Loukoianov, G. Tranquilli, M. Helguera, T. Fahima et al., 2003. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo, A., A. Sebastian, J. Talag, Y. Yu, H. Kim et al., 2007. Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol. Biol. 7 152. [DOI] [PMC free article] [PubMed] [Google Scholar]