Abstract
While the population genetics of inbreeding is fairly well understood, the effects of inbreeding on the physiological and biochemical levels are not. Here we have investigated the effects of inbreeding on the Drosophila melanogaster metabolome. Metabolite fingerprints in males from five outbred and five inbred lines were studied by nuclear magnetic resonance spectroscopy after exposure to benign temperature, heat stress, or cold stress. In both the absence and the presence of temperature stress, metabolite levels were significantly different among inbred and outbred lines. The major effect of inbreeding was increased levels of maltose and decreased levels of 3-hydroxykynurenine and a galactoside [1-O-(4-O-(2-aminoethyl phosphate)-β-d-galactopyranosyl)-x-glycerol] synthesized exclusively in the paragonial glands of Drosophila species, including D. melanogaster. The metabolomic effect of inbreeding at the benign temperature was related to gene expression data from the same inbred and outbred lines. Both gene expression and metabolite data indicate that fundamental metabolic processes are changed or modified by inbreeding. Apart from affecting mean metabolite levels, inbreeding led to an increased between-line variation in metabolite profiles compared to outbred lines. In contrast to previous observations revealing interactions between inbreeding and environmental stress on gene expression patterns and life-history traits, the effect of inbreeding on the metabolite profile was similar across the different temperature treatments.
INBREEDING is characterized by an increase in homozygosity resulting in increased expression of recessive deleterious alleles (partial dominance hypothesis) and/or reduced opportunity to express heterozygote superiority (overdominance hypothesis) (Charlesworth and Charlesworth 1999). The deleterious effects of inbreeding, especially on fitness traits, are well known and are of considerable concern in human genetics, animal breeding, and conservation biology. Only fairly recently, molecular and physiological investigations on the effects of inbreeding have been accomplished (Kristensen et al. 2005a, 2006; Pedersen et al. 2005). Results from these studies indicate that an increase in homozygosity caused by inbreeding has strong effects on gene expression and that expression of specific proteins is differentially regulated in inbred relative to outbred control populations across biological replicates. The directional changes observed across independent inbred lines suggest that differences between inbred and outbred control lines cannot be explained by genetic drift alone as drift would lead to line-specific fixation of alleles. Hypothetically, the molecular and biochemical changes caused by inbreeding could be due to a general disturbance of cellular homeostasis (Lerner 1954) or it could be due to more specific changes targeted toward single proteins or pathways and regulatory systems (Birchler et al. 2005). Empirical studies on the effects of fixation of deleterious mutations on protein instability and disturbance of processes related to the protein quality control system have the potential to reveal causative explanations for biochemical changes induced by inbreeding (Sangster et al. 2004; Depristo et al. 2005). However, currently, our knowledge within this field is limited.
The level of inbreeding depression is often enhanced under harsh environmental conditions (Armbruster and Reed 2005). Thus, research on the deleterious consequences caused by inbreeding ideally should be combined with studies on the response of inbred and outbred control populations to suboptimal environmental conditions.
Postgenomic technologies such as proteomics and metabolomics might provide integrative approaches needed to obtain an understanding of the physiological and biochemical effects of inbreeding and environmental stress. Metabolomics investigates whole biological systems of low-molecular-weight metabolites. Changes in the metabolite profile are downstream of transcription, translation, protein level, and activity, thus providing a biochemical fingerprint of the integrated response of an organism to internal or external stimuli. Together with powerful multivariate statistical methods, metabolite fingerprinting therefore provides a rapid and nonbiased assessment of the metabolome, making it an attractive analytical method for defining metabolic phenotypes. This method has previously been successfully used to elucidate the responses to temperature stress in Drosophila melanogaster (Malmendal et al. 2006; Overgaard et al. 2007).
This study presents results on metabolite profiles in inbred and outbred control populations of male D. melanogaster after exposure to benign temperature, cold stress, or heat stress. Data on the metabolomic effect of inbreeding at benign temperature have been integrated with published data (Kristensen et al. 2005a, 2006) on the effect of inbreeding on gene expression patterns in the same inbred and outbred lines.
MATERIALS AND METHODS
Experimental flies and design:
A genetically diverse population was generated in 2002 by mixing populations of D. melanogaster from different geographical regions (for further details, see Bubliy and Loeschcke 2005). Five inbred lines (I1–I5) with an expected inbreeding level of 0.67 were generated through five generations of full-sib mating (for details on the inbreeding procedure, see Pedersen et al. 2005). When the inbred lines reached the desired inbreeding level, the population size was flushed to ∼1000 individuals over two generations and held at this size for all following generations. The high population size is expected to reduce further increase in the level of inbreeding. Five outbred control populations (C1–C5) were founded from the diverse mass population, each by ∼1000 individuals at the time when the inbreeding procedure was initiated. Flies from all lines were reared and maintained at standard laboratory conditions (25° ± 0.2°, 50% relative humidity, 12/12 hr light/dark cycle). The experiments reported here were performed in January 2007. All flies used in the experiment were reared under controlled density (30 larvae/vial) on 7 ml standard medium. Five replicates of 50 male flies (4 ± 0.5 days old) per line were exposed to three different temperature regimes: benign temperature, heat stress, or cold stress. For benign temperature and heat stress, flies were exposed to 25° or 36° for 1 hr in vials in water baths. Flies were stressed in empty vials and subsequently allowed to recover for 1 hr at 25° in vials containing medium at standard laboratory conditions. Flies were exposed to cold stress in empty vials in a water bath at 0° for 2 h followed by 7 h of recovery at 25° in vials containing medium at standard laboratory conditions. After recovery, all flies were transferred to 5-ml microtubes, frozen in liquid nitrogen, and stored at −80°. All experiments were carried out on the same day, and all samples were frozen at the same time (±1 hr) in the afternoon. The temperatures used for cold- and heat-stress exposures were chosen on the basis of a pilot experiment showing that these temperatures could be considered stressful, but nonlethal, for the flies (results not shown).
Sample preparation and nuclear magnetic resonance measurements:
Flies from each sample were mechanically homogenized with a Kinematica, Pt 1200 (Buch & Holm A/S) in 1 ml of ice-cold acetonitril (50%) and centrifuged for 30 min (4°). The supernatant was transferred to new tubes, lyophilized, and stored at −80°. Immediately before the nuclear magnetic resonance (NMR) measurements were taken, samples were rehydrated in 650 μl of 50 mm phosphate buffer (pH 7.4) in D2O, and 600 μl was transferred to a 5-mm NMR tube. The buffer contained 50 mg/liter of the chemical shift reference 3-(trimethylsilyl)-propionic acid-D4, sodium salt (TSP). The NMR measurements were performed at 25° on a Bruker Avance-2 700 spectrometer (Bruker Biospin), operating at a 1H frequency of 700.09 MHz, equipped with a 5-mm HCN triple resonance probe. 1H NMR spectra were acquired using a single-90°-pulse experiment with a Carr–Purcell–Meiboom–Gill (CPMG) delay added to attenuate broad signals from high-molecular-weight components. The total CPMG delay was 40 msec and the spin-echo delay was 200 μsec. The water signal was suppressed by presaturation of the water peak during the relaxation delay of 1.5 sec. A total of 256 transients of 16,384 data points spanning a spectral width of 24 ppm were collected, corresponding to a total experiment time of 10 min. For assignment purposes, a two-dimensional 1H-1H TOCSY (Braunschweiler and Ernst 1983; Bax and Davis 1985) spectrum with 80 msec mixing was acquired. To identify the galactoside, additional NMR spectra were recorded. DQF-COSY, NOESY, TOCSY, HSQC, and a HMBC with suppression of one-bond correlations were recorded at 20° on a Bruker Avance 800 instrument operating at a 1H frequency of 799.96 MHz, equipped with a 5-mm probe. Here the chemical shift scale was calibrated to acetone for proton (2.22 ppm) and carbon (30.89 ppm), giving a HDO signal at 4.82 ppm.
Data reduction/treatment:
The spectra were processed using iNMR (http://www.inmr.net). An exponential line broadening of 0.5 Hz was applied to the free-induction decay prior to Fourier transformation. All spectra were referenced to the TSP signal at −0.017 ppm, automatically phased, and baseline corrected. Data reduction was accomplished by dividing the spectrum into 0.005-ppm regions (bins) over which the signal was integrated to obtain the signal intensity. The region around the residual water signal (4.85–4.7 ppm) was removed not to compromise the analysis. The high- and low-field ends of the spectrum, where no signals except the reference signal from TSP appear, were also removed (i.e., leaving data between 9.5 and 0.5 ppm). The integrals were normalized to a total intensity of 1000 to suppress trivial separation on the basis of variations in the amount of sample.
Principal component analysis (PCA) was carried out on data sets including control and inbred flies at benign temperature only; control and inbred flies at benign temperature and after heat stress; control and inbred flies at benign temperature and after cold stress; and on the full data set. The PCA was performed using in-house R-scripts (the R project; http://www.r-project.org), based on a script generously provided by Karl-Heinz Ott. Data were scaled using VAST scaling (Keun et al. 2003) to obtain, on average, unit variance within each line at each temperature (i.e., each region/bin was divided by the average standard deviation of the integral of that region within all combinations of line and temperature) and then centered. This scaling reduces the weight of variations that are not related to the treatments; i.e., random variations between “identical” samples are reduced, and the analysis is not biased toward metabolites present at high concentrations. Initial PCA identified one cold-stressed control sample as a significant outlier due to traces of organic solvent. This sample was excluded. The number of significant PCs was assessed by cross-validation (Wold 1978). The scores from the PCA including only samples treated at benign temperature were subjected to one-way ANOVA for inbreeding or line, and those from PCA including temperature-treated samples were subjected to two-way ANOVA for inbreeding or line and high- or low-temperature treatments using ranked Bonferroni correction for multiple testing (significance level 0.01). Tests of line effects were performed on control and inbred lines together and separately. The effects of inbreeding on the between-line variance was tested using a jackknifing procedure where mean values for each line were calculated for all combinations with one or two samples left out, and the between-line variances for the control and inbred samples were calculated for all combinations of these mean values. The groups with one or two samples left out were treated separately. Significant effects were detected using Wilcoxon tests (d.f. = 48). Generally, only metabolite effects at a significance level of 0.01 are discussed. A dendogram was created using hierarchical cluster analysis (HCA) with projection to latent structures (PLS) discriminant analysis (PLS-DA) scores of VAST scaled data calculated using Simca-P 11.5 (Umetrics, Umeå, Sweden) as input. The number of axes for each PLS-DA model was determined by cross-validation. HCA was then carried out using complete linkages in R (http://www.r-project.org) by using the Euclidean distance between the PLS-DA scores for each treatment. Orthogonal projection to latent structures (OPLS) discriminant analysis (OPLS-DA) (Bylesjö et al. 2006) was carried out between control and inbred flies on data sets including benign temperature, heat-stressed flies, and cold-stressed flies to identify metabolites responsive to inbreeding and between benign temperature and heat- or cold-stressed flies on data sets including control or inbred flies to identify metabolites responsive to heat or cold stress. OPLS separates the variance in x correlated with y (y predictive) with the orthogonal (noncorrelated, y orthogonal) variance (Trygg and Wold 2002). In contrast to regular PLS, a single y will result in only one predictive component. Data were scaled to obtain unit variance and then centered. OPLS was performed using Simca-P 11.5 (Umetrics).
The metabolite signals were identified from a two-dimensional 1H-1H TOCSY spectrum, from STOCSY spectra (Cloarec et al. 2005), and by comparison with known metabolite chemical shifts (Fan 1996; Malmendal et al. 2006; Overgaard et al. 2007). The identities of maltose and 3-hydroxykynurenine were verified by spiking with the pure substance. The assignment of the signals of the galactoside [1-O-(4-O-(2-aminoethyl phosphate)-β-d-galactopyranosyl)-x-glycerol] was based on the combination of DQF-COSY and TOCSY spectra (see figure legend of supplemental Figure S1).
Integration of NMR and gene expression data:
Gene expression on the same lines was studied 40 generations earlier using Affymetrix gene chips (for methods and results, see Kristensen et al. 2005a, 2006). Association of our metabolomic data with transcriptomic data at benign temperature (25°) was done using a modification of the method suggested by Rantalainen et al. (2006). One of the five inbred lines used for gene expression analysis went extinct before samples were obtained for NMR measurements. Thus, NMR and gene expression data used in the comparison of transcriptome and metabolome effects were from five control and four inbred lines. Concentrations of maltose, 3-hydroxykynurenine and the galactoside were correlated to gene expression data using OPLS analysis of gene array data with the intensity of the metabolite peaks as the y variable (maltose: 5.41–5.38 ppm; 3-hydroxykynurenine: 7.44–7.42, 7.03–7.015, 6.69–6.675 ppm; the galactoside: 4.48–4.46, 4.20–4.175 ppm). OPLS scores for each line were correlated to line-specific variation in gene expression (see flow diagram in supplemental Figure S2). Genes that show a high correlation with OPLS scores for a metabolite (R2 > 0.6) and a higher correlation with OPLS scores for that metabolite than with OPLS-DA scores for inbreeding 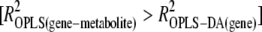 were assumed to be genes correlated to that metabolite. The criteria chosen are relatively stringent to avoid noise caused by intergenerational variation. This method, as with all correlation approaches, does not necessarily detect causal correlations, and it is not expected that all correlating genes are directly involved in processes and pathways related to the specific metabolites.
were assumed to be genes correlated to that metabolite. The criteria chosen are relatively stringent to avoid noise caused by intergenerational variation. This method, as with all correlation approaches, does not necessarily detect causal correlations, and it is not expected that all correlating genes are directly involved in processes and pathways related to the specific metabolites.
Lists of all genes observed to correlate to the specific metabolites were subjected to gene enrichment analysis using DAVID Functional Annotation Tool (Dennis et al. 2003). A gene enrichment analysis evaluates the probability of detecting the observed number of genes within a category by evaluating the proportion of observed genes in each gene list belonging to a category vs. the proportion of genes belonging to that category out of all the genes in the background gene list. The filtered data on gene expression were used as search background in the gene enrichment analysis. Significant groups within the gene-ontology themes “biological process,” “molecular function,” and “cellular compartment” are represented in supplemental Tables S2–S4. In addition, lists of either positively or negatively correlating genes subjected to gene enrichment analysis and the gene-ontology theme “biological process” are listed in supplemental Tables S5–S7. In the gene enrichment analysis, a high level of specificity was chosen for the gene-ontology themes (level 4) and the criteria for significance were (1) more than three genes within each group and (2) an EASE score P-value <0.1. Complete lists of genes correlating to the specific metabolites are in supplemental Tables S8–S10. The hierarchical construction of gene-ontology themes allows genes to be present in several groups (Dennis et al. 2003).
RESULTS
Overall effects of inbreeding and temperature stress:
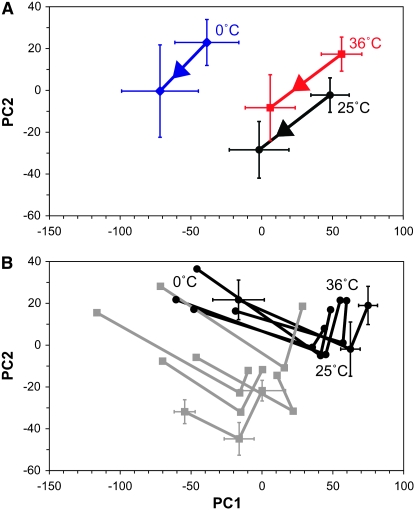
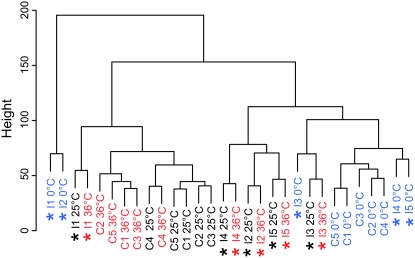
The relative similarities between the metabolomic responses to inbreeding, temperature stress, or the combination of the two were characterized by PCA (Figure 1) and HCA on the full data set (Figure 2). Many of the characteristics described by these analyses are described in more detail in the following sections. The two first principal components, PC1 and PC2, account for 38 and 17% of the total variance within the entire data set. These scores (Figure 1A) show that there are significant effects of inbreeding and temperature stress (P < 0.001, two-way ANOVA) for both PC1 and PC2. The similarity in the response to inbreeding after all temperature treatments indicates that the metabolite differences between inbred and control lines are independent of temperature treatment. Additionally, results presented in Figure 1A indicate that the response to cold stress is the largest compared to the response to heat stress and inbreeding. The between-line variance for the inbred lines was higher than for the control lines at all temperature treatments (P < 0.001, Wilcoxon test; Figure 1B). According to the scores, the metabolite profile of I1 is different from the other inbred lines (P < 0.01, two-way ANOVA) for both PC1 and PC2). The dendogram in Figure 2 shows that most of the control lines at benign temperature and heat stress form separate clusters while the inbred lines at benign temperature and heat stress cluster line specifically, indicating that inbreeding causes the major difference between samples at benign temperatures and after exposure to heat stress. Also here the I1 samples cluster closely to the control samples. All cold-stressed control lines cluster together while the cold-stressed inbred lines are scattered throughout the dendogram, verifying a strong interaction between line and cold stress in the inbred lines (see below).
Figure 1.—
Score plots showing the average scores from PCA on all NMR data where PC1 and PC2 account for 38 and 17% of the total variance. (A) The average scores for control and inbred lines after exposure to benign temperature, cold stress, and heat stress. (B) The average scores after exposure to cold, benign, and heat temperature treatment for individual lines. In A, black, blue, and red lines represent flies treated at benign temperature, cold stress, and heat stress, respectively, and arrows mark the direction of change from control to inbred flies. In B, black and gray lines and symbols represent control and inbred lines, respectively. The lines go from cold (left) to benign (middle) to heat (right) temperature treated and the temperatures for the control are labeled. For both A and B, the error bars correspond to 1 SD. In B, error bars are shown for only one representative control and inbred line.
Figure 2.—
Dendogram showing similarities between lines at benign temperature and after heat and cold stress. Black, blue, and red lines represent lines exposed to benign temperature, cold stress, and heat stress, respectively. Inbred lines are indicated by an asterisk. The length of the vertical axis is a measure of the dissimilarities between clusters/groups of samples.
Effects of inbreeding at benign temperature:
PCA was performed on control and inbred samples at benign temperature (i.e., 25° for 1 hr). Here, PC1 accounted for 34% of the variance in the samples and showed a significant effect of inbreeding (Table 1). None of the other six significant PCs showed effects of inbreeding (Table 1), but most (six) of the PCs showed a significant effect of line when all inbred and control lines were included (Table 1). The PCs showing a significant line effect account for 77% of the total variance. Three and four of the seven significant PCs (accounting for 63 and 70% of the total variance) showed a significant line effect for the control and inbred lines, respectively. For all significant PCs, except PC3, the between-line variance was higher for the inbred lines (P < 0.0001, Wilcoxon tests). Thus, the direct effect of inbreeding was a change in the levels of specific metabolites (see below) and an increased variability in metabolite levels between lines.
TABLE 1.
PCA statistics for control and inbred flies at benign temperatures (25°)
|
R2 (%)
|
Pb (inbreeding): |
Pb (line)
|
||||
|---|---|---|---|---|---|---|
| PC | Scaled dataa | Original data | All | All | Control | Inbred |
| 1 | 29.7 | 33.6 | 2.1 × 10−14c | 2.2 × 10−16c | 1.8 × 10−3c | 1.7 × 10−3c |
| 2 | 8.6 | 7.4 | 1.8 × 10−1 | 2.4 × 10−6c | 5.6 × 10−2 | 6.4 × 10−5c |
| 3 | 8.3 | 11.8 | 6.7 × 10−1 | 5.9 × 10−1 | 8.4 × 10−1 | 1.6 × 10−1 |
| 4 | 6.8 | 27.0 | 4.3 × 10−1 | 2.2 × 10−16c | 3.2 × 10−3c | 4.1 × 10−13c |
| 5 | 5.9 | 2.4 | 1.9 × 10−2 | 2.2 × 10−16c | 5.8 × 10−8c | 3.6 × 10−11c |
| 6 | 4.6 | 3.8 | 6.3 × 10−1 | 1.6 × 10−3c | 1.0 × 10−1 | 9.2 × 10−3 |
| 7 | 3.4 | 3.3 | 3.7 × 10−1 | 6.3 × 10−4c | 1.0 × 10−1 | 6.0 × 10−3 |
Explained variation (R2) and results from ANOVAs are presented. The effect of line was tested for all lines and for control and inbred lines separately. The seven significant PCs are shown.
Scaled data used in PCA.
The P-values shown are not corrected for multiple testing.
Significance (P = 0.01) using ranked Bonferroni correction (n = 7).
No detectable interaction between inbreeding and temperature stress:
To detect potential interactions between inbreeding and temperature on the metabolite profiles, PCAs were performed on inbred and control samples exposed to benign temperature/heat stress and benign temperature/cold stress and each PC was tested for interactions by ANOVA (significance level 0.01). For heat stress, none of the 14 significant PCs interacted with inbreeding (data not shown). Nor were there any interaction effects between heat stress and the individual lines (data not shown). For cold stress, none of the 15 significant PCs showed a significant interaction between cold stress and inbreeding (Table 2). Several types of preprocessing before PCA were tested and the VAST scaling used here showed some of the lowest P-values for tests of interactions between inbreeding and temperature stress. Potential interactions were thus not lost due to the preprocessing of data. Four of the 15 significant PCs, accounting for 40% of the variance, showed a significant interaction between cold stress and line (Table 2). A significant interaction between cold stress and line was seen within both control and inbred lines: One and 3 of the 15 significant PCs (accounting for 2 and 38% of the total variance) showed a significant interaction between line and cold stress in control and inbred lines, respectively (Table 2). To further test for an interaction between inbreeding and temperature stress, OPLS-DA was performed separately between control and inbred flies for the samples treated at benign temperature, heat stress, and cold stress. The regression coefficients for the temperature-stressed flies show a high correlation with the regression coefficients for the control flies (R2 = 0.92 for heat- and cold-stressed flies; data not shown). This confirms that the metabolomic effect of inbreeding is similar at benign and stressful temperatures.
TABLE 2.
PCA statistics for control and inbred flies at benign temperature and after exposure to cold stress
|
R2 (%)
|
Pb (inbreeding × treatment): |
Pb (line × treatment)
|
||||
|---|---|---|---|---|---|---|
| PC | Scaled dataa | Original data | All | All | Control | Inbred |
| 1 | 44.2 | 36.3 | 4.6 × 10−2 | 6.6 × 10−7c | 9.6 × 10−2 | 1.3 × 10−3c |
| 8 | 2.1 | 2.0 | 5.8 × 10−1 | 9.6 × 10−4c | 6.4 × 10−4c | 4.6 × 10−1 |
| 9 | 1.8 | 1.6 | 4.1 × 10−1 | 3.4 × 10−5c | 4.1 × 10−2 | 3.4 × 10−4c |
| 12 | 1.2 | 0.6 | 3.5 × 10−2 | 1.4 × 10−3c | 1.5 × 10−1 | 8.4 × 10−4c |
Explained variation and ANOVA results for the effect of temperature treatment and inbreeding or line are shown. Only PCs showing significant changes in interaction terms are presented.
Scaled data used in PCA.
P-values shown are not corrected for multiple testing.
Significance (P = 0.01) using ranked Bonferroni correction (n = 15).
Identification of metabolites associated with inbreeding:
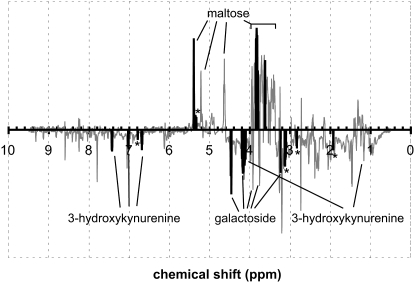
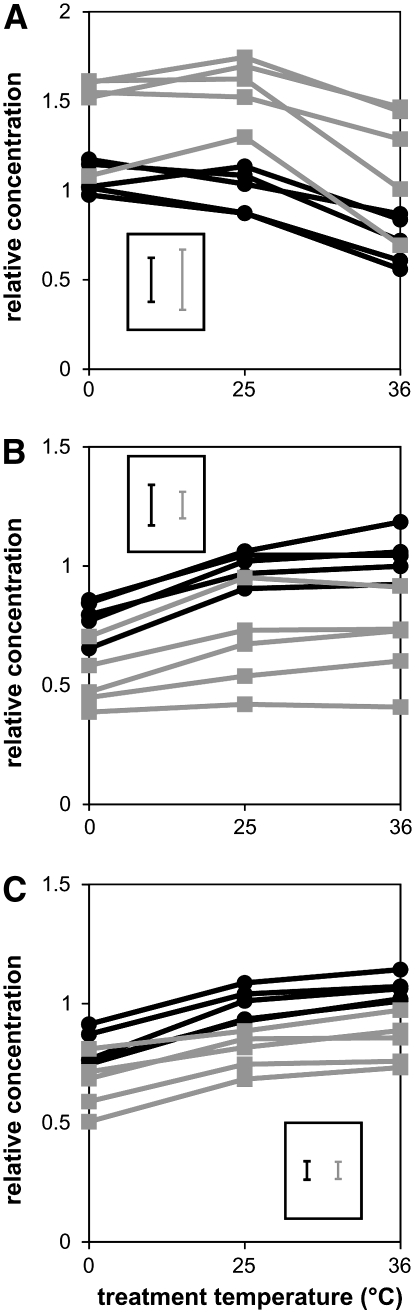
Because the metabolomic effect of inbreeding is similar across temperatures, the most prominent effects of inbreeding were identified using the average OPLS-DA output from the three temperature treatments. The highest correlations (R2 > 0.5) were obtained for increased levels of maltose and decreased levels of 3-hydroxykynurenine and 1-O-(4-O-(2-aminoethyl phosphate)-β-d-galactopyranosyl)-x-glycerol in the inbred lines (Figure 3). The latter is a galactoside synthesized exclusively in the accessory glands in males of some Drosophila species (Chen et al. 1977). Additional unassigned signals with high correlations (R2 > 0.5) were found at 6.79, 5.34, 3.13, 2.84, and 1.92 ppm (Figure 3). Line-specific relative concentrations of maltose, 3-hydroxykynurenine, and the galactoside at benign temperature and after exposure to temperature stress are shown in Figure 4. A significant effect of the breeding regime for concentrations of all three metabolites was observed (P < 0.0001, one-way ANOVA), Figure 4).
Figure 3.—
OPLS-DA regression coefficients between inbred and control samples. Signals above the horizontal line represent metabolites that are present at higher concentrations in the inbred samples, and signals below the horizontal line represent metabolites that are present at lower concentrations in the inbred samples. To minimize noise, the averages from the benign and two temperature-stressed conditions were used. Solid peaks have a predictive OPLS coefficient (R2) >0.5 and therefore are the most important in discriminating between inbred and control samples. Intensities are scaled as I/√|I| where I is the intensity. Signals from maltose, 3-hydroxykynurenine, and the galactoside are labeled. Unassigned signals with R2 >0.5 are marked with an asterisk.
Figure 4.—
Relative concentrations of (A) maltose, (B) 3-hydroxykynurenine, and (C) the galactoside for the different lines at different temperature treatments. Black and gray lines represent control and inbred lines, respectively. The concentration scale was normalized relative to the average concentration for the control lines treated at 25°. Error bars for each data point are not shown, but the average standard deviations for control and inbred lines are indicated.
Identification of metabolites associated with heat and cold stress:
The most prominent effects of temperature stress were identified using the OPLS-DA outputs for control and inbred lines. The outputs from inbred and control lines were generally similar, with the exception that lower correlations were observed for the inbred lines relative to the control lines. The control lines were therefore used for further analysis of metabolites associated with temperature stress. For heat stress, the highest correlations (R2 > 0.5) were obtained for increased levels of tyrosine, glutamate, glutamine, valine, and isoleucine and for decreased levels of several sugars. For cold stress, the highest correlations (R2 > 0.5) were obtained for increased levels of maltose, glucose, and trehalose and for decreased levels of NAD+, β-alanine, fatty acid methyl groups, 3-hydroxykynurenine, and the galactoside. These changes are in agreement with previous results (Malmendal et al. 2006; Overgaard et al. 2007), except identification of the galactoside, which is not expected to be detected in the female flies used in the previous studies.
Correlation of NMR and gene array data:
The concentrations of the three metabolites that show the highest correlation with inbreeding were correlated to gene expression data published in Kristensen et al. (2005a, 2006). Integration of transcriptomic and metabolomic data was performed only for flies exposed to benign temperature. A Venn diagram showing the distribution of the genes correlating with maltose, 3-hydroxykynurenine, and the galactoside is shown in supplemental Figure S3. Within the list of genes correlating to maltose several gene-ontology themes are significantly overrepresented. Among those are processes related to “cellular carbohydrate metabolic process” represented by 13 genes (supplemental Table S2). Other overrepresented processes are “larval feeding behavior,” “cellular respiration,” and “coenzyme metabolic process.” Genes related to the mitochondria and other organelle membranes were also overrepresented within the list of genes correlating to maltose (supplemental Table S2). When only negatively correlated genes were subjected to gene enrichment analysis, significant biological processes are, for example, the “cellular carbohydrate metabolic process,” the “carbohydrate catabolic process,” and the “monosaccharide metabolic process” (supplemental Table S5). The heat-shock protein 60 and two genes related to degradation of misfolded or incomplete synthesized endoplasmic reticulum proteins (Hsp60D, septin-interacting protein 3, Derlin-1) were observed to be positively correlated to variation in maltose (supplemental Table S8).
Genes correlated to 3-hydroxykynurenine are related mainly to metabolic processes involving nitrogen-containing compounds (e.g., the “amine biosynthetic process,” the “nitrogen compound biosynthetic process,” the “nucleobase metabolic process,” and the “nucleotide biosynthetic process”). No significant gene-ontology themes were found within the list of genes positively correlated to 3-hydroxykynurenine. The majority of genes related to processes involving nitrogen-containing compounds are negatively correlated with 3-hydroxykynurenine, indicating that these genes are upregulated in inbred lines. Genes previously shown to be related to processes involving 3-hydroxykynurenine (Rosy, Dorothy; Real and Ferré 1990) are found to correlate to 3-hydroxykynurenine (supplemental Table S9). Four genes correlating to 3-hydroxykynurenine are related to DNA replication and repair mechanisms (CG7769, Okra, DNA-replication-related element factor, structure-specific recognition protein). All four genes are negatively correlated to variation in 3-hydroxykynurenine (supplemental Table S9). Selenide, water dikinase, was found to be negatively correlated to 3-hydroxykynurenine (supplemental Table S9).
The galactoside correlates with genes that are related to amine and nitrogen compound biosynthetic processes. Furthermore, genes involved in meiosis were found to be overrepresented and negatively correlated to variation in the galactoside. Genes related to telomere maintenance and splicing (telomere fusion, splicing factor 1) were also correlated to variation in the galactoside (supplemental Table S10). The gene Takeout, which is known to be associated with male courtship behavior, was positively correlated with the galactoside (supplemental Table S10). The gene Thor, which is associated with diverse processes such as response to oxidative stress, defense response, immune response, and life span, was negatively correlated to the galactoside (supplemental Table S10).
DISCUSSION
The data presented here show that inbreeding strongly affects the metabolome. Both the mean level of specific metabolites and the variability of the metabolite profiles were different in inbred flies relative to flies from the control breeding regime (Figures 1 and 4, Table 1). The three metabolites that showed the highest correlation (R2 > 0.5) with the general effect of inbreeding were maltose, 3-hydroxykynurenine, and 1-O-(4-O-(2-aminoethyl phosphate)-β-d-galactopyranosyl)-x-glycerol, a galactoside specific to Drosophila males.
Maltose:
Maltose was found in higher concentrations in inbred lines (Figure 4A). Maltose is a disaccharide formed by hydrolysis of starch and can be further hydrolyzed to glucose. Different scenarios that result in accumulation of maltose can be suggested. Either hydrolysis of starch is increased or hydrolysis of maltose is decreased. Adult D. melanogaster have been shown to use starch as an energy resource and induction of starch metabolism has been linked to stimulation of growth and fitness (Yamazaki and Matsuo 1984; Fujimoto et al. 1999). A shortage of glucose has been shown to induce genes required for the use of maltose as an energy source (Hu et al. 2000). However, decreased levels of glucose in inbred flies were not observed. Associating metabolomic and gene expression data showed that a group of genes related to carbohydrate metabolic processes was found to be negatively correlated to maltose, indicating an upregulation in response to inbreeding (e.g., glycogen phosphorylase, phosphoglycerate kinase, enolase, trehalose-6-phosphate synthase). However, amylase and maltase, which are known to be involved in the regulation of maltose, were not observed to correlate with maltose in this experiment, indicating that the higher level of maltose observed in inbred lines may be regulated by alternative processes. Genes related to larval feeding behavior and cellular respiration correlated to maltose, indicating that inbreeding may affect energy metabolism and thus affect behavior related to feeding. Knowledge is accumulating that signifies that individuals with increased levels of homozygosity do have an altered metabolism, supporting the results found here (Mitton et al. 1986; Myrand et al. 2002). Genes associated with carbohydrate metabolism were also identified as differentially expressed in inbred and control lines in the gene expression study (Kristensen et al. 2005a, 2006). Six of these genes (Black, CG1140, Lethal(1)G0334, CG10932, NAD-dependent methylene–tetrahydrofolate dehydrogenase, Pugilist) are found to correlate with variation in maltose.
Maltose also plays an important role in relation to coping with temperature stress where its role is to protect the function of proteins and membranes (Kaplan and Guy 2004; Pereira and Hünenberger 2006). Increased maltose levels have been measured in response to rapid cold hardening and cold shock in D. melanogaster (Overgaard et al. 2007), and this was also observed in this study. Accumulation of maltose has also been demonstrated in response to heat and cold exposure due to a stress-induced increase of amylase activity (Kaplan and Guy 2004). Specifically, membrane-associated processes are suggested to benefit from the protective role of maltose (Kaplan and Guy 2004). An overrepresentation of genes correlating with maltose was associated with the mitochondria and other organelle membranes. General membrane and mitochondrial functions may be especially vulnerable to inbreeding, and higher levels of maltose in inbred lines may be required to protect these functions.
Interactions between maltose and molecular chaperones such as the heat-shock protein hsp90 have been observed (Bali et al. 2003), and a maltose-binding protein has been suggested as having chaperone properties, interacting with unfolded and denatured proteins (Richarme and Caldas 1997). The maltose-binding protein is hypothesized to act as a chaperone buffer preventing aggregation of unfolded proteins, suggesting that maltose may be associated with the protein quality control system and protein homeostasis. This indicates that processes associated with protein folding and degradation may be disturbed in inbred individuals. Fixation of deleterious mutant alleles due to inbreeding may cause decreased protein stability, resulting in increased unfolding and non-native conformational changes potentially resulting in protein aggregation, which is known to be harmful for cellular functions (Sangster et al. 2004; Depristo et al. 2005). Increased levels of maltose in inbred flies could help these unstable proteins to preserve their functions. Hsp60, which is involved in mitochondrial “de novo” protein folding and protein targeting to mitochondria, was found to be positively correlated with maltose variation. As a direct interaction between maltose and Hsp60 have not been reported, it is more plausible that a correlation between Hsp60 and maltose is due to both being induced by disturbance of protein homeostasis. The correlation with gene expression data showed that two genes associated with degradation of the endoplasmic reticulum (ER) proteins were positively correlated to maltose, indicating a role for maltose also in processes related to degradation of ER proteins. Overall, our data suggest a role for maltose in processes involved in protection, degradation, and folding of proteins and that these processes are upregulated in inbred lines. Previous results showing higher levels of Hsp70 in inbred populations (Kristensen et al. 2002; Pedersen et al. 2005; Cheng et al. 2006) corroborate that inbreeding may influence protein homeostasis.
3-hydroxykynurenine:
The metabolite 3-hydroxykynurenine was found at lower levels in inbred relative to control flies. Decreased 3-hydroxykynurenine levels in response to cold shock were found previously (Overgaard et al. 2007) and in this study. Genes correlating to 3-hydroxykynurenine were related mainly to processes involving nitrogen-containing compounds. Nitrogen-containing compounds are essential, as they provide components of the nucleic acids, DNA and RNA, and the energy currency of the cell, ATP. Hydroxykynurenine is an intermediate of tryptophan catabolism and this pathway was shown by Kristensen et al. (2006) to be differentially expressed between inbred and control lines at the gene expression level with most genes in the pathway being upregulated in inbred flies (such as CG9629 and glutamate oxaloacetate transaminase 2). Tryptophan catabolism has been suggested to be involved in aging, oxidative stress, and synthesis of brown-eye pigmentation in Drosophila (Okuda et al. 1998; Savvateeva-Popova et al. 2003; Sas et al. 2007). Reactive oxygen species (ROS) are known to be easily formed from oxidation of 3-hydroxykynurenine resulting in oxidative stress (Okuda et al. 1998; Han et al. 2007). ROS can damage important cell structures such as DNA, proteins, and lipids (Okuda et al. 1998; Landis et al. 2004). Preventing accumulation of 3-hydroxykynurenine is essential to minimize ROS production, suggesting that this is an adaptive response in inbred lines for avoiding oxidative stress. Genes associated with DNA repair and oxidative stress were observed to be negatively correlated with 3-hydroxykynurenine, indicating an upregulation of these genes in response to inbreeding. Selenide, water dikinase, which is associated with sensitivity to oxidative stress (Morey et al. 2003) and involved in production of selenoproteins that may act as cellular antioxidants, was also negatively correlated to 3-hydroxykynurenine. The observation that genes related to methyltransferase activity correlate to 3-hydroxykynurenine is interesting because methylation patterns are known to be important for DNA protection and gene regulation. The existence of DNA methylation in D. melanogaster has been debated; however, methylation of DNA is now known to occur in D. melanogaster (Lyko et al. 2000). Overall, our data indicate either that inbred individuals are oxidatively stressed or that the effect of inbreeding bears similarities to responses to oxidative stress (Landis et al. 2004; Kristensen et al. 2005a). However, the mechanistic relation between inbreeding and oxidative stress obviously requires further studies.
The galactoside:
The Drosophila-specific galactoside [1-O-(4-O-(2-aminoethyl phosphate)-β-d-galactopyranosyl)-x-glycerol] detected at lower concentrations in inbred flies is synthesized and believed to accumulate exclusively in the accessory glands in males of some Drosophila species, including D. melanogaster (Chen et al. 1977). It is transferred from male to female flies in the seminal fluid during copulation (Chen et al. 1977; Chen 1984). Variation in the galactoside was found to correlate with genes involved in meiosis and a gene involved in male courtship behavior (e.g., Mei-218, Takeout). Additionally, genes associated with telomere maintenance and splicing correlate with the variation in the galactoside. The gene Thor, which is related to defense response, immune response, oxidative stress, and life span, was negatively correlated to the galactoside, indicating an upregulation in the inbred lines. Thor is thought of as a general stress-responsive gene, as are the majority of heat-shock proteins; this indicates that inbreeding may induce general stress-responsive mechanisms. Knowledge is scarce on the biochemical function of this galactoside. It was originally suggested to be the major agent for elevation of oviposition and repression of sexual receptivity in females following copulation (Chen et al. 1977). Even if it has now been shown that the major role is played by the so-called sex peptide (Chen et al. 1988; Liu and Kubli 2003), lower concentrations of the galactoside and similar agents in inbred flies might have a negative effect on reproduction, which is a trait normally suffering from inbreeding depression. It is of interest in this context that a proteome analysis of the same inbred and control lines revealed lower levels of accessory gland proteins related to male mating success (K. S. Pedersen, M. C. Codrea, E. Bendixen, P. Berg, T. N. Kristensen and V. Loeschcke, unpublished results). The observation of lower levels in inbred lines of agents related to reproductive fitness is extremely interesting in relation to finding candidate genes and identifying processes particularly vulnerable to inbreeding. Interestingly, a decreased level of the galactoside in response to cold stress was also observed in this study.
Common stress responses:
The finding that all three metabolites that showed the highest correlation with inbreeding correlate strongly and in the same direction also with cold stress (maltose concentrations were higher, whereas 3-hydroxykynurenine and the galactoside concentrations were lower in both inbred and cold-stressed flies) suggests that the consequences of the distinct types of stress elicit similar stress responses. It has previously been shown that intrinsic stresses such as inbreeding, aging, and oxidative stress lead to a larger-than-expected overlap of genes being differentially expressed in all three groups (Kristensen et al. 2005a). This study shows that different stresses also may induce common responses at the metabolomic level.
Between-line variability:
Another important general finding of this study is the increased between-line variation in metabolite profiles in inbred relative to control flies. The additive genetic variance between lines is expected to increase with inbreeding (Falconer and Mackay 1996). Assuming other genetic and environmental variance components to be unaffected by inbreeding, it is expected that the phenotypic variance between lines/populations will behave in a similar manner. On the basis of these assumptions the observed increase in between-line variance of the metabolite profiles confirms the expectations and emphasizes the variable nature of inbreeding on this biological level. Within-gene variation of gene expression (Kristensen et al. 2005a, 2006) and genetic variance components of a neutral quantitative morphological character (sternopleural bristles, Kristensen et al. 2005b) have previously been estimated in the same lines as used in this study. Also at these levels inbreeding results in an increased between-line variance. All together, the data support, at several biological levels, that inbreeding results in increased phenotypic variance between lines. However, despite the observed increase in between-line variance, our results show general responses to inbreeding across independent biological replicates at the metabolite and transcriptomic levels (Kristensen et al. 2005a, 2006).
No observed interaction between inbreeding and environmental stress:
In contrast to other studies of inbreeding and its consequences at the phenotypic level, we did not observe strong interactions between inbreeding and the environment (different temperatures) on the metabolome. The metabolite fingerprint of inbreeding was stable across the three temperature conditions investigated. The explanation for the differences in inbreeding-by-environment interactions at different biological levels is not known. However, one reason for the lack of interactions observed here could be that the dominating metabolite responses to inbreeding and temperature stresses occur in different cell or tissue types, which do not show any interaction at the metabolite level, and because our samples are pools of whole flies, we are not able to separate these different metabolomic responses. Another reason for the lack of interactions could also be that genetic drift, purging, and accumulation of mutations have altered the genetic makeup of the investigated lines since the gene expression study was performed. However, the inbred and control lines were investigated for effects of interactions between inbreeding and temperature stress on the life-history trait “egg-to-adult viability” at approximately the same time as our metabolomic study was performed. This study showed clear-cut evidence for inbreeding depression being temperature dependent (Kristensen et al. 2008).
Interactions between line and cold stress were observed, while no significant interaction effects were detected between line and heat stress. Indeed, the metabolomic responses to heat and cold stress are different in composition and timescale (Malmendal et al. 2006; Overgaard et al. 2007). The cold stress applied in this study was more severe than the heat stress: flies were knocked out during cold stress, which was not the case with the heat-stress treatment. Moreover, cold stress has previously been shown to affect metabolite processes more strongly than heat stress (Malmendal et al. 2006; Overgaard et al. 2007). This may explain why significant interactions between line and temperature were observed for exposure to cold stress but not to heat stress.
Conclusion:
Overall, the metabolite fingerprints of inbred and control flies were clearly distinguishable. Maltose, 3-hydroxykynurenine, and a galactoside specific to male Drosophila were the metabolites that showed the highest correlation with the metabolomic effect of inbreeding. Inbred lines had increased levels of maltose and decreased levels of 3-hydroxykynurenine and the galactoside. Integration of information across multi-“omic” technologies has the potential to broaden the limited window provided by individual technologies, thus enhancing the understanding of the biological systems. The approach used in this study to associate metabolite and gene expression levels is suggested to be generally applicable to integrating multivariate data from different “omic” technologies (Rantalainen et al. 2006). In spite of the 40 generations separating the transcriptomic and metabolomic data, the attempts to integrate data from different biological levels revealed several new and interesting advances in understanding consequences of inbreeding. In summary, we speculate that processes related to basic metabolism and protein homeostasis (degradation and folding), and especially processes associated with membrane functions, are distorted in inbred flies and that inbreeding may influence processes also known to be induced in response to oxidative stress. Metabolite profiles were found to vary more between inbred relative to between control lines, confirming other empirical results. Contrary to most other studies investigating inbreeding-by-environment interactions, we found no interaction between inbreeding and temperature on the metabolome. In conclusion, the NMR technique used in this study proved to be efficient in distinguishing between different breeding regimes (inbred and control) and in identifying between-line variation. NMR technologies particularly benefit from the fact that prior information on the genome is not a necessity for obtaining useful results. We advocate that the method may have applications in fields where it is not usually used, such as environmental stress research, population genetics, and evolutionary research.
Acknowledgments
We are grateful to Doth Andersen for laboratory assistance; to Karl-Heinz Ott for the PCA R-script; to Ian Max Møller, Simon Metz Pedersen, and Johannes Overgaard for comments and discussions on the manuscript and on design issues; to Eric Kubli, Mariana Wolfner, and Tracey Chapman for information about the galactoside; and to Marius Cosmin Codrea for summation of gene data. This work was funded by the Danish Natural Research Council with centre and frame grants (V.L.), the Danish Veterinary and Agricultural Research Council (T.N.K.), the Graduate School of Environmental Stress Studies (K.S.P.), the Danish National Research Foundation (A.M. and N.C.N.), and the Danish Biotechnological Instrument Centre (N.C.N.). The spectra at 800 MHz were obtained on the Bruker Avance 800 spectrometer of the Danish Instrument Center for NMR Spectroscopy of Biological Macromolecules.
References
- Armbruster, P., and D. H. Reed, 2005. Inbreeding depression in benign and stressful environments. Heredity 95 235–242. [DOI] [PubMed] [Google Scholar]
- Bali, M., B. Zhang, K. A. Morano and C. A. Michels, 2003. The Hsp90 molecular chaperone complex regulates maltose induction and stability of the Saccharomyces MAL gene transcription activator Mal63p. J. Biol. Chem. 278 47441–47448. [DOI] [PubMed] [Google Scholar]
- Bax, A., and D. G. Davis, 1985. MLEV-1-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65 355–360. [Google Scholar]
- Birchler, J. A., N. C. Riddle, D. L. Auger and R. A. Veitia, 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 21 219–226. [DOI] [PubMed] [Google Scholar]
- Braunschweiler, L., and R. R. Ernst, 1983. Coherence transfer by isotopic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 53 521–528. [Google Scholar]
- Bubliy, O. A., and V. Loeschcke, 2005. Correlated responses to selection for stress resistance and longevity in a laboratory population of Drosophila melanogaster. J. Evol. Biol. 18 789–803. [DOI] [PubMed] [Google Scholar]
- Bylesjö, M., M. Rantalainen, O. Cloarec, J. K. Nicholson, E. Holmes et al., 2006. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 20 341–351. [Google Scholar]
- Charlesworth, B., and D. Charlesworth, 1999. The genetic basis of inbreeding depression. Genet. Res. 74 329–340. [DOI] [PubMed] [Google Scholar]
- Chen, P. S., 1984. The functional morphology and biochemistry of insect male accessory glands and their secretions. Annu. Rev. Entomol. 29 233–255. [Google Scholar]
- Chen, P. S., H. M. Fales, L. Levenbook, E. A. Sokoloski and H. J. Yeh, 1977. Isolation and characterization of a unique galactoside from male Drosophila melanogaster. Biochemistry 16 4080–4085. [DOI] [PubMed] [Google Scholar]
- Chen, P. S., E. Stumm-Zollinger, T. Aigaki, J. Balmer, M. Bienz et al., 1988. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell 54 291–298. [DOI] [PubMed] [Google Scholar]
- Cheng, P. H., X. Liu, G. F. Zhang and Y. W. Deng, 2006. Heat-shock protein70 gene expression in four hatchery Pacific abalone Haliotis discus hannai Ino populations using marker-assisted selection. Aquac. Res. 37 1290–1296. [Google Scholar]
- Cloarec, O., M. E. Dumas, A. Craig, R. H. Barton, J. Trygg et al., 2005. Statistical total correlation spectroscopy: an explorative approach for latent biomarker identification from metabolic H-1 NMR data sets. Anal. Chem. 77 1282–1289. [DOI] [PubMed] [Google Scholar]
- Dennis, G., Jr., B. T. Sherman, D. A. Hosack, J. Yang and W. Gao, 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4 R60. [PubMed] [Google Scholar]
- DePristo, M. A., D. M. Weinreich and D. L. Hartl, 2005. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat. Rev. Genet. 6 678–687. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Longman, New York.
- Fan, W. M. T., 1996. Metabolite profiling by one- and two-dimensional NMR analysis of complex mixtures. Prog. Nucl. Magn. Reson. Spectrosc. 28 161–219. [Google Scholar]
- Fujimoto, J., C. Kanou, Y. Eguchi and Y. Matsou, 1999. Adaptation to a starch environment and regulation of α-amylase in Drosophila. Biochem. Genet. 37 53–62. [DOI] [PubMed] [Google Scholar]
- Han, Q., B. T. Beerntsen and J. Li, 2007. The tryptophan oxidation pathway in mosquitoes with emphasis on xanthurenic acid biosynthesis. J. Insect Physiol. 53 254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z., Y. Yue, H. Jiang, B. Zhang, P. W. Sherwood et al., 2000. Analysis of the mechanism by which glucose inhibits maltose induction of MAL gene expression in Saccharomyces. Genetics 154 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, F., and C. L. Guy, 2004. β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 135 1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keun, H. C., M. D. T. Ebbels, H. Antti, M. E. Bollard, O. Beckonert et al., 2003. Improved analysis of multivariate data by variable stability scaling: application to NMR-based metabolic profiling. Anal. Chim. Acta 490 265–276. [Google Scholar]
- Kristensen, T. N., J. Dahlgaard and V. Loeschcke, 2002. Inbreeding affects Hsp70 expression in two species of Drosophila even at benign temperatures. Evol. Ecol. Res. 4 1209–1216. [Google Scholar]
- Kristensen, T. N., P. Sørensen, M. Kruhøffer, K. S. Pedersen and V. Loeschcke, 2005. a Genome-wide analysis on inbreeding effects on gene expression in Drosophila melanogaster. Genetics 171 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen, T. N., A. C. Sørensen, D. A. Sorensen, K. S. Pedersen and V. Loeschcke, 2005. b A test of quantitative genetic theory using Drosophila: effects of inbreeding and rate of inbreeding on heritabilities and variance components. J. Evol. Biol. 18 453–465. [DOI] [PubMed] [Google Scholar]
- Kristensen, T. N., P. Sørensen, K. S. Pedersen, M. Kruhøffer and V. Loeschcke, 2006. Inbreeding by environmental interactions affect gene expression in Drosophila melanogaster. Genetics 173 1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen, T. N., J. S. F. Barker, K. S. Pedersen and V. Loeschcke, 2008. Extreme temperatures increase the deleterious consequences of inbreeding under laboratory and semi-natural conditions. Proc. Biol. Sci. 275 2055–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis, G. N., D. Abdueva, D. Skvortsov, J. Yang, B. E. Rabin et al., 2004. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 101 7663–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, I. M., 1954. Genetic Homeostasis. Oliver & Boyd, Edinburgh.
- Liu, S., and E. Kubli, 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko, F., B. H. Ramsahoye and R. Jaenisch, 2000. DNA methylation in Drosophila melanogaster. Nature 408 538–540. [DOI] [PubMed] [Google Scholar]
- Malmendal, A., J. Overgaard, J. G. Bundy, J. G. Sørensen, N. C. Nielsen et al., 2006. Metabolomic profiling of heat stress: hardening and recovery of homeostasis in Drosophila. Am. J. Physiol. Regul. Integr. Comp. Physiol. 291 R205–R212. [DOI] [PubMed] [Google Scholar]
- Mitton, J. B., C. Carey and T. D. Kocher, 1986. The relation of enzyme heterozygosity to standard and active oxygen-consumption and body size of tiger salamanders, Ambystoma tigrinum. Physiol. Zool. 59 574–582. [Google Scholar]
- Morey, M., S. Florenci and C. Montserrat, 2003. Halving the selenophosphate synthase gene dose confers hypersensitivity to oxidative stress in Drosophila melanogaster. FEBS Lett. 534 111–114. [DOI] [PubMed] [Google Scholar]
- Myrand, B., R. Tremblay and J. M. Sevgny, 2002. Selection against blue mussel (Mytilus edulis L.) homozygotes under various stressful conditions. J. Hered. 93 238–248. [DOI] [PubMed] [Google Scholar]
- Okuda, S., N. Nishiyama, H. Saito and H. Katsuki, 1998. 3-Hydroxykynurenine, an endogenous oxidative stress generator, causes neuronal cell death with apoptotic features and region selectivity. J. Neurochem. 70 299–307. [DOI] [PubMed] [Google Scholar]
- Overgaard, J., A. Malmendal, J. G. Sørensen, J. G. Bundy, V. Loeschcke et al., 2007. Metabolomic profiling of rapid cold hardening and cold shock in Drosophila melanogaster. J. Insect Physiol. 53 1218–1232. [DOI] [PubMed] [Google Scholar]
- Pedersen, K. S., T. N. Kristensen and V. Loeschcke, 2005. Effects of inbreeding and rate of inbreeding in Drosophila melanogaster: Hsp70 expression and fitness. J. Evol. Biol. 18 756–762. [DOI] [PubMed] [Google Scholar]
- Pereira, C. S., and P. H. Hünenberger, 2006. Interaction of the sugars trehalose, maltose and glucose with a phospholipid bilayer: a comparative molecular dynamics study. J. Phys. Chem. 110 15572–15581. [DOI] [PubMed] [Google Scholar]
- Rantalainen, M., O. Cloarec, O. Beckonert, I. D. Wilson, D. Jackson et al., 2006. Statistically integrated metabonomic-proteomic studies on a human prostate cancer xenograft model in mice. J. Proteome Res. 5 2642–2655. [DOI] [PubMed] [Google Scholar]
- Real, M. D., and J. Ferré, 1990. Biosynthesis of xanthurenic acid 8-β-D-glucoside in Drosophila. J. Biol. Chem. 265 7407–7412. [PubMed] [Google Scholar]
- Richarme, G., and T. D. Caldas, 1997. Chaperone properties of the bacterial periplasmic substrate-binding proteins. J. Biol. Chem. 272 15607–15612. [DOI] [PubMed] [Google Scholar]
- Sangster, T. A., S. Lindquist and C. Queitsch, 2004. Under cover: causes, effects and implications of hsp90 mediated genetic capacitance. BioEssays 26 348–362. [DOI] [PubMed] [Google Scholar]
- Sas, K., H. Robotka, J. Toldi and L. Vécsei, 2007. Mitochondria, metabolic disturbances, oxidative stress and the kynurenine system, with focus on neurodegenerative disorders. J. Neurol. Sci. 257 221–239. [DOI] [PubMed] [Google Scholar]
- Savvateeva-Popova, E. V., A. V. Popov, T. Heinemann and P. Riederer, 2003. Drosophila mutants of the kynurenine pathway as a model for ageing studies. Adv. Exp. Med. Biol. 527 713–722. [DOI] [PubMed] [Google Scholar]
- Trygg, J., and S. Wold, 2002. Orthogonal projections to latent structures (OPLS). J. Chemom. 16 119–128. [Google Scholar]
- Wold, S., 1978. Cross-validatory estimation of the number of components in factor and principal components models. Technometrics 20 397–405. [Google Scholar]
- Yamazaki, T., and Y. Matsuo, 1984. Genetic analysis of natural populations of Drosophila melanogaster in Japan. III. Genetic variability of inducing factors of amylase and fitness. Genetics 108 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]