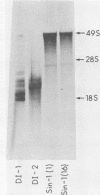
Abstract
Sindbis virus generates defective interfering (DI) particles during serial high-multiplicity passage in cultured cells. These DI particles inhibit the replication of infectious virus and can be an important factor in the establishment and maintenance of persistent infection in BHK cells. In an effort to understand how these DI particles are generated and how they interfere with the replication of standard virus, we performed a partial sequence analysis of the RNA obtained from two independently isolated populations of DI particles and from two Sindbis virus variants and compared these with the RNA of the parental wild-type virus. The 3'-terminal regions of the RNAs were sequenced by the dideoxy chain terminating method. Internal regions of the RNA were examined by restriction endonuclease digestion of cDNA's made to the various RNAs and by direct chemical sequencing of 5' end-labeled restriction fragments from cDNA made to the DI RNAs. One of the variant viruses examined was originally derived from cells persistently infected with Sindbis virus for 16 months and is resistant to interference by the DI strains used. In the 3'-terminal region of the RNA from this variant, only two base changes were found; one of these occurs in the 20-nucleotide 3'-terminal sequence which is highly conserved among alphaviruses. The DI RNA sequences were found to have been produced not by a single deletional event, but by multiple deletion steps combined with sequence rearrangements; all sequences examined are derived from the plus strand of Sindbis virion RNA. Both DI RNAs had at least 50 nucleotides of wild-type sequence conserved at the 3' terminus; in addition, they both contained conserved and perhaps amplified sequences derived from the non-26S region of the genome which may be of importance in their replication and interference ability.
Full text
PDF



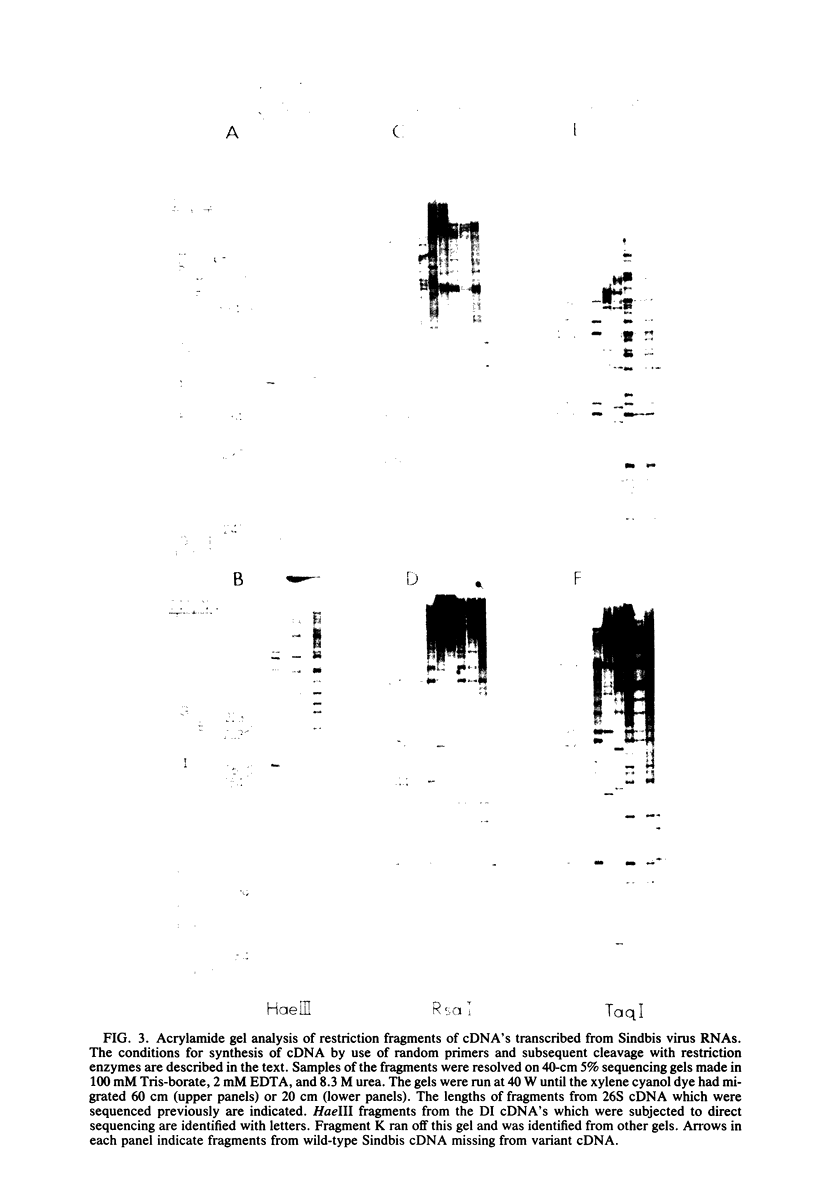



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carmichael G. G., McMaster G. K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. doi: 10.1016/s0076-6879(80)65049-6. [DOI] [PubMed] [Google Scholar]
- Dohner D., Monroe S., Weiss B., Schlesinger S. Oligonucleotide mapping studies of standard and defective Sindbis virus RNA. J Virol. 1979 Feb;29(2):794–798. doi: 10.1128/jvi.29.2.794-798.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Frischauf A. M., Simons K., Lehrach H., Delius H. Nucleotide sequence of cdna coding for Semliki Forest virus membrane glycoproteins. Nature. 1980 Nov 20;288(5788):236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- Garoff H., Frischauf A. M., Simons K., Lehrach H., Delius H. The capsid protein of Semliki Forest virus has clusters of basic amino acids and prolines in its amino-terminal region. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6376–6380. doi: 10.1073/pnas.77.11.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guild G. M., Stollar V. Defective interfering particles of Sindbis virus. III. Intracellular viral RNA species in chick embryo cell cultures. Virology. 1975 Sep;67(1):24–41. doi: 10.1016/0042-6822(75)90400-6. [DOI] [PubMed] [Google Scholar]
- Guild G. M., Stollar V. Defective interfering particles of Sindbis virus. V. Sequence relationships between SVSTD 42 S RNA and intracellular defective viral RNAs. Virology. 1977 Mar;77(1):175–188. doi: 10.1016/0042-6822(77)90416-0. [DOI] [PubMed] [Google Scholar]
- Kennedy S. I., Bruton C. J., Weiss B., Schlesinger S. Defective interfering passages of Sindbis virus: nature of the defective virion RNA. J Virol. 1976 Sep;19(3):1034–1043. doi: 10.1128/jvi.19.3.1034-1043.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S. I. Sequence relationships between the genome and the intracellular RNA species of standard and defective-interfering Semliki Forest virus. J Mol Biol. 1976 Dec;108(2):491–511. doi: 10.1016/s0022-2836(76)80132-5. [DOI] [PubMed] [Google Scholar]
- Laskey R. A. The use of intensifying screens or organic scintillators for visualizing radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65(1):363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- Lehtovaara P., Söderlund H., Keränen S., Pettersson R. F., Käriäinen L. 18S defective interfering RNA of Semliki Forest virus contains a triplicated linear repeat. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5353–5357. doi: 10.1073/pnas.78.9.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ou J. H., Strauss E. G., Strauss J. H. Comparative studies of the 3'-terminal sequences of several alpha virus RNAs. Virology. 1981 Mar;109(2):281–289. doi: 10.1016/0042-6822(81)90499-2. [DOI] [PubMed] [Google Scholar]
- Pettersson R. F. 5'-Terminal nucleotide sequence of Semliki forest virus 18S defective interfering RNA is heterogeneous and different from the genomic 42S RNA. Proc Natl Acad Sci U S A. 1981 Jan;78(1):115–119. doi: 10.1073/pnas.78.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2062–2066. doi: 10.1073/pnas.78.4.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Synthesis, cleavage and sequence analysis of DNA complementary to the 26 S messenger RNA of Sindbis virus. J Mol Biol. 1981 Aug 15;150(3):315–340. doi: 10.1016/0022-2836(81)90550-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Calvo J. M. Nucleotide sequence of the E coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980 May 24;8(10):2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Kennedy S. I. The generation and propagation of defective-interfering particles of Semliki Forest virus in different cell types. Virology. 1978 Aug;89(1):285–299. doi: 10.1016/0042-6822(78)90060-0. [DOI] [PubMed] [Google Scholar]
- Weiss B., Rosenthal R., Schlesinger S. Establishment and maintenance of persistent infection by Sindbis virus in BHK cells. J Virol. 1980 Jan;33(1):463–474. doi: 10.1128/jvi.33.1.463-474.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B., Schlesinger S. Defective interfering particles of Sindbis virus do not interfere with the homologous virus obtained from persistently infected BHK cells but do interfere with Semliki Forest virus. J Virol. 1981 Feb;37(2):840–844. doi: 10.1128/jvi.37.2.840-844.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]