Abstract
It is generally considered that meiotic recombination rates increase with temperature, decrease with age, and differ between the sexes. We have reexamined the effects of these factors on meiotic recombination in the nematode Caenorhabditis elegans using physical markers that encompass >96% of chromosome III. The only difference in overall crossover frequency between oocytes and male sperm was observed at 16°. In addition, crossover interference (CI) differs between the germ lines, with oocytes displaying higher CI than male sperm. Unexpectedly, our analyses reveal significant changes in crossover distribution in the hermaphrodite oocyte in response to temperature. This feature appears to be a general feature of C. elegans chromosomes as similar changes in response to temperature are seen for the X chromosome. We also find that the distribution of crossovers changes with age in both hermaphrodites and females. Our observations indicate that it is the oocytes from the youngest mothers—and not the oldest—that showed a different pattern of crossovers. Our data enhance the emerging hypothesis that recombination in C. elegans, as in humans, is regulated in large chromosomal domains.
MEIOTIC recombination establishes a physical link between homologs that helps ensure segregation to opposite poles during the first meiotic division. Thus, failure to recombine can lead to chromosome missegregation and aneuploid gametes. Accordingly, crossover formation is tightly regulated to ensure that each chromosome (chr) receives at least one crossover, known as the obligate crossover. In many organisms, including Caenorhabditis elegans, chromosome pairs receive the obligate crossover and very few additional crossovers. When additional exchanges do occur, they tend to be widely distributed and evenly spaced, a phenomenon known as crossover interference (CI).
Meiotic crossovers are induced by programmed double-strand breaks (DSBs) catalyzed by the topoisomerase-like protein, Spo11. DSBs occur nonrandomly along chromosomes and chromatin architecture plays a fundamental role in determining the break sites. (reviewed in de Massy 2003). The physical positions of crossovers are regulated locally, with hotspots and coldspots corresponding to DNAse-sensitive, open chromatin and DNAse-insensitive, closed chromatin, respectively (Robine et al. 2007). Exchanges are thought to occur preferentially in chromatin loops (Blat et al. 2002) away from the chromosome axis that is involved in both synaptonemal complex formation and chromatin cohesion (Glynn et al. 2004). Chromatin loops may also contribute to the crossover landscape as population studies in yeast, mice, and humans indicate that exchanges fall into large, coordinately regulated chromosomal blocks (Baudat and Nicolas 1997; Borde et al. 1999; Gerton et al. 2000; Daly et al. 2001; Gabriel et al. 2002). In addition, telomeres and centromeres establish recombinationally repressed regions (Stern 1926; Lambie and Roeder 1986; Blitzblau et al. 2007). How these crossover domains are established and regulated remains an outstanding question (Dorman et al. 2007; Fukuda et al. 2008).
As in other organisms, the organization of the genes on the C. elegans chromosomes has supported the suggestion that recombination domains exist, although concrete evidence for their existence has been lacking. The central region of each autosome is gene rich and relatively “cold” with close to a fivefold lower frequency of crossovers/kilobase than the chromosome arms. In contrast, the X chromosome has a more uniform distribution of genes and recombination frequencies (Barnes et al. 1995). The gene-dense clusters appear to have an inherent property that makes them refractory to recombination as exchanges were repressed even when these domains were relocated closer to the end of the chromosome (Hillers and Villeneuve 2003).
C. elegans has been thought to demonstrate an extreme example of CI, with each chromosome having just one crossover in almost all meioses (Hodgkin et al. 1979; Hillers and Villeneuve 2003). Hillers and Villeneuve (2003) elegantly showed that end-to-end chromosome fusions comprising nearly half the genome (three chromosomes) still received approximately one crossover per meiosis. Furthermore, genetic analyses suggest that there is only a single pathway for crossover formation in C. elegans, making it an attractive system to understand crossover regulation (Kelly et al. 2000).
Recent work in C. elegans has provided insight into CI. The chromosome fusion experiments described above indicated that CI acts chromosome-wide and depends on a contiguous chromosome axis (Hillers and Villeneuve 2003). This work has been supported by the identification of mutations in axial element components that abrogate interference (Hillers and Villeneuve 2003; Borner et al. 2004; Fung et al. 2004; Nabeshima et al. 2004). Furthermore, mutations in dpy-28, a dosage compensation complex subunit that resembles a condensin subunit, lead to double and triple crossovers implicating chromatin structure as a major determinant of CI (Tsai et al. 2008). Another determinant of CI is prophase progression. Mutations in him-8 prevent pairing of the X chromosome and cause meiotic nuclei to stall the cell cycle during early pachytene, the time at which DSBs are made. This delay results in an increased level of double crossovers (DCOs) and a loss of CI on the (paired) autosomes (Carlton et al. 2006).
In addition to chromatin, chromosome context and CI, recombination rates are also affected by parental age (Stern 1926) and sex (reviewed in Lenormand and Dutheil 2005), as well as temperature (Plough 1917; Lamb 1969; Rose and Baillie 1979; Saleem et al. 2001), radiation (Mavor and Svenson 1923; Muller 1925; Kovalchuk et al. 1998), and other stresses (Schewe et al. 1971; Barth et al. 2000). In most cases, the effects of these factors on recombination have been determined for specific intervals on a chromosome with different chromosome regions responding differently to each stress. How these factors influence CI and exchanges along an entire chromosome are only now beginning to be analyzed in the genetic systems (Barth et al. 2000; Paigen et al. 2008).
In part, such studies have been hampered by the fact that they require analysis of large populations to determine how recombination rates across the population are influenced by different environmental conditions. With the advent of new genotyping technologies for mapping single nucleotide polymorphisms (SNPs), the ability to genotype hundreds of animals at tens (to hundreds) of positions along a chromosome is making it feasible to obtain such population data under varying conditions. Indeed a recent study in mice pinpointed CI as a major factor driving sex-specific differences in recombination rates (Petkov et al. 2007). This is unlikely to be the case in C. elegans where CI is extremely high, although sex differences in CI have been reported (Meneely et al. 2002). Instead, the analyses of sex, temperature, and age effects on recombination in C. elegans have led to the suggestion that domain-specific changes in recombination rates underlie many of the differences (Zetka and Rose 1990).
We have explored this question in more detail by building a detailed recombination map of chr III at three temperatures from both male sperm and hermaphrodite oocytes. In addition, we further explore whether results we obtain for chr III hold true for the X chromosome and whether the effects of temperature can be generalized to other stresses, specifically to aging.
MATERIALS AND METHODS
Genetic crosses:
Strains were grown according to the standard procedures (Brenner 1974). Strains used were: Bristol N2; Hawaiian strain CB4856; EG1285 lin-15(n765) oxIs12 [Punc-47∷GFP; lin-15(+)]X; dpy-18(e364) III; unc-45(e286) dpy-18(e364) III; dpy-18(e364) unc-64(js115) III; and tra-2(q122gf) II. To measure recombination in oocytes: dpy-18 hermaphrodites were crossed with CB4856 males to obtain non-DPY heterozygous N2/Hawaiian hermaphrodites. These F1 progeny were crossed to unc-47∷GFP (X) males and GFP-positive, L4 hermaphrodites were collected, individually plated, grown to starvation, and harvested for genomic DNA according to established protocols (Wicks et al. 2001). The F1 animals were plated individually and clonally expanded prior to isolation of genomic DNA. This expansion step was necessary to obtain the quantities of DNA needed for genotyping multiple SNPs, but does not change the representation of each SNP in the lysate (Wicks et al. 2001).
For recombination rates from male sperm, Bristol N2 hermaphrodites were crossed with Hawaiian males. The heterozygous male offspring were crossed with dpy-18 hermaphrodites and non-DPY L4 cross progeny were grown for genomic DNA as described above. All crosses were done at the temperatures being tested: 16°, 20°, or 23°. For both oocytes and male sperm, the L4 cross progeny collected were from the first 4–4.5 days of egg laying. During this time >95% of all eggs are laid.
To determine whether there is a difference between the genetic and SNP map of chromosome III, we assayed markers that span 96% of the chromosome by crossing unc-45 dpy-18 hermaphrodites to Hawaiian or N2 males, collecting non-Unc non-Dpy cross progeny. These heterozygous cross progeny were selfed and transferred every 2 days until the extinction of egg laying. All progeny were scored for wild-type, Dpy, Unc, and Dpy Unc phenotypes. Total progeny and map size were calculated according to Brenner (1974).
For analysis of recombination rates in oocytes of young and old hermaphrodites, crosses were set up as described above, but the hermaphrodites were moved to new plates 24 hr after the onset of egg laying (as ascertained by visual inspection). These plates became the source of the day 0/1 samples. On day 6, fresh males were provided to increase progeny production (Hughes et al. 2007). Adults were removed after 24 hr and this plate of progeny became the day 6/7 samples. Since very few eggs are laid in both of these time periods, ∼15 crosses with four hermaphrodites and seven males each were set up to enable the collection of sufficient cross progeny. For females, we used tra-2(q122gf) females instead of dpy-18 hermaphrodites in the first set of crosses. Lysates were made from hermaphrodite cross progeny of the second cross: tra-2/Hawaiian females X GFP+ (X) males.
SNP analysis:
Most polymorphisms were analyzed by real-time PCR procedure of Wang et al. (2005) with slight modifications described below. In brief, allele-specific PCR primers were designed with unique 6-mer or 14-mer GC-rich tails to discriminate PCR products on the basis of differences in melting temperature. Primers for real-time PCR were designed using the C. elegans SNP database (http://genome.wustl.edu/genome/celegans/celegans_snp.cgi) based on described specifications (Wang et al. 2005). For a list of all primers used, see supplemental Table 1.
Real-time PCR mixes were as follows: 1 μl of lysate to 14 μl of PCR mix (0.3 μl of each primer (10 mm), 1.5 μl real-time buffer (0.1 m Tris pH 8.0, 0.4 m KCl, 0.25 m MgCl2), 0.075 μl 10 mm dNTPs, 0.3 μl 10× SYBR Green, 0.3 μl Rox reference dye, 2.0 μl 25% dimethyl sulfoxide (DMSO), 0.37 μl 100% glycerol, 5.88 μl H2O and 2.0 μl Stoffel conjugate (1.8 μl 10× Stoffel buffer and 0.2 μl AmpliTaq DNA polymerase, Stoffel fragment). PCR reaction setup was done in 96-well low-profile multiplates and sealed with Microseal “B” adhesive film. Product was initially heat activated at 95° for 12 min and followed by 40 cycles of DNA amplification (20 sec at 95°, 1 min at 60°, and 30 sec at 72°) in a MT Research PTC-225 Peltier thermal cycler. Melting curve analysis was performed from 70° to 95° using the DNA engine Opticon continuous fluorescence detector. For primer sets chr III P20 and chr X P5, dilution buffer for JumpStart Taq antibody, JumpStart Taq antibody, and AmpliTaq DNA polymerase, Stoffel fragment in the ratio of 8:4:1 was incubated at room temperature for 10 min before 2 μl of conjugate were added to the PCR mix. For primer sets chr III P1, chr III P6, and chr III P16, a 5:1 ratio of Hawaiian forward primer to N2 forward primer was used in the reaction. Primers from cosmid W06F12 (Davis et al. 2005) at physical position 13.72 Mb were used in lieu of chr III P20 for the aging analysis.
Statistical analysis:
Because the analysis is based on SNP mapping, all crossover frequencies were calculated using raw data. Chi-square tests were performed to test for significant changes in frequency and position of crossovers between sexes and temperature. Whole chromosomal map units (MU) for each sex and temperature were calculated using the formula MU = (no. of single crossovers (SCOs) + 2(no. of DCOs))/sample size × 100. MU for specific intervals in a chromosome were calculated using the formula MU = no. of COs in interval/no. of COs in chromosome X MU for chromosome.
To calculate interference, we first calculated the coefficient of coincidence (COC) for any two intervals using the formula COC = observed no. of DCOs/expected number of DCOs. Interference (I) was calculated as I = 1 − COC. The expected number of DCOs for any two intervals was calculated as
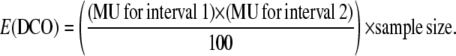 |
Class I has two crossovers that occur in interval 0.22–5.43 Mb and 5.43–13.44 Mb, respectively. Class II has two crossovers that occur in interval 6.64–10.54 Mb and 10.54–13.44 Mb, respectively. Class III has two crossovers that occur in interval 0.22–1.33 Mb and 1.33–3.92 Mb, respectively. The MU used to calculate E(DCO) for all classes and temperatures are summarized in supplemental Table 2.
RESULTS
Chromosome-wide mapping of recombination in C. elegans:
We measured crossover frequencies on chr III from male sperm and hermaphrodite oocytes at three different growth temperatures, 16°, 20°, and 23°. C. elegans grow optimally at 20° (Lewis and Fleming 1995). Fecundity at higher or lower temperatures is decreased, although significant numbers of progeny are attained at 16° and 23°. This contrasts with 13° and 26° at which C. elegans growth and fecundity are significantly impacted (Hirsh et al. 1976). We reasoned that these temperatures would allow us to examine how temperature and sex affect recombination without the confounding effect of severe stress to the organism. Heterozygotes of the two polymorphic wild-type strains were outcrossed to marked (N2) strains to obtain cross progeny. These animals are genotyped to determine whether they have acquired any of the Hawaiian strain polymorphic markers from the heterozygous sperm or oocyte (see materials and methods).
We found that the method of genotyping with Tm-shift primers (Wang et al. 2005) was amenable to large-scale genotyping in 96-well plates and was cheaper than other methods described for C. elegans (Wicks et al. 2001). This method requires PCR amplification with three primers, a common primer for both N2 and Hawaiian and two allele-specific primers, one of which is designed with a 6-mer tag, the other with a 14-mer tag. The PCR products are quantified in a real-time PCR machine, which can discriminate the melting temperatures of the two products and which gives a readout of corresponding peaks. This method can be adapted for use at almost any polymorphism and thus we were able to design primers that extended close to the ends of the chromosome.
Previous studies determined that strain polymorphisms between Bristol (N2) and Hawaiian did not interfere with crossover formation for a small region of the X chromosome (Wicks et al. 2001). We confirmed that this held true for the whole of chr III using genetic markers that span ∼93% of the chromosome (Table 1). Thus, we have confirmed that the differences between the Bristol and Hawaiian strains, which include single nucleotide polymorphisms, insertions, and deletions, do not interfere with recombination between the strains.
TABLE 1.
Recombination frequencies and map size for chr III
| Segregation from unc-45 dpy-18/+ +
|
Segregation from dpy-18 unc-64/+ +
|
|||||
|---|---|---|---|---|---|---|
| Genotype | Recombination frequency | Map size (MU) | χ2 | Recombination frequency | Map size (MU) | χ2 |
| N2/N2 | 3318/9593 | 44.5 | 0.4* | 580/4421 | 14.1 | 0.2* |
| N2/Hawaiian | 2243/6572 | 43.6 | 531/3969 | 14.4 | ||
*P > 0.5.
An overlay of the genetic and physical maps of chr III with the marker positions used in this study is shown in Figure 1A. The physical markers used in this study encompass 96% of chr III, more than any of the previous studies (Zetka and Rose 1990; Meneely et al. 2002; Davis et al. 2005; Hammarlund et al. 2005). Given the repression near the telomeres (see below), the domains that we analyzed actually harbor 99% of all crossovers. We attribute our ability to detect a significant number of DCOs to the comprehensive coverage of the chromosome. Like all the C. elegans autosomes, chr III has a central gene-rich cluster that is recombinationally suppressed. This cluster, which extends from near the pal-1 gene at 4.81 Mb to glp-1 at 9.09 Mb, occupies only 2.61 cM of the genetic map. This is flanked on both sides by ∼4.7 Mb of gene-poor sequence, which has elevated recombination and encompasses ∼24 cM on each side (Wicks et al. 2001). Thus, the rates of recombination on the arms vs. in the cluster differ almost 10-fold (∼5.11 MU/Mb vs. ∼0.54 MU/Mb, respectively). Extending from both arms toward the telomere are more gene dense regions, which are also recombinationally active (Wicks et al. 2001). Since telomeric sequences in other systems are known to repress recombination (Blitzblau et al. 2007), we examined the 60–360 kb from the right end of chr III. This region has 6-fold fewer crossovers than expected (supplemental Table 3), supporting the conclusion that the telomeres establish a domain repressive for crossover formation. Further analysis of the recombination data for the interval 20–500 kb from the IIIL telomere between par-2 and unc-45 (http://www.wormbase.org) revealed that this interval is also recombinationally suppressed. Thus, it appears that telomeric suppression of recombination is conserved in C. elegans. The organization of the autosomes into gene-rich cluster and gene-poor arms is poorly understood.
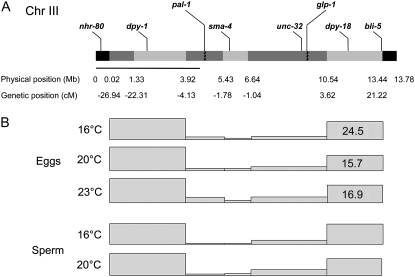
Figure 1.—
Positions of crossovers on chr III differ with sex and temperature. (A) Superimposition of the physical and genetic map of chr III. The locations of the genetic markers across the chromosome are shown above the chromosome. pal-1 and glp-1 mark the ends of the central, gene-rich cluster. The physical markers (Mb) that we used in this study are shown below the chromosome with different colored shading demarcating the regions analyzed. The pairing center is demarcated by the line under the chromosome. (B) Single and double crossover positions have been mapped to five intervals on chr III. The size of the genetic map on the basis of crossover distribution is depicted by shaded squares. The map size for intervals that differ significantly with temperature are written in the respective boxes.
Sex-specific difference in genetic map:
Our data for crossover frequencies on chr III are shown in Table 2. We obtained a map size of ∼54 MU in oocytes and ∼52 MU in sperm at 20°. Our results are in good agreement with previous analyses with regard to the overall map size and interference levels in the hermaphrodite (Zetka and Rose 1990, 1995; Meneely et al. 2002). However, the similarity between the oocyte and male sperm maps at 20° (and 23°) contrasts with previous studies (Zetka and Rose 1990; Meneely et al. 2002) which observed a significantly smaller genetic map in sperm (∼31 MU). The differences between our data and those of Zetka and Rose (1990) can be explained if different autosomes have dramatically different map sizes, as they analyzed chr I and we have analyzed chr III. Differences in the recombination rates between autosomes have been observed in C. elegans, albeit in mutant backgrounds with altered recombination rates (Hodgkin et al. 1979; Carlton et al. 2006; Tsai et al. 2008). The differences between our observations and those of Meneely et al. (2002) can be explained by the size of their data set. A random sampling of even 90 animals can falsely lead to an inaccurate map if that set of animals were predominantly nonrecombinant (smaller map) or recombinant (expanded map). We observed such skewing during the course of data collection. Alternatively, the difference between this study and others may be explained by position and coverage of the markers used.
TABLE 2.
Effects of temperature on crossover frequencies in eggs and sperm on chr III
| Noncrossover
|
Single crossover
|
Double crossover
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | Total | Map size (MU) | ||
| Eggs | 16° | 215 | 44.0 | 272 | 55.6 | 2 | 0.4 | 489 | 56.4 |
| 20° | 297 | 47.1 | 325 | 51.6 | 8 | 1.3 | 630 | 54.1 | |
| 23° | 121 | 46.5 | 131 | 50.4 | 8 | 3.1 | 260 | 56.5 | |
| Sperm | 16° | 286 | 54.9 | 232 | 44.5 | 3 | 0.6 | 521 | 45.7 |
| 20° | 91 | 50.6 | 85 | 47.2 | 4 | 2.2 | 180 | 51.7 | |
| 23° | 223 | 49.0 | 225 | 49.5 | 7 | 1.5 | 455 | 52.5 | |
We observed an overall difference in the genetic map size of chr III between sperm developed at 16° and 23° [χ2(1, N = 462) = 8.41, P < 0.005] and between eggs and sperm developed at 16° [χ2(1, N = 524) = 24.14, P < 0.005].
Previous studies of crossover frequencies in oocyte and male sperm observed more recombination as temperatures were raised. We wanted to determine whether the temperature-dependent changes were due to an effect on the overall frequency of crossing over, which is reflected in the size of the genetic map. Therefore, we compared the genetic map of chr III from oocytes and sperm at 16°, 20°, and 23° (Table 2). The sperm developed at 16° had the smallest map size (45.7 MU). This differed significantly from sperm at 20° and 23° (51.7 and 52.5), indicating that sperm respond to higher temperatures by increasing the total number of crossovers per chromosome.
The temperature-dependent changes in map size observed in male sperm are not seen in oocytes. Rather, it appears that the genetic map is stable in oocytes, 56.4 MU at 16° compared to 56.5 MU at 23°. At the higher temperatures, this map size is not statistically different from that in sperm, suggesting the two germ lines may regulate recombination frequencies in the same manner at these temperatures. The differences between the genetic maps of oocytes and sperm at 16°, however, suggests that at the lower temperature the mechanism for regulating recombination frequencies is different between the germ lines. Thus, the change in the genetic map in sperm and not in oocytes suggests that recombination rates in the two germ lines respond differentially to changes in temperature.
Temperature globally affects recombination in sperm:
The observed temperature-dependent change in the map size of male sperm (45.7 MU at 16° to 52.5 MU at 23°) could be due to a chromosome-wide effect on crossover formation, to the local activation of a recombination hotspot at higher temperatures, or to the formation of a coldspot at lower temperatures. The latter two models predict that the changes in recombination frequency could be mapped to a single chromosomal domain whereas the former predicts that the temperature-dependent changes would be shared across the chromosome.
To distinguish between these alternative models, we assayed whether crossover position changes in response to temperature by mapping the positions of all single and double crossovers from our SNP analyses (Figure 1, Table 3). As described previously (Barnes et al. 1995), we also observed that the middle of chr III is repressed for recombination and the terminal ∼4 Mb of each chromosome share ∼90% of all crossovers at 20°. Comparison of five different intervals showed no statistically significant change in crossover distribution between sperm at all three temperatures. However, we noted that some intervals could account for the increase in map size more than other intervals. For example, the interval between 0.02 and 3.92 Mb shows a difference of ∼5 MU (P < 0.07) between 16° and 23°. A larger sample size could resolve whether the bias in this region is biologically relevant. Nevertheless, since the overall pattern of recombination is the same in sperm at all temperatures, we suggest that the increased map size at 20° and 23° is due to a global effect on the chromosome.
TABLE 3.
Crossover distribution on chr III
| Interval (Mb)
|
||||||
|---|---|---|---|---|---|---|
| 0.02–3.92 | 3.92–5.43 | 5.43–6.64 | 6.64–10.54 | 10.54–13.44 | ||
| Eggs | 16° | 29.4 (144) | 0.8 (4) | 0.2 (1) | 1.4 (7) | 24.5 (120) |
| 20° | 34.9 (220) | 0.6 (4) | 0.5 (3) | 2.4 (15) | 15.7 (99) | |
| 23° | 34.6 (90) | 1.9 (5) | 0.4 (1) | 2.7 (7) | 16.9 (44) | |
| Sperm | 16° | 22.1 (115) | 0.2 (1) | 0.2 (1) | 2.5 (13) | 20.7 (108) |
| 20° | 22.2 (40) | 0.6 (1) | 0.6 (1) | 1.1 (2) | 27.2 (49) | |
| 23° | 27.2 (124) | 0.2 (1) | 0.4 (2) | 4.0 (18) | 20.6 (94) | |
Numbers shown are MU (no. of crossover in interval). No statistical differences between sperm at any temperature are observed except for eggs developed at 16° and 20° [χ2(5, N = 276) = 29.55, P < 0.005] and for eggs developed at 16° and 23° [χ2(5, N = 147) = 16.34, P < 0.01].
Crossover distribution changes with temperature in oocytes:
Although we observed no temperature-dependent change in the map size in oocytes, previous work had suggested that recombination frequencies in some chromosomal intervals increased with temperature (Rose and Baillie 1979). We reasoned that chromosomal domains may be differentially sensitive to changes in temperature so that a loss of crossovers in one domain would appear to be compensated for by an increase in other domains. Thus, we examined crossover positions from oocytes reared at 16°, 20°, and 23°. In contrast to sperm, oocytes display dramatic changes in crossover distribution in response to temperature (Figure 1, Table 3). Crossovers are nearly equally distributed on the right and left ends of the chromosome in oocytes developed at 16°, with a slight bias toward the right end (8.4 MU/Mb on IIIR vs. 7.9 MU/Mb on IIIL). In contrast, at 20° and 23°, almost two-thirds of all crossovers on chr III occur near the left end of the chromosome. These results suggest that in C. elegans oocytes chromosomal domains may be differentially sensitive to changes in temperature.
The temperature-dependent differences in crossover distribution could be explained by the activation of coldspots or hotspots (depending on the reference temperature). To determine whether a coldspot/ cold domain is activated on the right end of the chromosome at temperatures >16° (interval 10.54–13.44 Mb), we further analyzed the crossover distributions in each of the four other intervals on chr III. Since the map size of the chromosome does not change with temperature, the crossovers that are lost by activation of a coldspot should be redistributed proportionally to the remainder of the chromosome. In effect, the rest of the chromosome would become “hotter” with each interval receiving a proportionate increase in the crossovers relative to the map at the lower temperature. When we analyzed the oocyte 20° and 23° data in this manner, they revealed that crossovers are redistributed across the chromosome with values near to proportional (see supplemental Table 4). Therefore, it seems likely that the domain at the right end of chr III is repressed for crossover formation at higher temperatures.
Changes in crossover distribution are a general feature of C.elegans chromosomes in oocytes:
The loss of crossovers on the right end of chr III and redistribution toward the left end is striking because the pairing center (PC) localizes to the left end of the chromosome (see Figure 1). The PC is a specialized chromosomal domain required to both establish and maintain homolog alignment. Synapsis starts at the PC and proceeds processively down the chromosome. One tantalizing possibility for the shift toward the pairing center is that crossovers adjacent to the PC could stabilize homolog interactions that would be more labile at higher temperatures. To determine whether temperature-dependent changes in crossover distribution are a general feature of all chromosomes and influenced by the PC, we extended our studies to the X chromosome. The sex chromosome has several features that distinguish it from autosomes, including differences in transcriptional states in the germ line, recombination patterns, chromatin organization, and gene distribution (Goldstein 1982; Zetka and Rose 1990; Garvin et al. 1998; Reinke et al. 2000; Kelly et al. 2002). Figure 2A shows the genetic and physical maps with the marker positions used in the study. The PC is on the left end of the X chromosome. Our analysis of X chromosome recombination focused on oocytes developed at 16° and 20° since the changes we observed in recombination distribution on chr III occurred only in oocytes between these two temperatures (Figure 1B).
Figure 2.—
Positions of crossovers on the X chromosome differ with temperature. (A) Superimposition of the physical and genetic map of the X chromosome. The locations of the genetic markers across the chromosome are shown above the chromosome. The physical markers (Mb) that we used in this study are shown below the chromosome with different colored shading demarcating the regions analyzed. The pairing center is demarcated by the line under the chromosome. (B) Single and double crossover positions have been mapped to six intervals on X for oocytes at 16° and 20°. The relative map size for each interval on the basis of crossover distribution is depicted by shaded squares. The map size for intervals that differ with temperature are written in the respective boxes.
Similar to previous observations, crossovers on the X chromosome are more uniformly distributed across the chromosome (Barnes et al. 1995; Figure 2B, Table 5), reflecting the inherent differences in the genetic map between the X and autosomes (Brenner 1974). As with chr III, the map size of the X chromosome in oocytes remains the same at different temperatures, ∼51 MU (Table 4). Also similar to chr III, X chromosome crossover distribution changes with temperature. We observed a cold region from 2.34–4.13 Mb (Figure 2B, Table 5). However, the increases in crossovers on the rest of the chromosome to compensate for this cold domain were not proportional as on chr III. Instead there appears to be a bias toward the 6.88- to 10.64-Mb interval, which is further away from the X chromosome PC (Figure 2, supplemental Table 5). Although we cannot rule out that the X chromosome and autosomes might regulate crossovers differently, this result suggests that it is unlikely that the PC regulates the temperature-dependent changes in crossover distribution. Furthermore, changes in crossover distribution in oocytes are a shared feature of multiple chromosomes and the underlying mechanism appears to operate on large chromosomal domains.
TABLE 5.
Crossover distribution on the X chromosome
| Interval (Mb)
|
|||||||
|---|---|---|---|---|---|---|---|
| 0.06–2.34 | 2.34–4.13 | 4.13–6.88 | 6.88–10.64 | 10.64–14.26 | 14.26–17.22 | ||
| Eggs | 16° | 8.1 (22) | 16.1 (44) | 6.6 (18) | 2.2 (6) | 10.6 (29) | 9.5 (26) |
| 20° | 7.3 (27) | 9.5 (35) | 6.5 (24) | 7.6 (28) | 9.8 (36) | 10.3 (38) | |
Numbers shown are MU (no. of crossovers in interval). The change in crossover positions with temperature is statistically significant: χ2(6, N = 141) = 23.35, P < 0.005.
TABLE 4.
The effects of temperature on crossovers on the X chromosome in eggs
| Noncrossover
|
Single crossover
|
Double crossover
|
Map size (MU) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | (%) | No. | % | No. | % | Total | |||
| Eggs | 16° | 130 | 47.6 | 141 | 51.7 | 2 | 0.7 | 273 | 52.4 |
| 20° | 180 | 48.9 | 188 | 51.1 | 0 | 0.0 | 368 | 51.1 | |
No difference between samples: χ2(1, N = 275) = 0.31, P > 0.5.
Regulation of double crossovers differs between the sexes:
Domain specific regulation of crossovers has been proposed to explain recombination maps in multiple systems (International Hapmap Consortium 2007; Coop et al. 2008; Fukuda et al. 2008). One of the governing principles for crossover regulation in most systems is CI. Previous studies have suggested that CI is almost 100% in C. elegans (Hodgkin et al. 1979; Hillers and Villeneuve 2003), meaning that one and only one crossover occurs on each chromosome. When multiple crossovers are seen, they follow the “rules” of CI in mapping to distant chromosomal domains (Hillers 2004). Since we were able to screen >96% of chr III, we observed an unprecedented number of DCOs in both oocyte and sperm. As shown in Table 2, we observed as many as 3.1% of all crossovers manifest as DCOs on chr III in oocytes when hermaphrodites were raised at 23° and almost 1.5% of crossovers as DCOs in sperm. We note that due to the rare occurrence of these events within such a large sample size that we cannot conclude whether the number of DCOs achieved at any temperature between or within germ lines is statistically significant. Nevertheless, the appearance of DCOs in our assays allowed us to ask whether CI is regulated in the same fashion in the two germ lines.
CI can be reflected in two components: the number of crossovers per chromosome and the position of crossovers (the crossover landscape). When we examined crossover position along the chromosome, we observed that DCOs in sperm can be more closely spaced than in oocytes (Figure 3, Fisher's exact test, P < 0.02). At least 4 of the 14 mapped DCOs in sperm occurred in adjacent intervals (classes II and III, as summarized in Figure 3), whereas no such DCOs were observed in oocytes (Figure 3). Instead, in oocytes, all DCOs fell into class I, with one crossover mapping to each half of the chromosome. Thus, values for interference along the chromosome are lower in sperm (Table 6). These results reinforce that recombination is regulated differentially in oocytes and sperm and suggests that the mechanisms that control crossover position along the chromosome can be regulated independently of the mechanisms that determine crossover number.
Figure 3.—
Positions of double crossovers on chromosome III. The physical markers analyzed are shown at the top. The patterns of observed DCOs are indicated by the altered shadowing in each graphic. The percentage (and number) of DCOs with each pattern in oocyte and male sperm are shown at right. Classes of crossovers are used to calculate interference as described in the materials and methods.
TABLE 6.
Interference between sexes and temperatures
| Class Ia
|
Class IIa
|
Class IIIa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Expected DCO | Observed DCO | Interference | Expected DCO | Observed DCO | Interference | Expected DCO | Observed DCO | Interference | ||
| Eggs | 16° | 39 | 2 | 0.9 | 2 | 0 | 1.0 | — | — | — |
| 20° | 42 | 8 | 0.8 | 2 | 0 | 1.0 | — | — | — | |
| 23° | 19 | 8 | 0.6 | 1 | 0 | 1.0 | — | — | — | |
| Sperm | 16° | 27 | 1 | 1.0 | 3 | 1 | 0.6 | 5 | 1 | 0.8 |
| 20° | 12 | 4 | 0.7 | 1 | 0 | 1.0 | 1 | 0 | 1.0 | |
| 23° | 31 | 5 | 0.8 | 4 | 1 | 0.7 | 5 | 1 | 0.8 | |
Refer to Figure 3 and materials and methods for descriptions of classes.
Almost all instances of DCOs are rare, with different chromosomes showing different levels of CI. In our experiments, chr III is more permissive to DCOs than the X (compare Table 2 to Table 4), which is in line with previous studies (Brenner 1974; Hodgkin et al. 1979; Zetka and Rose 1990; Meneely et al. 2002). Further, recent analysis of chr IV suggests that ∼10% of all recombinants had DCOs (J. Henzel and K. Hillers, unpublished data). It is intriguing to speculate that the differences between the X and the autosomes may reflect the inherent differences in the chromatin organization of these chromosomes. In the germ line, the X is maintained in a predominantly silent, heterochromatic state (Kelly and Fire 1998; Kelly et al. 2002; Bender et al. 2004, 2006) whereas the autosomes are transcriptionally active. Although there is no known functional link between the transcriptional state and recombination in C. elegans as in other organisms (Nicolas 1998; Coop et al. 2008), it may be that the chromatin status regulates both transcriptional activity levels and crossover levels.
The effect of age on oocyte recombination:
The observations described above suggest that crossover distribution is influenced by growth environment. Growth at high or low temperatures is thought to exert stress upon the organism, which responds not only by altering patterns of recombination (shown here, and (Rose and Baillie 1979) but also by affecting the aging process and the ability to respond to other stresses (Klass 1977; Lithgow et al. 1994). The links between the stress and aging pathways led us to reexamine whether the effects of age on recombination parallel the effects of temperature. Previous experiments analyzing chr I in hermaphrodites suggested that recombination rates decline dramatically with age in C. elegans (Rose and Baillie 1979). However, changes either in the total number of crossovers (the genetic map size) or in the position of the crossovers could have led to the observed decrease in crossing over in the intervals tested.
Since SNP analysis allows us to simultaneously address crossover frequency and position for a given chromosome, we were able to determine whether the number or the pattern of recombination is changed in young vs. old mothers. C. elegans are fertile for almost 9 days under ideal growth conditions and good mating (Hughes et al. 2007). During the first 24 hr of egg laying (days 0/1) at 20°, ∼2% of eggs are laid; during days 6/7, <1% of eggs are laid (J. Lim and J. Yanowitz, unpublished data). We thus compared day 0/1 to day 6/7 at the onset and decline of fertility. As shown in Table 7, the map size in young and old oocytes is statistically indistinguishable, 50.8 vs. 48.2 MU in hermaphrodites. Thus, at least on C. elegans chr III, age-related changes in nondisjunction are not due to the inability to establish crossovers.
TABLE 7.
Effects of age on crossover frequency on C. elegans chr III
| Noncrossover
|
Single crossover
|
Double crossover
|
Map size (MU) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | Total | |||
| Hermaphrodite | Day 0/1 | 230 | 49.5 | 234 | 50.3 | 1 | 0.2 | 465 | 50.8 |
| Day 6/7 | 193 | 52.3 | 174 | 47.2 | 2 | 0.5 | 369 | 48.2 | |
| Female | Day 0/1 | 146 | 52.3 | 133 | 47.7 | 0 | 0.0 | 279 | 47.7 |
| Day 6/7 | 133 | 48.4 | 136 | 49.5 | 6 | 2.2 | 275 | 53.8 | |
When crossover positions from young and old mothers were analyzed, a dramatic change in crossover position was observed. The oocytes from young mothers showed a pattern of recombination similar to that described above for sperm at 20° with an equal distribution of crossovers to the left and right ends of chr III (Figure 4, Table 8). In the oocytes from old mothers, the pattern appeared more representative of the hermaphrodite pattern of crossovers at 20° (described above, Figure 1) with over two-thirds of crossovers occurring on the left end of the chromosome (Figure 4, Table 8). This altered crossover distribution with age may reflect an inherent change in the regulation of recombination in aging oocytes. For example, age-dependent changes in chromatin organization could affect accessibility or activity of the Spo11 cleavage complex. Alternatively, the change in crossover distribution may be due to the switch from the sperm mode to the oocyte mode of development in the hermaphrodite germ line early in development.
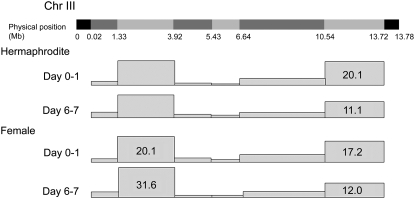
Figure 4.—
Effects of aging on recombination in oocytes. Physical markers on chromosome III are shown at the top. (Bottom) A pictograph of the map size for each interval for oocytes from the first 24 hr of egg laying (day 0/1) to near the final days of egg laying (day 6/7).
TABLE 8.
Crossover distribution in aging worms on chr III
| Chr III interval (Mb)
|
|||||||
|---|---|---|---|---|---|---|---|
| 0.02–1.33 | 1.33–3.92 | 3.92–5.43 | 5.43–6.64 | 6.64–10.54 | 10.54–13.72 | ||
| Hermaphrodite | Day 0/1 | 4.5 (21) | 21.4 (100) | 0.9 (4) | 0.2 (1) | 3.4 (16) | 20.1 (94) |
| Day 6/7 | 6.9 (26) | 24.6 (93) | 1.9 (7) | 0.5 (2) | 2.1 (8) | 11.1 (42) | |
| Female | Day 0/1 | 3.9 (11) | 20.1 (56) | 1.4 (4) | 0.4 (1) | 4.7 (13) | 17.2 (48) |
| Day 6/7 | 5.8 (16) | 31.6 (87) | 0.7 (2) | 0.7 (2) | 2.9 (8) | 12.0 (33) | |
Numbers shown are MU (no. of crossover in interval). The change in crossover positions with age is statistically significant for hermaphrodite eggs [χ2(6, N = 178) = 30.83, P < 0.005] and female eggs [χ2(6, N = 133) = 26.19, P < 0.005].
To distinguish between these possibilities, we reexamined the aging phenomenon in C. elegans females—XX hermaphrodites whose germ line has been feminized by a dominant gain-of-function allele of tra-2. No statistical differences in map size or crossover distribution were seen between hermaphrodites and female oocytes at either age (Tables 7 and 8, Figure 4). Thus, the change in crossover distribution between young and old oocytes suggests that the first oocytes laid fundamentally differ from older oocytes.
DISCUSSION
We have used a set of SNPs spanning >96% of C. elegans chr III to study how temperature, sex, and age affect recombination rates. We show that the germ lines respond to temperature differently. In male sperm, temperature alters the ratio between noncrossover and crossover chromosomes; whereas in oocytes, the position of crossovers changes with temperature. Further, the two germ lines show differential crossover interference. Male sperm have DCOs that are spaced more closely, often in adjacent intervals. These results suggest that different pathways regulate how each germ line affects crossover frequencies.
Differences in recombination rates between sexes has been reported in many systems, but the extent and direction of the difference are species specific (Lenormand and Dutheil 2005). Recently, genomic mapping of humans and mice has led to the conclusion that differences between males and females occur at all genomic scales (International Hapmap Consortium 2007; Coop et al. 2008). Globally, differences in chromatin loop organization have been suggested to explain differences in CI between the sexes (Petkov et al. 2007). More locally, differences in hotspot usage can explain variation between individuals of both sexes (Coop et al. 2008). In C. elegans, we observe differences in CI between the sexes, suggesting that CI may be a linchpin for sexual dimorphism in recombination.
We also observed that the sexes respond differently to varying environmental conditions. Although temperature and stress have long been known to affect recombination rates (Plough 1917; Mavor and Svenson 1923), this is the first study to directly compare chromosome-wide recombination rates for both sexes under different growth conditions. We observed that the size of the genetic map changes in male sperm in response to temperature. The increased frequency of crossovers at higher temperatures is not due to the activity of a recombination hotspot, as the distribution of crossovers across the chromosome was strikingly similar at all temperatures analyzed. Rather, one possible explanation is that the complexes that exert a choice between the crossover and noncrossover pathways may be regulated differently in each germ line by temperature. For example, the components required for the crossover pathway may be more highly expressed in sperm at higher temperatures, thereby increasing the number of these complexes on chromosomes. Alternatively, this complex may be more stable or more active at higher temperatures. However, since we know little about how the noncrossover/crossover decision is made, particularly in C. elegans, distinguishing between these possibilities will need to await further characterization of these pathways.
One of the most striking observations that we made is that crossover positions change dramatically in oocytes in response to temperature and age (Figures 2, 3, and 4). Since earlier studies used genetic markers to map recombination in specific intervals, they would not have been able to detect a change in the pattern of crossing over along the chromosome (Rose and Baillie 1979; Zetka and Rose 1990). Consequently, region-specific differences would have been seen as increases or decreases in crossover frequencies. Thus, we believe that the apparent differences between our data and others can be resolved by invoking a global mechanism for chromosome-wide changes in crossover distribution in response to temperature, age, and sex.
The regulation of crossovers in large chromosomal domains is shared across many organisms, including humans (International Hapmap Consortium; Coop et al. 2008), suggesting that the mechanisms that establish these domains may be conserved. We envision that domain-specific regulation of crossovers is mediated by large-scale chromatin packaging mechanisms. Recent work from the Meyer lab (Tsai et al. 2008) has suggested that components of the C. elegans dosage compensation complex, which resemble the condensin complex, play important roles in regulating crossover interference and crossover position. In the dpy-28 mutant, they observed an increase in crossovers in the same domain where we observed a temperature-dependent increase in crossovers on the X chromosome (Figure 2). It will be interesting to determine whether we could observe differences in condensin complex localization at different temperatures. Recent work in Saccharomyces cerevisiae identified condensin binding sites every 10–50 kb—which is two orders of magnitude more frequent than crossovers (Wang and Strunnikov 2008). Thus, additional levels of regulation would need to be imposed on condensin (or any of the known chromatin complexes) to regulate a chromosome-wide phenomenon at megabase distances.
At the center of an emerging picture for higher order chromosome organization are the nuclear envelope and lamina. During meiosis, attachment of telomeres or PCs to the nuclear periphery facilitates pairing (Harper et al. 2004; Scherthan 2007) and their release promotes proper meiotic progression (Chikashige et al. 2006). The nuclear lamina has been shown to interact with numerous transcription and replication factors, chromatin-associated proteins, and RNA processing proteins and accordingly is thought to play a central role in organizing hetero- vs. euchromatin (reviewed in (Sexton et al. 2007; Wagner and Krohne 2007). Recently, the association between the nuclear envelope and DNA silencers in Drosophila (Dorman et al. 2007) suggested that chromatin loops may be organized into larger domains by association with the nuclear periphery. We find it tempting to speculate that such foci at the nuclear periphery could establish crossover domains.
In humans, it has been suggested that the more distal crossovers are responsible for the increase in nondisjunction rates as women age (Hassold et al. 2000). Our data suggest in C. elegans, crossover position does not contribute significantly to the increased nondisjunction frequencies. Rather, we suggest that other molecular pathways play a more fundamental role in age-related meiotic dysfunction. These may include the integrity of the spindle checkpoint (Lacefield and Murray 2007), or the strength of cohesion (Hodges et al. 2005), or as yet unknown pathways that act on the homologs at late meiotic stages.
Our data show that the major difference in crossover regulation in C. elegans oocytes is seen between the youngest mothers (first day of egg laying) and the oldest mothers. Recent observations have shown that these first oocytes transit more rapidly through prophase I than older oocytes (Jaramillo-Lambert et al. 2007). Together, these data suggest that C. elegans young adults may represent a juvenile period of development that was heretofore unappreciated. It will be interesting to determine whether the changes in cell cycle progression are responsible for the differences in crossover distribution.
Acknowledgments
The authors wish to thank Margaret Hoang for extensive reading and discussion of the manuscript. We also thank Joe Gall, Bruce Vogel, and Cynthia Wagner for poignant and thoughtful commentary. This work was supported by the Carnegie Institution of Washington and National Institutes of Health grant 1K01AG031296-01 to J.L.Y.
References
- Barnes, T. M., Y. Kohara, A. Coulson and S. Hekimi, 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, S., A. E. Melchinger, B. Devezi-Savula and T. Lubberstedt, 2000. A high-throughput system for genome-wide measurement of genetic recombination in Arabidopsis thaliana based on transgenic markers. Funct. Integr. Genomics 1 200–206. [DOI] [PubMed] [Google Scholar]
- Baudat, F., and A. Nicolas, 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA 94 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, L. B., R. Cao, Y. Zhang and S. Strome, 2004. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr. Biol. 14 1639–1643. [DOI] [PubMed] [Google Scholar]
- Bender, L. B., J. Suh, C. R. Carroll, Y. Fong, I. M. Fingerman et al., 2006. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133 3907–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat, Y., R. U. Protacio, N. Hunter and N. Kleckner, 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111 791–802. [DOI] [PubMed] [Google Scholar]
- Blitzblau, H. G., G. W. Bell, J. Rodriguez, S. P. Bell and A. Hochwagen, 2007. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr. Biol. 17 2003–2012. [DOI] [PubMed] [Google Scholar]
- Borde, V., T. C. Wu and M. Lichten, 1999. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol. 19 4832–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G. V., N. Kleckner and N. Hunter, 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 29–45. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton, P. M., A. P. Farruggio and A. F. Dernburg, 2006. A link between meiotic prophase progression and crossover control. PLoS Genet. 2 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige, Y., C. Tsutsumi, M. Yamane, K. Okamasa, T. Haraguchi et al., 2006. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125 59–69. [DOI] [PubMed] [Google Scholar]
- Coop, G., X. Wen, C. Ober, J. K. Pritchard and M. Przeworski, 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319 1395–1398. [DOI] [PubMed] [Google Scholar]
- Daly, M. J., J. D. Rioux, S. F. Schaffner, T. J. Hudson and E. S. Lander, 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29 229–232. [DOI] [PubMed] [Google Scholar]
- Davis, M. W., M. Hammarlund, T. Harrach, P. Hullett, S. Olsen et al., 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy, B., 2003. Distribution of meiotic recombination sites. Trends Genet. 19 14–22. [DOI] [PubMed] [Google Scholar]
- Dorman, E. R., A. M. Bushey and V. G. Corces, 2007. The role of insulator elements in large-scale chromatin structure in interphase. Semin. Cell Dev. Biol. 18 682–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda, T., K. Kugou, H. Sasanuma, T. Shibata and K. Ohta, 2008. Targeted induction of meiotic double-strand breaks reveals chromosomal domain-dependent regulation of Spo11 and interactions among potential sites of meiotic recombination. Nucleic Acids Res. 36 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, J. C., B. Rockmill, M. Odell and G. S. Roeder, 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116 795–802. [DOI] [PubMed] [Google Scholar]
- Gabriel, S. B., S. F. Schaffner, H. Nguyen, J. M. Moore, J. Roy et al., 2002. The structure of haplotype blocks in the human genome. Science 296 2225–2229. [DOI] [PubMed] [Google Scholar]
- Garvin, C., R. Holdeman and S. Strome, 1998. The phenotype of mes-2, mes-3, mes-4, and mes-6, maternal-effect genes required for survival of the germline in Caenorhabditis elegans, is sensitive to chromosome dosage. Genetics 148 167–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn, E. F., P. C. Megee, H. G. Yu, C. Mistrot, E. Unal et al., 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2 E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, P., 1982. The synaptonemal complexes of Caenorhabditis elegans: pachytene karyotype analysis of male and hermaphrodite wild-type and him mutants. Chromosoma 86 577–593. [DOI] [PubMed] [Google Scholar]
- Hammarlund, M., M. W. Davis, H. Nguyen, D. Dayton and E. M. Jorgensen, 2005. Heterozygous insertions alter crossover distribution but allow crossover interference in Caenorhabditis elegans. Genetics 171 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, L., I. Golubovskaya and W. Z. Cande, 2004. A bouquet of chromosomes. J. Cell Sci. 117 4025–4032. [DOI] [PubMed] [Google Scholar]
- Hassold, T., S. Sherman and P. Hunt, 2000. Counting cross-overs: characterizing meiotic recombination in mammals. Hum. Mol. Genet. 9 2409–2419. [DOI] [PubMed] [Google Scholar]
- Hillers, K. J., 2004. Crossover interference. Curr. Biol. 14 R1036–R1037. [DOI] [PubMed] [Google Scholar]
- Hillers, K. J., and A. M. Villeneuve, 2003. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 13 1641–1647. [DOI] [PubMed] [Google Scholar]
- Hirsh, D., D. Oppenheim and M. Klass, 1976. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 49 200–219. [DOI] [PubMed] [Google Scholar]
- Hodges, C. A., E. Revenkova, R. Jessberger, T. J. Hassold and P. A. Hunt, 2005. SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat. Genet. 37 1351–1355. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, S., K. Evason, C. Xiong and K. Kornfeld, 2007. Genetic and pharmacologic factors that influence reproductive aging in nematodes. PLoS Genet. 3 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium, 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert, A., M. Ellefson, A. M. Villeneuve and J. Engebrecht, 2005. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308 206–221. [DOI] [PubMed] [Google Scholar]
- Kelly, K. O., A. F. Dernburg, G. M. Stanfield and A. M. Villeneuve, 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, W. G., and A. Fire, 1998. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, W. G., C. E. Schaner, A. F. Dernburg, M. H. Lee, S. K. Kim et al., 2002. X-chromosome silencing in the germline of C. elegans. Development 129 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass, M. R., 1977. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech. Ageing Dev. 6 413–429. [DOI] [PubMed] [Google Scholar]
- Kovalchuk, I., O. Kovalchuk, A. Arkhipov and B. Hohn, 1998. Transgenic plants are sensitive bioindicators of nuclear pollution caused by the Chernobyl accident. Nat. Biotechnol. 16 1054–1059. [DOI] [PubMed] [Google Scholar]
- Lacefield, S., and A. W. Murray, 2007. The spindle checkpoint rescues the meiotic segregation of chromosomes whose crossovers are far from the centromere. Nat. Genet. 39 1273–1277. [DOI] [PubMed] [Google Scholar]
- Lamb, B. C., 1969. Related and unrelated changes in conversion and recombination frequencies with temperature in Sordaria fimicola, and their relevance to hybrid-DNA models of recombination. Genetics 62 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie, E. J., and G. S. Roeder, 1986. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics 114 769–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand, T., and J. Dutheil, 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3 e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J. A., and J. T. Fleming, 1995. Basic culture methods in Caenorhabditis elegans, pp. 3–29 in Modern Biological Analysis of an Organism, Vol. 48, edited by H. F. Epstein and D. C. Shakes. Academic Press, San Diego.
- Lithgow, G. J., T. M. White, D. A. Hinerfeld and T. E. Johnson, 1994. Thermotolerance of a long-lived mutant of Caenorhabditis elegans. J. Gerontol. 49 B270–B276. [DOI] [PubMed] [Google Scholar]
- Mavor, J. W., and H. K. Svenson, 1923. X-Rays and crossing over. Science 58 124–126. [DOI] [PubMed] [Google Scholar]
- Meneely, P. M., A. F. Farago and T. M. Kauffman, 2002. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics 162 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1925. The regionally differential effect of X rays on crossing over in autosomes of Drosophila. Genetics 10 470–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima, K., A. M. Villeneuve and K. J. Hillers, 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168 1275–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, A., 1998. Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc. Natl. Acad. Sci. USA 95 87–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen, K., J. P. Szatkiewicz, K. Sawyer, N. Leahy, E. D. Parvanov et al., 2008. The recombinational anatomy of a mouse chromosome. PLoS Genet. 4 e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov, P. M., K. W. Broman, J. P. Szatkiewicz and K. Paigen, 2007. Crossover interference underlies sex differences in recombination rates. Trends Genet. 23 539–542. [DOI] [PubMed] [Google Scholar]
- Plough, H. H., 1917. The effect of temperature on linkage in the second chromosome of Drosophila. Proc. Natl. Acad. Sci. USA 3 553–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, V., H. E. Smith, J. Nance, J. Wang, C. Van Doren et al., 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6 605–616. [DOI] [PubMed] [Google Scholar]
- Robine, N., N. Uematsu, F. Amiot, X. Gidrol, E. Barillot et al., 2007. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 27 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A. M., and D. L. Baillie, 1979. The effect of temperature and parental age on recombination and nondisjunction in Caenorhabditis elegans. Genetics 92 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem, M., B. C. Lamb and E. Nevo, 2001. Inherited differences in crossing over and gene conversion frequencies between wild strains of Sordaria fimicola from “Evolution Canyon.” Genetics 159 1573–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan, H., 2007. Telomere attachment and clustering during meiosis. Cell. Mol. Life Sci. 64 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schewe, M. J., D. T. Suzuki and U. Erasmus, 1971. The genetic effects of mitomycin C in Drosophila melanogaster. II. Induced meiotic recombination. Mutat. Res. 12 269–279. [DOI] [PubMed] [Google Scholar]
- Sexton, T., H. Schober, P. Fraser and S. M. Gasser, 2007. Gene regulation through nuclear organization. Nat. Struct. Mol. Biol. 14 1049–1055. [DOI] [PubMed] [Google Scholar]
- Stern, C., 1926. An effect of temperature and age on crossing-over in the first chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 12 530–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C. J., D. G. Mets, M. R. Albrecht, P. Nix, A. Chan et al., 2008. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 22 194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, N., and G. Krohne, 2007. LEM-domain proteins: new insights into lamin-interacting proteins. Int. Rev. Cytol. 261 1–46. [DOI] [PubMed] [Google Scholar]
- Wang, B. D., and A. Strunnikov, 2008. Transcriptional homogenization of rDNA repeats in the episome-based nucleolus induces genome-wide changes in the chromosomal distribution of condensin. Plasmid 59 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., K. Chuang, M. Ahluwalia, S. Patel, N. Umblas et al., 2005. High-throughput SNP genotyping by single-tube PCR with Tm-shift primers. Biotechniques 39 885–893. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28 160–164. [DOI] [PubMed] [Google Scholar]
- Zetka, M. C., and A. M. Rose, 1990. Sex-related differences in crossing over in Caenorhabditis elegans. Genetics 126 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka, M. C, and A. M. Rose, 1995. The genetics of meiosis in Caenorhabditis elegans. Trends Genet. 11 27–31. [DOI] [PubMed] [Google Scholar]