Abstract
To determine whether recombination and/or sister-chromatid cohesion affect the timing of meiotic prophase events, the horsetail stage and S phase were analyzed in Schizosaccharomyces pombe strains carrying mutations in the cohesin genes rec8 or rec11, the linear element gene rec10, the pairing gene meu13, the double-strand-break formation genes rec6, rec7, rec12, rec14, rec15, and mde2, and the recombination gene dmc1. The double-mutant strains rec8 rec11 and rec8 rec12 were also assayed. Most of the single and both double mutants showed advancement of bulk DNA synthesis, start of nuclear movement (horsetail stage), and meiotic divisions by up to 2 hr. Only mde2 and dmc1 deletion strains showed wild-type timing. Contrasting behavior was observed for rec8 deletions (delayed by 1 hr) compared to a rec8 point mutation (advanced by 1 hr). An hypothesis for the role of cohesin and recombination proteins in the control of the G1-to-S transition is proposed. Finally, differences between azygotic meiosis and two other types of fission yeast meiosis (zygotic and pat1-114 meiosis) are discussed with respect to possible control steps in meiotic G1.
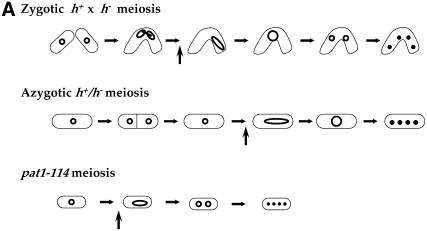
IN sexually reproducing eukaryotes, meiosis results in haploid gametes, which fuse to form the diploid zygote. In organisms with a diploid life cycle, this zygote will give rise to a colony of diploid vegetative cells, or a multicellular body consisting of somatic cells. In Schizosaccharomyces pombe (haploid life cycle), the gametes differentiate into spores (endurance state), which germinate on nutrient-rich media to form colonies of haploid cells. Under nutritional stress (especially nitrogen starvation), haploid cells of opposite mating type differentiate and fuse, and the resulting zygote usually undergoes meiosis immediately (zygotic meiosis; see Figure 1A). However, when returned to rich media before commitment to meiosis, zygotes can resume vegetative growth and form colonies of diploid cells. Under nitrogen starvation, diploid cells heterozygous for mating type will undergo azygotic meiosis (Figure 1A) (Egel 1973; Egel and Egel-Mitani 1974). Azygotic meiosis is more synchronous than zygotic meiosis, but less synchronous than pat1-114 meiosis (Figure 1A).
Figure 1.—
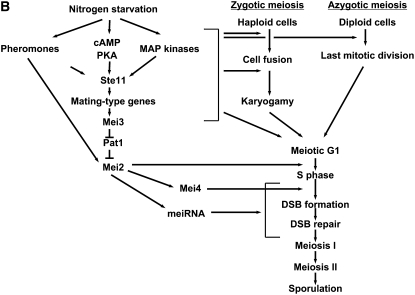
Fission yeast meiosis and its regulation. (A) The three frequently analyzed types of meiosis in S. pombe are presented schematically. Natural zygotic meiosis involves the mating of haploid cells. After cell fusion and karyogamy the zygote immediately proceeds to meiosis and sporulation. It is used for crosses and analysis of recombination. Azygotic meiosis is started in diploid cells heterozygous for the mating types h+ and h− by starvation. Due to its better synchrony, azygotic meiosis is the choice for cytological analysis. A shorter and more synchronous meiosis is induced by temperature shift in haploid or diploid strains carrying the mutation pat1-114. It does not require the presence of both mating types and is frequently applied to the study of gene expression and to physical analysis of recombination intermediates. The vertical arrows indicate the approximate onset of meiotic DNA replication. (B) A simplified scheme of meiosis regulation in zygotic and azygotic meiosis. A selection of steps and regulators are presented. On the left are the major signaling pathways for initiation of cell mating, meiosis, and recombination that are shown on the right. The brackets indicate that a number of interactions are known, but not always the exact targets within the specific cell cycle stages. Mei2 activity is required before S phase (mechanism unknown), before DSB formation after S phase, and perhaps at later steps before the MI division. For more information see the text and recent reviews (Nielsen 2004; Yamamoto 2004; Harigaya and Yamamoto 2007).
A single round of DNA replication followed by two divisions is one major difference between meiosis and mitosis (for review see Page and Hawley 2003). The second meiotic division (MII) is equational, comparable to mitosis. During prophase I, homologous chromosomes pair and recombine. In many organisms, a proteinaceous structure called the synaptonemal complex (SC) is formed between homologous chromosomes. Recombination is initiated by DNA double-strand-break (DSB) formation, catalyzed by a topoisomerase-like protein called Spo11 in Saccharomyces cerevisiae and other eukaryotes and Rec12 in S. pombe (Bergerat et al. 1997; Keeney et al. 1997; Davis and Smith 2001). In both fission and budding yeast, proteins in addition to Spo11/Rec12 are required for DSB formation, several of them without conservation of amino acid sequences between species (Davis and Smith 2001; Keeney 2001). Some of the DSBs are processed into crossovers, which, in combination with sister-chromatid cohesion (SCC), physically link homologous chromosomes. After SC disassembly, the microscopically discernible chiasmata form. They are required for proper segregation of the homologous chromosomes during meiosis I (MI), the reductional division. The sister chromatids of the chromosomes are then segregated during MII.
In contrast to other eukaryotes, S. pombe forms linear elements (LE), which resemble the axial elements, but not SCs (Bahler et al. 1993). During prophase I, the fission yeast nucleus elongates to form the “horsetail” (HT) nucleus, which moves back and forth in the cell (Chikashige et al. 1994; Svoboda et al. 1995). The bouquet structure involves clustering of the telomeres at the spindle pole body at the leading end of the HT nucleus. It is maintained throughout prophase in S. pombe (Kohli and Bahler 1994; Hiraoka and Chikashige 2004). Telomere clustering, attachment to the spindle pole body, and HT movement contribute to full pairing of homologous chromosomes and wild-type levels of recombination (Cooper et al. 1998; Yamamoto et al. 1999; Niwa et al. 2000; Chikashige et al. 2006).
Obviously, the many complex processes required for successful cell mating and meiosis are in need of coordination. Also, regulation of cell mating and meiosis are linked to a large extent, probably as an evolutionary consequence of the haploid life cycle of S. pombe. Some of the early steps of meiosis initiation are well understood (Nielsen 2004; Yamamoto 2004; Harigaya and Yamamoto 2007). Several signal-transduction pathways that are also involved in stress response and nutrient sensing (nitrogen starvation), together with mating-pheromone signaling, are important for the initiation of cell mating, karyogamy, and meiosis (Figure 1B). For progression beyond karyogamy and meiotic G1 into meiotic S phase, the Pat1 kinase, a mitotic repressor of meiosis, has to be inactivated. This leads to activation of Mei2, an RNA-binding protein that controls entry into meiotic S phase. However, the specific target of Mei2 for the mediation of DNA replication is not known. Mei2 together with meiRNA (transcript of the sme2 gene) is again required for a step after DNA replication (Figure 1B) (Yamamoto 2004). For the study of molecular events, a third shorter and more synchronous type of fission yeast meiosis is frequently applied. It is based on the temperature-sensitive mutant pat1-114 (Iino and Yamamoto 1985; Nurse 1985). When haploid or diploid vegetative cells with pat-114 are shifted to the restrictive temperature, meiosis occurs without the need for many of the signal-transduction mechanisms required for induction of zygotic and azygotic meiosis (Yamamoto 2004). Under specific conditions, pat1-114 meiosis may even be induced from G2 (Watanabe et al. 2001). Application of whole-genome arrays for transcript analysis in pat1-114 and azygotic meiosis led to the identification of four successive waves of gene expression: (1) response to nutritional changes (starvation- and pheromone-induced genes), (2) meiotic S phase and recombination genes (early), (3) meiotic divisions (middle) under control of the transcription factor Mei4 (Abe and Shimoda 2000), and (4) spore formation (late) (Mata et al. 2002).
In fission yeast, less is known about the regulatory mechanisms coordinating progression from meiotic S phase through prophase I. In all eukaryotes, SCC is established during mitotic and meiotic DNA replication (Nasmyth 2001). The protein Rec8 was discovered to be required for meiotic SCC first in S. pombe (Molnar et al. 1995) and is found in meiosis of most eukaryotes. In addition, the fission yeast gene rec11 codes for another meiotic cohesin subunit (Kitajima et al. 2003), which is conserved in some, but not all, eukaryotes (Nasmyth and Schleiffer 2004). In wild-type cells, DNA replication has to be completed in a chromosome segment before recombination can be initiated by DSB formation (Borde et al. 2000; Tonami et al. 2005).
In fission yeast, a screen for recombination-deficient mutants led to identification of the genes rec6, rec7 (whose gene product is homologous to S. cerevisiae Rec114), rec14 (homologous to S. cerevisiae Ski8), and rec15 among other ones (Ponticelli and Smith 1989; De Veaux et al. 1992). These genes are required for DSB formation (Cervantes et al. 2000; Ellermeier and Smith 2005). Recent additions to the list of these accessory proteins are rec24, rec25, rec27 (Martin-Castellanos et al. 2005), and mde2 (Gregan et al. 2005). Interestingly, Mde2 is not expressed as early as all other known recombination proteins. Instead, it is under control of the middle-wave transcription factor Mei4 (Abe and Shimoda 2000), adding a novel, unexpected control on DSB formation (Gregan et al. 2005). The rec10 gene codes for a component of the LEs (Molnar et al. 2003; Lorenz et al. 2004). Its deletion leads to a strong reduction in DSB formation and meiotic recombination (Ellermeier and Smith 2005). Its distant S. cerevisiae homolog Red1 is a component of axial elements, and deletion of RED1 reduces, but does not eliminate, interhomolog recombination (Rockmill and Roeder 1990). The gene meu13, HOP2 in S. cerevisiae (Leu et al. 1998) is expressed as an early gene like rec12, and its deletion leads to partial loss of chromosome pairing and recombination (Nabeshima et al. 2001). After DSBs have been processed, repair leads to conversions and crossovers, for which many proteins in different pathways are required (for review see Paques and Haber 1999). Among them, the meiosis-specific RecA homolog Dmc1, occurring in S. cerevisiae, S. pombe, and many other eukaryotes, forms filaments on DNA and catalyzes strand transfer and hybrid DNA formation (Bishop et al. 1992; Sauvageau et al. 2005; Murayama et al. 2008). While deletion of DMC1 in some budding-yeast strain backgrounds leads to checkpoint blockage of prophase progression (Lydall et al. 1996), deletion of dmc1 or of any other recombination gene does not block prophase progression, meiotic divisions, or sporulation in S. pombe. In S. cerevisiae mutation of the early recombination genes RAD50, REC102, REC104, or REC114 was reported to confer earlier MI (Galbraith et al. 1997; Jiao et al. 1999; Malone et al. 2004). The analysis of S-phase length in rec102 revealed no differences to wild type (Slater et al. 1977; Brewer et al. 1984; Cha et al. 2000). It was concluded that DSBs are not the signal for the delay of MI in wild type and that events earlier than MI were not affected (Malone et al. 2004).
This report indicates that proteins required for DSB formation, LE assembly, chromosome pairing, and sister-chromatid cohesion are involved in the timing of the G1-to-S transition in azygotic meiosis of S. pombe. FACS analysis and 4′,6-diamidino-2-phenylindole (DAPI) staining of null mutant cells progressing through azygotic meiosis exhibit a significant advancement of meiotic DNA synthesis, the HT stage, and the meiotic divisions. In contrast, a delay in the timing of these landmarks was observed in rec8 deletion strains. This novel regulation of the G1-to-S transition is discussed with respect to possible mechanisms and in the context of the S. pombe life cycle. It may explain the shortening of meiotic G1 in pat1-114 meiosis. We suggest that in zygotic meiosis a checkpoint-like regulation may protect the cells undergoing mating and karyogamy from precocious progression into meiotic S phase and beyond.
MATERIALS AND METHODS
Strains, media, and standard genetic methods:
S. pombe strains used in this study are listed in Table 1. The standard media and general methods were as described (Gutz et al. 1974; Moreno et al. 1991). Yeast extract agar (YEA) contained 0.5% Difco yeast extract, 3% glucose, and 1.8% agar; yeast extract liquid (YEL) 0.5% Difco yeast extract and 3% glucose; malt extract agar (MEA) 3% malt extract, 1.8% agar; minimal medium (MMA) 0.65% Difco nitrogen base without amino acids, 1% glucose, and 1.8% agar. YEA + 5 and MEA + 5 were supplemented with adenine, uracil, leucine, lysine, and histidine at 100 mg/liter. MMA was supplemented with nutrients according to experimental requirements at 100 mg/liter.
TABLE 1.
S. pombe strains
| Straina | Genotype | Diploidb |
|---|---|---|
| 1-21 | h+ ade6-M210 | Wild type |
| 1-24 | h−ade6-M216 | Wild type |
| ED45 | h+ ade6-M216 leu1-32 rec6-151∷LEU2 | rec6 |
| ED43 | h−ade6-M210 leu1-32 rec6-151∷LEU2 | rec6 |
| 82-3252 | h+ ade6-M216 ura4-D18 rec7∷ura4 | rec7 |
| 82-3253 | h−ade6-M210 ura4-D18 rec7∷ura4 | rec7 |
| ED79 | h+ ade6-M210 ura4-D18 rec8∷ura4 | rec8∷ura4 |
| ED80 | h−ade6-M216 ura4-D18 rec8∷ura4 | rec8∷ura4 |
| GO1 | h+ ade6-M210 ura4-D18 rec8∷kanMX | rec8∷kanMX |
| GH1 | h−ade6-M216 ura4-D18 rec8∷kanMX | rec8∷kanMX |
| ED81 | h+ ade6-M210 rec8-110 | rec8-110 |
| ED82 | h−ade6-M216 rec8-110 | rec8-110 |
| ED35 | h+ ade6-M216 leu1-32 rec10-155∷LEU2 | rec10 |
| ED36 | h−ade6-M210 leu1-32 rec10-155∷LEU2 | rec10 |
| ED46 | h+ ade6-M216 leu1-32 rec11-156∷LEU2 | rec11 |
| ED48 | h−ade6-M210 leu1-32 rec11-156∷LEU2 | rec11 |
| ED50 | h+ ade6-M216 leu1-32 rec12-152∷LEU2 | rec12 |
| ED38 | h−ade6-M210 leu1-32 rec12-152∷LEU2 | rec12 |
| ED56 | h+ ade6-M216 leu1-32 rec14-161∷LEU2 | rec14 |
| ED55 | h−ade6-M210 leu1-32 rec14-161∷LEU2 | rec14 |
| 118-4711 | h+ ade6-M216 rec15∷kanMX | rec15 |
| 118-4709 | h−ade6-M210 rec15∷kanMX | rec15 |
| ALP28 | h+ ade6-M210 meu13∷ura4 ura4-D18 | meu13 |
| ALP29 | h−ade6-M216 meu13∷ura4 ura4-D18 | meu13 |
| ED70 | h+ ade6-M216 leu1-32 ura4-D18 rec8∷ura4 rec11-156∷LEU2 | rec8 rec11 |
| ED69 | h−ade6-M210 leu1-32 ura4-D18 rec8∷ura4 rec11-156∷LEU2 | rec8 rec11 |
| ED82 | h+ ade6-M216 leu1-32 ura4-D18 rec12-152∷LEU2 rec8∷ura4 | rec8 rec12 |
| ED81 | h−ade6-M210 leu1-32 ura4-D18 rec12-152∷LEU2 rec8∷ura4 | rec8 rec12 |
| ED86 | h+ ade6-M216 mde2∷clonNat | mde2 |
| ED83 | h−ade6-M210 mde2∷clonNat | mde2 |
| AG58 | h+ ade6-M216 ura4-D18 dmc1∷ura4 | dmc1 |
| AG61 | h−ade6-M210 ura4-D18 dmc1∷ura4 | dmc1 |
| BS26 | h−smt-0 pat1-114 rad50S ura4-D18 ura4A-10 |
Strains marked with ED, GH, and GO were constructed for this study from strains in the Berne collection. Strains with other designations were taken directly from the collection. The rec6, rec8-11, rec10, rec11, rec12, and rec14 mutants were originally obtained from Gerald Smith, Hutchinson Cancer Research Center, Seattle; meu13 from Alexander Lorenz, University of Vienna; and mde2 from Juraj Gregan, University of Vienna.
The strains of different mating types placed in adjacent rows were mated for formation of the diploids homozygous for the recombination gene alleles indicated.
Crosses were carried out on MEA + 5 at 25°. YEA + 5 was used as standard growth medium at 30°. For meiotic time-course experiments, S. pombe synthetic minimal medium (PM) and PM-N (PM without NH4Cl) were used (Beach et al. 1985; Watanabe et al. 1988). Random spore analysis, dissection of asci, and interrupted mating for construction of diploid strains were performed as described previously (Gutz et al. 1974). Diploids are prototrophic and white on YEA medium as a result of interallelic complementation between the ade6-M216 and ade6-M210 mutations (Moreno et al. 1991). Since diploid strains are rather unstable, they were freshly constructed from the parental haploid strains (Table 1). In some cases, diploids were obtained by protoplast fusion of the parental haploid strains (Sipiczki and Ferenczy 1977).
Time-course experiments with azygotic meiosis:
These experiments were carried out as published (Bahler et al. 1993). The h+/h− strains were streaked onto YEA plates and incubated for 4 days at 30°. Four single colonies were picked for inoculation of the first YEL precultures. After incubation for 24 hr at 30°, 50–100 μl of each preculture were dropped onto MEA + 5 plates and again incubated for 24 hr at 30°. In addition, the second precultures were inoculated into 10 ml YEL and grown at 30° for 24 hr. The sporulation capacity of the cells on the MEA + 5 plates was checked microscopically and by iodine staining. The second preculture of the best sporulating clone was chosen for inoculation of 100 ml PM medium (1:50 and 1:100) and grown at 30° for 16–18 hr. The cell titer of the PM cultures was determined, and care was taken to achieve a concentration of 1–2 × 107. The cells were collected by centrifugation, washed with water, and resuspended in 1 liter PM-N in a 2-liter Erlenmeyer flask. Cultures were incubated at 30° on a table shaker at 180 rpm. For DAPI and FACS analysis, 1-ml samples were taken hourly from t = 0 hr up to 10 hr, and final samples were taken at 24 hr. The samples were centrifuged and resuspended in ice-cold 70% ethanol. For each strain at least three independent experiments were carried out.
Monitoring of cells by DAPI staining, cell titer determination, and FACS analysis:
A solution of 1 μg DAPI per milliliter water was prepared. A 5-μl cell suspension and 5-μl DAPI solution were mixed on a slide and covered with a coverslip, and the edges were sealed with nail polish to avoid evaporation. The cells were then examined by fluorescence microscopy. For every time point at least 200 cells were classified. For monitoring the last mitotic division, the cell titer was determined at t = 0 hr, t = 1 hr, t = 2 hr, and when necessary, also at t = 3 hr, by counting in a hemocytometer (Table 2).
TABLE 2.
Ascus formation, last mitotic division, and length of the horsetail stage
| Fate of input cellsc
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % asci at 0 hr | Cell titer at:a
|
HT stage (min)b | % asci with spores
|
|||||||
| Strains | 0 hr | 1 hr | 2 hr | 3 hr | 2 | 3 | 4 | % cells | ||
| Wild type | 0.3 | 1 | 1 | 2 | 54 ± 3 | 0.6 | 0.0 | 87.5 | 11.9 | |
| rec8 | 0.2 | 1 | 1 | 2 | 29 ± 1 | 0.8 | 5.4 | 80.7 | 13.1 | |
| rec8-110 | 0.1 | 1 | 1 | 2 | 41 ± 2 | 3.3 | 1.0 | 85.8 | 9.9 | |
| rec11 | 2.6 | 1 | 1 | 1.6 | 2 | 68 ± 10 | 4.0 | 6.7 | 72.4 | 16.9 |
| rec8 rec11 | 8 | 1 | 1 | 1.6 | 2 | 45 ± 1 | 11.1 | 7.7 | 73.5 | 7.7 |
| meu13 | 3 | 1 | 1 | 2 | 61 ± 7 | 1.2 | 5.0 | 86.0 | 7.8 | |
| rec10 | 1.6 | 1 | 1 | 2 | 65 ± 10 | 2.4 | 4.0 | 77.1 | 16.5 | |
| rec12 | 5.5 | 1 | 1 | 2 | 55 ± 1 | 4.2 | 3.3 | 83.8 | 8.7 | |
| rec8 rec12 | 50 | 1 | 1 | 1.6 | 49 ± 8 | ND | ND | ND | ND | |
| rec14 | 0.8 | 1 | 1 | 2 | 79 ± 7 | 11.4 | 12.5 | 55.9 | 20.2 | |
| rec6 | 2.4 | 1 | 1 | 1.6 | 2 | 69 ± 3 | 22.7 | 9.0 | 52.0 | 16.3 |
| rec7 | 3.1 | 1 | 1 | 2 | 57 ± 6 | 4.7 | 19.0 | 63.2 | 13.1 | |
| rec15 | 1.9 | 1 | 1.5 | 2 | 70 ± 1 | 5.7 | 12.5 | 62.1 | 19.7 | |
| mde2 | 0.2 | 1 | 1 | 2 | 40 ± 1 | 25.1 | 7.6 | 54.0 | 13.3 | |
| dmc1 | 0 | 1 | 1 | 2 | 48 ± 5 | 1.6 | 0.0 | 89.6 | 8.8 | |
All the values are the mean from three to five independent experiments, and the standard error is indicated for the HT-stage length. ND, not determined due to sporulation throughout prophase.
The cell titer at time 0 hr was set as standard (1 = 100%) for the values at the time points 1, 2, and 3 hr. At the latest time point, 3 hr, the titer had doubled (2 = 200%).
The length of the HT stage was calculated as described in materials and methods.
Not all cells formed asci (for explanation see text). Asci with one or five spores were extremely rare and thus not considered.
For FACS analysis the samples were washed twice in 50 mm Na-Citrate (pH 7), and the cell titer was adjusted to 4 × 106 cells/ml. A total of 25 μl RNAse A (10 mg/ml) was added to the 1-ml samples and incubated for 1 hr at 37°. The samples were then transferred to 5-ml Falcon tubes, 1 ml propidium iodide solution was added (2.5 ng/ml in 50 mm Na-Citrate, pH 7), and stored in the dark at 4° for 3 days to assure full diffusion of propidium iodide into the cells. Before measuring with a FACS Scan, the samples were sonicated at 10% for 8 × 0.5 sec with 0.5 sec breaks after each cycle with a Branson sonifier. The FACS profiles for DNA content were collected and analyzed up to t = 10 hr and analyzed with the CellQuest program.
Determination of cumulative curves of S phase and the HT stage and the length of the HT stage:
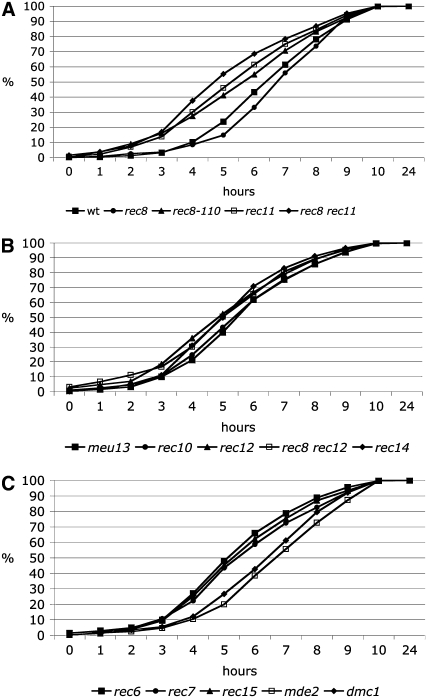
With the few exceptions described below, the procedures were carried out as described before in detail (Cha et al. 2000). Cumulative curves for the HT stage were derived from the percentages of cells with elongated nuclei (DAPI staining) at any time point of the time-course experiments. For calculation of the MI cumulative curves the percentages of cells with more than one nucleus were used, but only from the time points after completion of the last mitotic divisions. For wild type this was the case at t = 5 hr, when the overall curve for cells with more than one nucleus reached its minimum (Figure 2A). For a given time point of a cumulative curve, the percentage from that time point was added to the values from all former time points. This sum was then divided by the final cumulative sum obtained at the latest time point (t = 24 hr).
Figure 2.—
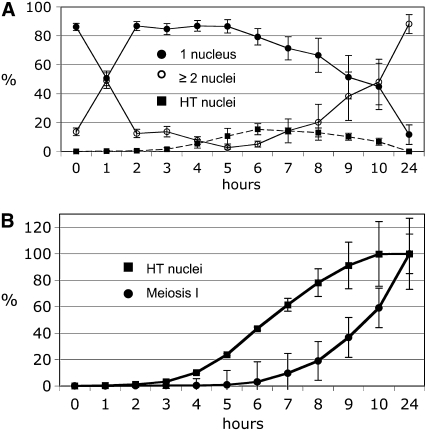
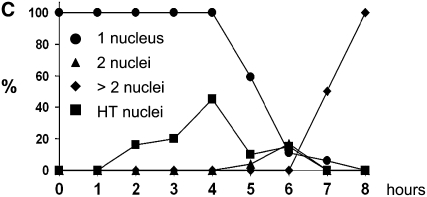
Time-course analysis of azygotic and pat1-114 meiosis. (A) Azygotic meiosis was induced in the h+/h− diploid formed from the strains 1-21 and 1-24 (Table 1) and aliquots collected at intervals of 1 hr up to 10 hr and finally after 24 hr. The cells of five independent time-course experiments were stained with DAPI and classified by microscopy. Means and standard errors are shown. The peak of cells with two nuclei 1 hr after induction results from the last mitosis. The majority of cells have completed cytokinesis at t = 3 hr, and almost all by t = 5 hr. The subsequent rise of the curve corresponds to completion of meiosis I. Meiotic DNA replication starts after t = 4 hr (Egel 1973; Egel and Egel-Mitani 1974). For further explanation, see the legends of Figures 3–5 and the text. (B) The passage of the HT and MI stages is visualized by cumulative curves. The values at the time points indicate the percentage of cells that have reached or passed the HT stage or MI. The standard errors are shown. One hundred percent (100%) corresponds to the sum of cells that had passed the corresponding stages and had sporulated after 24 hr. For detailed description of the calculations, see materials and methods. (C) The analysis of DAPI-stained cells from a pat1-114 meiosis time course of strain BS26 (Sakem 2005). After induction the cells proceed directly to DNA replication and the HT stage. The start of DNA synthesis occurs between t = 1 and 2 hr, as demonstrated by FACS analysis (see, for example, Martin-Castellanos et al. 2005).
Estimates of the average length of S phase were calculated as published (Cha et al. 2000), although the parameters of azygotic meiosis in S. pombe (partial overlap with the last mitotic division) are not comparable with the more favorable situation in S. cerevisiae. Since the resulting data were not considered to be fully reliable, they are not shown. For determination of the average length of the HT stage, the noncumulative curve of the percentage of cells in the HT stage was used directly (see example for the diploid shown in Figure 2A).
RESULTS
This work was provoked by the observation that in azygotic meiosis a rec15 deletion mutant enters the meiotic divisions earlier than wild type (Doll et al. 2005). Thus, a systematic study of recombination and cohesin mutants was initiated. The timing of meiotic DNA replication, the appearance and disappearance of HT nuclei, and the occurrence of the meiotic divisions in wild type (Figure 2) were compared to the scheduling of these events in the mutants (Figures 3–5). In addition, other features of azygotic meiosis were investigated: precocious sporulation, timing of the last mitotic division, the length of the HT stage, and ascus morphology (Table 2). The overview in Figure 6 groups the mutants in approximate order of execution of the functions controlled by the respective gene products: Rec8 and Rec11 (sister-chromatid cohesion); Meu13 (homologous pairing); Rec10 (LE formation); Rec6, Rec7, Rec12, Rec14, Rec15, Mde2 (DSB formation); and Dmc1 (DSB repair).
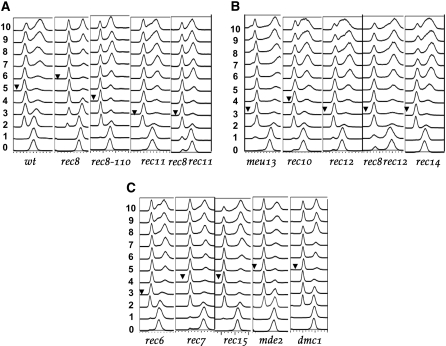
Figure 3.—
Analysis of meiotic DNA replication in azygotic meiosis. The FACS profiles from cells harvested at hourly intervals (t = 0–10) are shown. At induction of meiosis (t = 0 hr) the cells were in G2 with a 4C DNA content (peak at right). The last mitotic division then reduced DNA content to 2C (peak at left), followed by a shift back to 4C as a result of DNA replication. For each strain at least three independent experiments were analyzed and a typical one is shown. The arrowheads indicate the time point with the smallest 4C peak as a marker for the “start of bulk DNA synthesis,” as defined in the text. (A) Phenotypes of alleles of the two meiosis-specific cohesin genes in comparison to wild type. While the rec8 deletion results in a delay of bulk DNA replication, the rec8-110 allele shows advancement. A more pronounced advancement was observed for the rec11 deletion and the rec8 rec11 double deletion strains. (B) The effects of deletion of the pairing gene meu13, of a C-terminal truncation of the linear element component gene rec10, and of full deletions of the genes rec12 and rec14 required for DSB formation. Also included is the double deletion rec8 rec12. (C) The genes rec6, rec7, and rec15 are required for DSB formation. They are expressed earlier than the mde2 gene that is also required for DSB formation, while dmc1 is required only at the later stage of DSB repair. In contrast to the first three mutants, mde2 and dmc1 deletion did not lead to advancement of the onset of bulk DNA replication.
Figure 4.—
Comparison of the timing of the horsetail stage in wild type and mutants. Cumulative curves were constructed as described in the legend to Figure 2B and in the materials and methods. The result of statistical analysis was omitted for better visibility of the curves, but standard errors were similar for the mutants as shown for wild type in Figure 2B. The values at the time points indicate the percentage of cells that have reached or passed the HT stage. The strains are ordered in A–C as in Figure 3. (A) Wild type is compared with mutants of the cohesin genes rec8 and rec11. The rec8 deletion strain shows a moderate delay, while all other strains showed advancement. (B) All the mutants showed advancement of the HT stage: deletion of the pairing gene meu13, truncation of the linear element gene rec10, and deletion of the genes rec12 and rec14 required for DSB formation and of the double mutant rec8 rec12. (C) Advancement also results from deletion of the DSB formation genes rec6, rec7, and rec15, but not from deletion of mde2 (late DSB formation gene) and also not from deletion of the DSB repair gene dmc1.
Figure 5.—
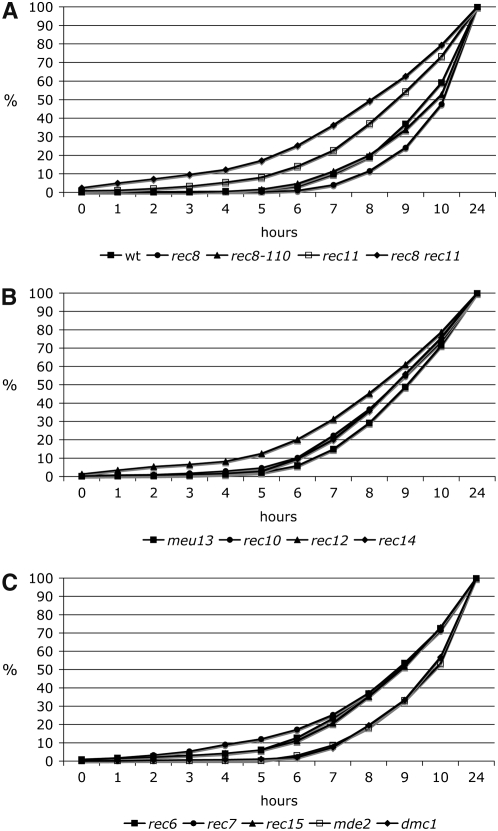
Timing of the meiosis I division in wild type and mutants. For derivation of the cumulative curves and other details, see the legends to Figure 4 and Figure 2B and materials and methods. The values at the time points indicate the percentage of cells that have passed MI. The precocious asci detected at t = 0 hr were included, but the cells with two nuclei at t = 1–4 hr were excluded (last mitotic division; see Figure 2A and Table 2). The strains are ordered into A, B, and C as in Figure 4. The mutants showed similar changes of MI timing (or lack thereof) as for the HT stage (Figure 4). For further details of comparison, see Figure 6.
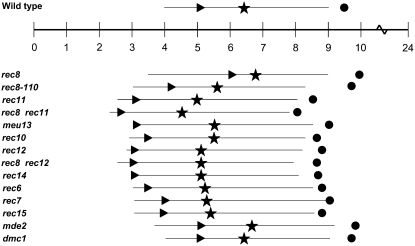
Figure 6.—
Overview of the timing of events in azygotic meiosis. The landmarks of meiosis are shown schematically on the basis of results displayed in Figures 3–5. Above the time scale, wild-type timing is shown and below the time scale that of the mutants (same order as in Figures 3–5). While in Figure 3 one example of FACS analysis is shown, here the approximate average time for onset of bulk DNA replication was derived from all experiments (indicated by arrowheads). A star indicates that 50% of cells were in the HT stage or had passed it, as judged from the cumulative curves in Figure 4. The horizontal line starts when 10% of cells are in the HT stage and ends when 90% of the cells have passed it. The length of the line does not indicate the average length of the HT stage given in Table 2. The circle indicates the time at which 50% of MI has been completed, as derived from the cumulative curves in Figure 5. For evaluation of the data, see results and discussion.
The timing of meiotic events in azygotic wild-type meiosis:
In the mitotic cell cycle of S. pombe, DNA synthesis cannot be analyzed easily since it is short and starts in the daughter nuclei before cytokinesis occurs. As a consequence, the resulting FACS signal corresponds to the G2 DNA content throughout the cell cycle. With the protocol used before (Beach et al. 1985; Bahler et al. 1993) and applied in this study (see materials and methods), diploid cells are in the G2 phase at induction of azygotic meiosis. Then they undergo the last mitotic division (Figure 2A), as judged from DAPI staining and fluorescence microscopy. A peak of cells with two nuclei was observed at t = 1 hr, while at t = 2 hr the fraction of cells with two nuclei was down to a low level (Figure 2A). Thus, meiotic G1 starts in many cells shortly before or after t = 1 hr. Two hours after induction, the cell number had doubled (Table 2), as judged from counting of DAPI-stained cells. A low percentage of cells still had two nuclei at t = 3 and 4 hr, indicating that some cells showed a delay of cytokinesis. At t = 5 hr, the frequency of cells with two nuclei was very low, making the subsequent increase from t = 6 hr onward clearly attributable to passage of meiosis I. At t = 3 hr, the very first HT nuclei appeared, and cells at this stage were found up to 10 hr, when about half of the cells had undergone MI already. This shows that prophase passage of cells in azygotic meiosis occurs with poor synchrony, complicating the analysis. The appearance of the HT nuclei with their altered shape and movement has been approximately correlated with the start of meiotic DNA synthesis (Beach et al. 1985; Bahler et al. 1993). The summation curves shown in Figure 2B were derived by quantification of cell types in Figure 2A. About 50% of the cells have entered or passed the HT stage 6.5 hr after induction and completed the MI division (two or more nuclei per cell) after 9.5 hr. MII division (more than two nuclei per cell, appearing first at t = 7 hr) was also monitored (data not shown). The MII summation curve parallels the MI curve with displacement of the 50% value to ∼11 hr. Twenty-four hours after induction close to 90% of the cells had completed meiosis and spore formation (Table 2).
Meiotic DNA synthesis was monitored by FACS analysis (Figure 3A). At time points t = 0 and 1 hr only a 4C signal was found, which then decreased, while a 2C signal appeared at t = 2 hr and then transiently increased. The 4C peak started growing again after t = 5 hr. Since the proceeding of cells through azygotic meiosis is not very synchronous, the reduction of the 4C peak by cytokinesis and the increase of DNA to 4C by DNA replication overlap. Therefore, it is not possible to determine when the first cells enter meiotic S phase. But since the 4C peak reproducibly starts growing only after t = 5 hr, we conclude that the majority of cells start DNA replication at or after 5 hr and in the following refer to the time point with the smallest 4C peak as the “start of bulk DNA synthesis.” This was then used for qualitative comparison of wild-type and mutant meiosis. The results of FACS analysis for the time points t = 0–10 hr are shown in Figure 3A. It has to be kept in mind that the residual 2C peak at the later time points cannot be taken as an estimate for the fraction of cells not entering DNA replication, since cells undergoing spore formation and asci with mature spores are no longer scored as cells with 4C content. The accumulating spore-wall material shows a strong natural fluorescence. This leads to displacement of these cells from the window set for fluorescence scoring of 2C and 4C cells. The percentage of cells not undergoing meiotic divisions and spore formation was estimated by DAPI staining to be ∼12% in wild type (Table 2). Thus 12% is also the upper limit for estimation of cells not undergoing DNA synthesis, but the relative size of the 2C and 4C peaks at t = 10 hr (Figure 3A) indicates that a good part of the nonsporulating cells actually underwent DNA replication.
The pretreatment procedure of cells before induction of meiosis is not highly reproducible and leads sometimes to poor sporulation. On the basis of experience, time-course experiments with <45% of cells carrying two nuclei at t = 1 hr or a final sporulation yield <70% were discarded. Further features of wild-type and mutant meiosis are shown in Table 2 and discussed after presentation of the analysis of different types of mutants with respect to start of bulk DNA synthesis and the kinetics of the HT and meiosis I stages.
The timing of events in mutants defective in meiotic sister-chromatid cohesion, homologous pairing, or LE formation:
Deletion of the genes rec8 and rec11 coding for meiosis-specific cohesin subunits (Parisi et al. 1999; Watanabe and Nurse 1999; Kitajima et al. 2003) leads to reduction of DSB formation (Young et al. 2004) and reduced recombination frequencies in a region-specific manner (Deveaux and Smith 1994; Krawchuk et al. 1999; Parisi et al. 1999). The same strongly aberrant LE morphology has been recorded for the rec8-110 mutant (Molnar et al. 1995) and for double deletion of rec8 and rec11 (Molnar et al. 1995, 2003). In contrast to these common phenotypes, the cohesin mutants differ strongly with respect to the start of bulk DNA synthesis and later events. The rec11 mutant had already started bulk DNA synthesis at 3 hr after meiotic induction, 2 hr earlier than wild type (Figure 3A). In contrast to rec11 and to all other mutants tested, the rec8 deletion strain showed a delay of ∼1 hr. Interestingly, the rec8 rec11 double mutant started bulk DNA synthesis similar to rec11 or even a bit earlier. Corresponding to the alteration of the start of bulk DNA replication, the HT stage and the occurrence of meiosis I were advanced in the rec11 deletion and in the double mutant and somewhat delayed in strains with rec8 deletions (Figures 4A and 5A). The estimated delays of the HT stage and MI in the rec8 deletion were not as pronounced as the delay of meiotic S phase (see the overview in Figure 6).
The delay of events observed in the rec8 deletion contrasts markedly with the reported advancement of meiosis in the rec8-110 mutant (Molnar et al. 1995). Therefore, rec8-110 strains with the same background as the other mutants were constructed (Table 1), and the timing of meiotic events was examined. Bulk DNA replication was advanced by ∼1 hr (Figure 3), as well as the HT stage (Figure 4A), while MI was close to wild type (Figure 5A). The rec8-110 strain carries two mutations in the rec8 gene: a G-to-A transition at position 853 (aspartate-to-glycine substitution) and a G-to-A transition at position 1334, altering a tryptophan codon into a stop codon. The resulting C-terminal truncation of the protein amounts to about a third of its length (Halvarsson 1999).
Deletion of meu13 leads to reduced homologous pairing early in prophase and to reduced recombination (Nabeshima et al. 2001). In h+/h− azygotic meiosis, the meu13 mutant started bulk DNA synthesis 2 hr before wild type (Figure 3B). The HT stage and MI were somewhat delayed in comparison with other mutants showing an advancement of bulk DNA synthesis, but clearly earlier than in wild type (Figures 4B and 5B). This indicates that prophase in the meu13 mutant is somewhat longer than in wild type. This was also observed for other mutants, but less pronounced (for overview see Figure 6).
The rec10 gene codes for an LE component (Molnar et al. 2003; Lorenz et al. 2004). While deletion of rec10 strongly reduces DSB formation and recombination (Ellermeier and Smith 2005), some rec10 point mutants retain reduced recombination (Deveaux and Smith 1994; Pryce et al. 2005; Wells et al. 2006). The insertion mutation rec10-155∷LEU2 used in this study truncates the gene for 103 codons at the 3′-end and retains reduced meiotic recombination (Lin and Smith 1995; Wells et al. 2006). Meiotic S phase was advanced by ∼1.5 hr in this mutant (Figures 3B and 6), and the HT stage and MI by >1 hr (Figures 4B and 5B).
The timing of meiotic events in mutants blocked in DSB formation:
Strains with deletions of rec12 and rec14 show an altered pattern of LE formation; the rec6, rec7, and rec15 mutants form regular LEs (Molnar et al. 2003). DSB formation is abolished in all these mutants (Young et al. 2004). Bulk DNA synthesis was advanced by 1 hr in the rec7 and rec15 mutants, by ∼1.5 hr in rec6, and by 2 hr in rec12 and rec14 (Figure 3, B and C). The midpoint of the HT stage was advanced by ∼1 hr (Figure 4, B and C), and 50% of the meiocytes had passed MI ahead of wild type by up to 1 hr (Figure 5, B and C).
After having made the observations with the rec8 mutants described above, we attempted to study a rec8 rec12 double deletion strain. Usually time-course experiments were started at a cell titer of 1–2 × 107 cells/ml. Due to premature sporulation of the cells, the rec8 rec12 double mutant could not be forced to reach this titer. Thus, the time courses were started at a titer of ∼0.8 × 107 cells/ml, and at t = 0 hr asci already made up 50% of the population. The regular duplication of the cell titer after meiotic induction was not observed in the double mutant because many cells formed asci directly, soon after t = 0 hr (Table 2). The HT stage was analyzed in the mutant with DAPI staining (Figure 4B), but the progression through MI could not be followed. Although the FACS profile of the rec8 rec12 strain was different from that of other mutants, showing a 2C peak already at t = 0 hr (Figure 3B), the 4C peak was at its lowest at t = 3 hr, as in the rec12 single mutant. The HT stage was advanced by ∼2 hr, and MI completion by ∼1 hr.
Recently, the mde2 mutant defective in DSB formation was described (Gregan et al. 2005). Its expression increases later than that of the other meiotic mutants (Mata et al. 2002). Most of the recombination genes analyzed here show a peak in their expression 4 hr after induction of meiosis. Only rec6, rec7, and rec14 are exceptions. The former two already show an enhanced expression at t = 2 hr (Mata et al. 2002), while rec14 is expressed in vegetative cells as well (Evans et al. 1997). Expression of the mde2 gene is increased significantly only at t = 4 hr, like that of its regulator mei4 (Abe and Shimoda 2000). Peak expression of both genes occurs at 5–9 hr, when expression of the early genes (including dmc1) has already decreased (Mata et al. 2002). FACS analysis of an mde2 deletion strain showed no difference in comparison to wild type (Figure 3C). The HT stage and MI occurred with normal timing (Figures 4C and 5C).
The timing of meiotic events in the DSB repair mutant dmc1:
For comparison with the early recombination mutants affecting DSB formation, the DSB-proficient dmc1 mutant (Young et al. 2004) was also assayed. Like its homologs in other organisms, Dmc1 is a meiosis-specific strand invasion protein (Fukushima et al. 2000; Grishchuk et al. 2004; Murayama et al. 2008). Deletion of dmc1 reduces meiotic recombination fivefold (Grishchuk and Kohli 2003). For pat1-114 meiosis of a dmc1 deletion strain, a short delay of meiosis I has been reported (Shimada et al. 2002). No change of S phase (Figure 3C), the HT stage (Figure 4C), and MI (Figure 5C) was detected.
The length of meiotic G1:
To check whether an event before onset of bulk DNA synthesis may influence the timing of S phase, HT stage, and meiosis I, the last mitotic division after induction of meiosis was carefully monitored in all time-course experiments (Table 2). Most of the mutants showed the peak frequency of cells with two nuclei at t = 1 hr (data not shown for the mutants) similar to wild type (Figure 2A), but still the same cell number as at t = 0 hr (Table 2). In these cases, the number of cells had roughly doubled at t = 3 hr (Figure 2A and Table 2). But a low number of cells with two nuclei at t = 0 hr and at t = 4 and 5 hr were also found (Figure 2A). Among the mutants only rec15 showed slightly earlier mitosis and cytokinesis with more cells already at t = 1 hr. In contrast, the mutant strains rec11 and rec6 and the double mutant rec8 rec11 showed a slight delay of mitosis and cytokinesis with the cell number doubling at 3 hr instead of at 2 hr. Interestingly, these three strains showed an advancement of the later events by ∼2 hr compared to wild type (Figures 3–5). In conclusion, the timing of the last mitotic division is similar in all strains. Thus, advancement (or delay in the case of the rec8 deletion) of the later prophase events does not notably correlate with a time shift of the last mitosis and cytokinesis. If entry into G1 after meiotic induction is set at the time point with the highest frequency of cells with two nuclei (t = 1 hr in most strains, Table 2), and the lowest 4C peak as a measure of onset of bulk DNA replication (t = 5 hr in wild type, Figure 3), an approximate length of G1 of 4 hr results for wild type. Technical caveats influencing the estimates are presented below, but the results obtained for wild type and mutants are most easily explained by changes of the length of G1 in the mutants.
The length of the horsetail stage and additional phenotypes of the mutants:
The length of mitotic S phase is ∼23 min in the haploid wild-type strain 972 in minimal medium at 25°, as determined by autoradiography after pulse labeling with 3H-uracil (Nasmyth et al. 1979). It is known that, for S. cerevisiae and other organisms, S phase is longer in meiotic than in vegetative cells (Williamson et al. 1983; Cha et al. 2000; Strich 2004). It was attempted to use the data from FACS analysis (Figure 3) to calculate the length of meiotic S phase in wild type and mutants according to the method published for S. cerevisiae meiosis (Cha et al. 2000). For wild type, an average length of S phase of ∼40 min was obtained. Detailed results on wild type and mutants are not shown because of the following difficulties: In comparison with S. cerevisiae strain SK1 meiosis (Cha et al. 2000), S. pombe azygotic meiosis is less synchronous and the last mitotic cell division overlaps with the start of DNA replication.
The data on average length of the HT stage (Table 2) are more reliable since HT nuclei can be scored independently from other meiosis events. Six of the 13 mutants did not differ significantly from wild type (54 ± 3 min). Lengthening of the HT stage to maximally 79 min was observed in four mutant strains (rec6, rec11, rec14, rec15). Shortening of the HT stage to minimally 29 min was apparent in four mutant strains (rec8-110 and the deletions rec8, rec8 rec11, and mde2). But the low values for diploids carrying rec8 mutations may be artifactual. The lateral compaction of chromatin is reduced in rec8 mutants (Ding et al. 2006). As a consequence, the organization of the meiotic chromosomes is impaired, and the mass of the chromatin does not follow the spindle pole body to the cell ends during prophase nuclear movement (Molnar et al. 2001a). This may have led to scoring of rec8 HT nuclei as round non-HT nuclei.
At induction of wild-type meiosis (t = 0 hr) 0.3% of the cells had already sporulated and formed asci (Table 2). Most of the mutants showed a higher amount of precocious sporulation with exception of the mde2, dmc1, and rec8 mutant strains. Rather high values were detected in the rec12 mutant (5.5%) and the rec8 rec11 double mutant (8%). Since the rec8 rec12 double mutant showed an extreme amount of precocious asci (50%), some of the later events could not be quantified (see Table 2). Several of the mutants with higher frequencies of precocious asci carry the S. cerevisiae LEU2 gene as a marker of the respective gene disruptions (Table 1). It has been observed before that in some sequence contexts LEU2 does not fully complement the leu1-32 mutation. Thus, some of the strains may have been limited already for leucine during vegetative growth, which in turn might explain the observed precocious asci formation.
Finally, Table 2 contains the data on sporulation of all the strains. In all time-course experiments, some of the cells did not proceed to completion of meiosis and asci formation. The frequency of such cells was ∼12% in wild type and varied between 8 and 20% in the mutants. For the calculation of the summation curves shown in Figures 2, 4, and 5, these cells were excluded (for details see materials and methods). Except for wild type with only a few two-spored asci, azygotic meiosis in the mutants often led to asci with fewer than four spores, as expected for mutants defective in recombination. The frequency of asci with two, three, or four spores was quantified (Table 2). Asci with one or five spores were very rare and not considered. The most irregular mutants were rec6 and mde2 with up to 32% of asci carrying fewer than four spores. Ascus formation and spore number were quite regular in the dmc1 deletion.
Summary of the results:
A synopsis is given in Figure 6. Onset of bulk DNA replication in wild type was found to occur at t = 5 hr, which must be close to the start of DNA replication. Deletion of the two genes with later functions (mde2 for DSB formation and dmc1 for DSB repair) showed no difference in wild type, while most of the mutants with defects in earlier functions showed advancement of bulk DNA replication by up to 2 hr. Advancement by 1 hr was found for the rec8-110 allele carrying two point mutations, but for rec8 deletions a delay of 1 hr was detected. The same pattern was found for the timing of the HT stage. The data indicate that the HT stage and DNA replication occur more or less at the same time in wild type and mutants, with exception of the rec8 deletions, where 50% HT accumulation was close to the delayed onset of DNA replication. MI (completed to 50% at 9.5 hr in wild type) was advanced in many mutants, except for rec8-110 and the rec8 deletions. In general, the later events (HT stage, MI) were somewhat delayed in the mutants relative to onset of bulk DNA synthesis in comparison to wild type. The analysis of the timing of MII (data not shown) indicated that in wild type and mutants the MII division followed MI after a constant interval of 90–120 min.
DISCUSSION
A model on the timing of meiotic events in S. pombe:
The divergent behavior of different rec8 alleles and the fact that Rec8 and Rec11 are subunits of the meiotic cohesion complex (Parisi et al. 1999; Watanabe and Nurse 1999; Kitajima et al. 2003) puts these proteins at the center of the stage. The cohesin genes and most of the other investigated genes (including dmc1) are expressed early in meiosis (Mata et al. 2002). The only exception is the mde2 gene, which is required for DSB formation, but expressed later under the control of Mei4 (Abe and Shimoda 2000). The mde2 deletion strain behaved similar to wild type, as well as to a dmc1 deletion strain.
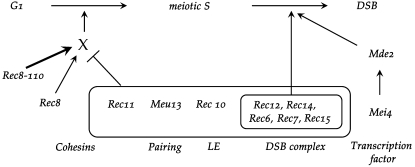
The regulation model is shown in Figure 7. During meiotic G1, the indicated proteins are formed and interact, probably at the chromosomes. The combined action of Meu13, Rec6, Rec7, Rec10, Rec12, Rec14, and Rec15 together with the cohesin subunit Rec11 is proposed to inhibit the activity of a regulator X (or a regulation pathway X) that enhances G1-to-S transition. The cohesin subunit Rec8 is proposed to activate X. The phenotypes of the rec8 rec11 and rec8 rec12 mutants confirm activation of X by Rec8. The mutant Rec8-110 protein is suggested to be an even stronger activator of X. The coordination of X activation and X inhibition activities may occur through interaction of the cohesin subunits at the chromatin.
Figure 7.—
Model of the regulation of the G1-to-S transition in meiosis. It is proposed that the hypothetical regulator X is transiently inhibited by the proteins grouped in the middle (cohesin Rec11, pairing protein Meu13, linear element protein Rec10, and most of the DSB-formation proteins) and that all are likely to be bound to chromatin. Rec8, and more strongly the mutant Rec8-110, are thought to stimulate X with respect to meiotic G1-to-S transition. The Mde2 protein expressed after activation of Mei4 is proposed to stimulate the DSB-formation complex with Rec12 as the catalytic subunit.
When all the indicated proteins are already present in the nucleus during meiotic G1, it is important to inhibit the activity of Rec12 (together with its likely associates Rec6, Rec7, Rec14, and Rec15) for DSB formation during G1 and S phase. Masked DSB formation activity has been demonstrated during S phase since an S-phase checkpoint mutant allows DSB formation before completion of DNA replication (Tonami et al. 2005; Ogino and Masai 2006). After completion of DNA replication, the inhibition of the DNA cleavage complex has to be released. Mde2 is a candidate for effecting this release, since it is required for DSB formation (Gregan et al. 2005), and deletion of mde2 does not advance S phase (Figure 6). Expression of the mde2 gene depends on the Mei4 transcription factor that itself is not expressed in early meiosis (Abe and Shimoda 2000). Mei4 expression in turn depends on activation of Mei2 (Harigaya and Yamamoto 2007), but it is not clear whether Mei2 has additional functions for promotion of MI (Figure 2B).
Evaluation of the regulation model:
Is there evidence for the early presence of the proteins in question? Of all the activities tested, only rec14 (like its S. cerevisiae homolog SKI8) is already expressed in vegetative cells (Evans et al. 1997; Mata et al. 2002). A GFP-tagged and functional version of the Rec7 protein has been visualized in nuclei of mating cells before completion of karyogamy (Molnar et al. 2001b), and in rec7 deletion strains the morphology of the HT nuclei differed from wild type at karyogamy (Molnar et al. 2001a). Fluorescence foci have been observed in homothallic haploid cells carrying Rec8-GFP after induction of mating before cell fusion (Grishchuk 2003). When azygotic meiotic nuclei were spread and stained with an antibody raised against Rec8 protein, the first foci were detected before onset of bulk DNA replication at t = 4 hr (Parisi et al. 1999).
The proteins suggested to activate or inhibit X are known or proposed to occur in complexes at the chromosomes. It is conceivable that the integrity of the DSB forming and the cohesin complexes are required for the transitory inhibition of the G1-to-S transition. This would explain the surprising finding that inactivation of many different recombination genes leads to advancement of meiotic events. Rec10 is a component of the LEs and is required for Rec7 localization to LEs (Lorenz et al. 2006). Together with Rec7, the entire DSB complex may be loaded. Meu13 has been shown to promote chromosome pairing (Nabeshima et al. 2001). Possibly it is located at the chromatin in contact with the DSB or cohesin complexes during G1. Colocalization of a part of the defined Rec8 (cohesin) binding sites on chromosomes (Ding et al. 2006) with a fraction of the strong Rec12 (DSB complex) binding sites has recently been documented on the basis of protein–DNA crosslinking and microarray analysis (Ludin et al. 2008). Aside from regulation through protein interactions as proposed in Figure 7, mechanisms of RNA processing such as mRNA degradation and stage-specific splicing may also be involved (Averbeck et al. 2005; Malapeira et al. 2005; Harigaya et al. 2006).
What may be the nature of the G1-to-S promoting activity X? A signal for inhibition of the onset of DNA replication may be directed to the preinitiation complex at the origins of DNA replication. The ORC, MCM, and Cdt1 complexes have been shown to be subject to regulation (for review see Masukata et al. 2004). Another regulated activity at the onset of S phase is the ribonucleotide reductase complex. Mutation of components of this machinery can lead to a block of meiosis at the onset of S phase (Yoshida et al. 2003; Holmberg et al. 2005). Since our findings involve components of the meiotic cohesin complex in G1-to-S transition, X may release inhibition of a cohesin-loading activity, for example, Mis4 (Tomonaga et al. 2000), and in this way activate S-phase-specific loading of cohesin complexes for DNA replication.
Another possible target of X may be Mei2, the meiosis regulator acting before and after meiotic S phase (see Figure 1B; Yamamoto 2004; Harigaya et al. 2006). Deletion of mei2 leads to a block before meiotic S phase, but the mechanism of Mei2 action at this step is unknown. It is interesting to note that Rep1, the inducer of Res2 belonging to the meiotic START complex required for initiation of DNA replication, is expressed in a cascade independent of Mei2 (Sugiyama et al. 1994). This may indicate that the effect of Mei2 in G1 is fed late in the transition to S phase after action of the START complex. A better understood role of Mei2, involving interaction with meiRNA, is required for events after S phase (Figure 2B) that are required before the MI division (Yamamoto 2004; Harigaya and Yamamoto 2007). Mei2 action is required for DSB formation through the expression of Mei4, which in turn is required for Mde2 expression (Abe and Shimoda 2000).
The activity of Cdc2–cyclin complexes is required for passage through the START regulation point in S. pombe G1 (Nurse and Bissett 1981). Components of the meiotic START complex have been characterized (Miyamoto et al. 1994; Sugiyama et al. 1994) and may be the target of X. The timing of transition through meiotic START is not known. Nevertheless, loss of X inhibition by the proteins indicated in Figure 7 may lead to earlier passage of START, explaining the shortening of G1 in the mutants. The two cyclins Cig2 and Rem1 associating with Cdc2 are required for initiation of meiotic DNA replication in azygotic meiosis (Borgne et al. 2002; Malapeira et al. 2005). Interestingly, the Rem1 function is expressed under the control of Mei4. The double mutants cig2 rem1 and cig2 mei4 are blocked before DNA replication in azygotic meiosis (Malapeira et al. 2005), but not in pat1-114 meiosis (J. Ayté, personal communication). These observations are in line with our interpretation that some steps in azygotic G1 are omitted in pat1-114 meiosis.
In the mutants showing advancement of bulk DNA replication by 2 hr, the length of G1 is reduced to ∼2 hr, which corresponds to the G1 length in pat1-114 meiosis (Figure 2C). pat1-114 meiosis is initiated by a temperature shift leading to inactivation of the Pat1 kinase, which inhibits Mei2 function. It may be that processes in G1 lasting for ∼2 hr in azygotic meiosis are bypassed by precocious Mei2 activation in pat1-114 meiosis. The omitted steps may be those that are subject to activation by X, which then would have no role in pat1-114 meiosis.
Is the proposed control of meiotic G1-to-S transition a checkpoint for completion of karyogamy and chromatin remodeling in zygotic meiosis?
It is surprising that in S. pombe no clear regulatory block of meiosis I has been described in response to a recombination defect, in particular not after deletion of dmc1, which in S. cerevisiae triggers the pachytene checkpoint (Lydall et al. 1996). Only for a meu13 deletion has a short checkpoint-function-dependent delay of MI been described in pat1-114 meiosis (Shimada et al. 2002; Perez-Hidalgo et al. 2003). This finding could not be confirmed for azygotic meiosis (Figure 6). But then, in contrast to S. cerevisiae, and due to the small chromosome number (n = 3), abolition of crossing over by deletion of one of the genes required for DSB formation reduces spore viability to only ∼25% of wild type (Davis and Smith 2001; Keeney 2001). This difference, and also the proposed control of G1-to-S transition (Figure 7), may be a consequence of the haploid life cycle of S. pombe. Fusion and karyogamy of haploid homothallic cells in natural zygotic meiosis are followed by meiosis without intervening mitotic divisions of the zygote (Figure 1A). Nitrogen starvation and the immediately following pheromone signaling induce many functions for cell fusion, karyogamy, and meiosis by the same signal-transduction pathways (Figure 1B). No coordination pattern of step-wise regulation of cell fusion, karyogamy, and the following meiotic G1 phase has been identified. In a population of homothallic cells, the occurrence of these steps in neighboring cells is likely to be asynchronous due to varying gradients of nutrient depletion, differences in cell cycle stage, cell orientation, and cell density. It may be an advantage for efficient completion of sporulation to derepress cell fusion, karyogamy, and meiotic G1 functions right after starvation is sensed. In such an “unordered” process, it will be important not to initiate critical later events such as DNA replication and recombination before earlier steps that are also critical for genome stability have been completed properly. In fact, in zygotic meiosis, G1 lasts much longer than in azygotic meiosis, as judged from time-course microscopy of single zygotes (Molnar et al. 2001a).
In addition to completion of karyogamy and the spatial reorganization of chromosomes, it seems that the chromatin has to be prepared for chromosome pairing and recombination before DNA replication. A pronounced transient decondensation of chromatin has been described 3 hr after induction of azygotic meiosis (Hartsuiker et al. 1998). At the same time chromatin remodeling involving the shifting of nucleosomes at the recombination hotspot ade6-M26 has been demonstrated (Mizuno et al. 1997). This chromatin remodeling is subject to the same signaling pathways (Figure 1B) responsible for the induction of cell mating and meiosis (Mizuno et al. 2001) and also involves histone acetylation in the ade6 region (Yamada et al. 2004). Thus, it is proposed that in S. pombe some of the preparations of chromatin for DSB formation and recombination may occur ahead of bulk DNA replication starting 2 hr later.
Acknowledgments
We thank K. Nabeshima (University of Osaka), G. Smith (Hutchinson Cancer Research Center, Seattle), and A. Lorenz (University of Vienna) for S. pombe strains. Thanks for helpful discussions and suggestions go to J. Ayté (Universitat Pompeu Fabra, Barcelona), M. Lichten (National Institutes of Health, Bethesda, MD), A. Lorenz (Oxford University), K. Nasmyth (Oxford University), K. Ohta (Tokyo University), V. Simanis (Swiss Institute for Cancer Research, Lausanne, Switzerland), and the editor and referees. This work was supported by grants from the Swiss National Science Foundation.
References
- Abe, H., and C. Shimoda, 2000. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics 154 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck, N., S. Sunder, N. Sample, J. A. Wise and J. Leatherwood, 2005. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol. Cell 18 491–498. [DOI] [PubMed] [Google Scholar]
- Bahler, J., T. Wyler, J. Loidl and J. Kohli, 1993. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol. 121 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach, D., L. Rodgers and J. Gould, 1985. RAN1+ controls the transition from mitotic division to meiosis in fission yeast. Curr. Genet. 10 297–311. [DOI] [PubMed] [Google Scholar]
- Bergerat, A., B. de Massy, D. Gadelle, P. C. Varoutas, A. Nicolas et al., 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386 414–417. [DOI] [PubMed] [Google Scholar]
- Bishop, D. K., D. Park, L. Xu and N. Kleckner, 1992. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69 439–456. [DOI] [PubMed] [Google Scholar]
- Borde, V., A. S. Goldman and M. Lichten, 2000. Direct coupling between meiotic DNA replication and recombination initiation. Science 290 806–809. [DOI] [PubMed] [Google Scholar]
- Borgne, A., H. Murakami, J. Ayte and P. Nurse, 2002. The G1/S cyclin Cig2p during meiosis in fission yeast. Mol. Biol. Cell 13 2080–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, B. J., E. Chlebowicz-Sledziewska and W. L. Fangman, 1984. Cell cycle phases in the unequal mother/daughter cell cycles of Saccharomyces cerevisiae. Mol. Cell. Biol. 4 2529–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes, M. D., J. A. Farah and G. R. Smith, 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5 883–888. [DOI] [PubMed] [Google Scholar]
- Cha, R. S., B. M. Weiner, S. Keeney, J. Dekker and N. Kleckner, 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 14 493–503. [PMC free article] [PubMed] [Google Scholar]
- Chikashige, Y., D. Q. Ding, H. Funabiki, T. Haraguchi, S. Mashiko et al., 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science 264 270–273. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., C. Tsutsumi, M. Yamane, K. Okamasa, T. Haraguchi et al., 2006. Meiotic proteins Bqt1 and Bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125 59–69. [DOI] [PubMed] [Google Scholar]
- Cooper, J. P., Y. Watanabe and P. Nurse, 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392 828–831. [DOI] [PubMed] [Google Scholar]
- Davis, L., and G. R. Smith, 2001. Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 98 8395–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux, L. C., and G. R. Smith, 1994. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes Dev. 8 203–210. [DOI] [PubMed] [Google Scholar]
- De Veaux, L. C., N. A. Hoagland and G. R. Smith, 1992. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics 130 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, D. Q., N. Sakurai, Y. Katou, T. Itoh, K. Shirahige et al., 2006. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J. Cell Biol. 174 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll, E., M. Molnar, Y. Hiraoka and J. Kohli, 2005. Characterization of rec15, an early meiotic recombination gene in Schizosaccharomyces pombe. Curr. Genet. 48 323–333. [DOI] [PubMed] [Google Scholar]
- Egel, R., 1973. Commitment to meiosis in fission yeast. Mol. Gen. Genet. 121 277–284. [Google Scholar]
- Egel, R., and M. Egel-Mitani, 1974. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 88 127–134. [DOI] [PubMed] [Google Scholar]
- Ellermeier, C., and G. R. Smith, 2005. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 102 10952–10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, D. H., Y. F. Li, M. E. Fox and G. R. Smith, 1997. A WD repeat protein, Rec14, essential for meiotic recombination in Schizosaccharomyces pombe. Genetics 146 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima, K., Y. Tanaka, K. Nabeshima, T. Yoneki, T. Tougan et al., 2000. Dmc1 of Schizosaccharomyces pombe plays a role in meiotic recombination. Nucleic Acids Res. 28 2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith, A. M., S. A. Bullard, K. Jiao, J. J. Nau and R. E. Malone, 1997. Recombination and the progression of meiosis in Saccharomyces cerevisiae. Genetics 146 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan, J., P. K. Rabitsch, B. Sakem, O. Csutak, V. Latypov et al., 2005. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr. Biol. 15 1663–1669. [DOI] [PubMed] [Google Scholar]
- Grishchuk, A., 2003. Functional characterization of the RecA-like proteins in fission yeast meiosis. Ph.D. Thesis, University of Berne, Berne, Switzerland.
- Grishchuk, A. L., and J. Kohli, 2003. Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination. Genetics 165 1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk, A. L., R. Kraehenbuehl, M. Molnar, O. Fleck and J. Kohli, 2004. Genetic and cytological characterization of the RecA-homologous proteins Rad51 and Dmc1 of Schizosaccharomyces pombe. Curr. Genet. 44 317–328. [DOI] [PubMed] [Google Scholar]
- Gutz, H., H. Heslot, U. Leupold and N. Loprieno, 1974. Schizosaccharomyces pombe, pp. 395–446 in Handbook of Genetics, edited by R. C. King. Plenum Press, New York.
- Halvarsson, G., 1999. Study of rec8 mutations in fission yeast Schizosaccharomyces pombe. Diploma Thesis, University of Berne, Berne, Switzerland.
- Harigaya, Y., and M. Yamamoto, 2007. Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res. 15 523–537. [DOI] [PubMed] [Google Scholar]
- Harigaya, Y., H. Tanaka, S. Yamanaka, K. Tanaka, Y. Watanabe et al., 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442 45–50. [DOI] [PubMed] [Google Scholar]
- Hartsuiker, E., J. Bahler and J. Kohli, 1998. The role of topoisomerase II in meiotic chromosome condensation and segregation in Schizosaccharomyces pombe. Mol. Biol. Cell 9 2739–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka, Y., and Y. Chikashige, 2004. Telomere organization and nuclear movement, pp. 191–205 in The Molecular Biology of Schizosaccharomyces pombe, edited by R. Egel. Springer-Verlag, Berlin.
- Holmberg, C., O. Fleck, H. A. Hansen, C. Liu, R. Slaaby et al., 2005. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 19 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino, Y., and M. Yamamoto, 1985. Mutants of Schizosaccharomyce spombe which sporulate in the haploid state. Mol. Gen. Genet. 198 416–421. [DOI] [PubMed] [Google Scholar]
- Jiao, K., S. A. Bullard, L. Salem and R. E. Malone, 1999. Coordination of the initiation of recombination and the reductional division in meiosis in Saccharomyces cerevisiae. Genetics 152 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52 1–53. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375–384. [DOI] [PubMed] [Google Scholar]
- Kitajima, T. S., S. Yokobayashi, M. Yamamoto and Y. Watanabe, 2003. Distinct cohesin complexes organize meiotic chromosome domains. Science 300 1152–1155. [DOI] [PubMed] [Google Scholar]
- Kohli, J., and J. Bahler, 1994. Homologous recombination in fission yeast: absence of crossover interference and synaptonemal complex. Experientia 50 295–306. [DOI] [PubMed] [Google Scholar]
- Krawchuk, M. D., L. C. DeVeaux and W. P. Wahls, 1999. Meiotic chromosome dynamics dependent upon the rec8(+), rec10(+) and rec11(+) genes of the fission yeast Schizosaccharomyces pombe. Genetics 153 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu, J. Y., P. R. Chua and G. S. Roeder, 1998. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell 94 375–386. [DOI] [PubMed] [Google Scholar]
- Lin, Y., and G. R. Smith, 1995. Molecular cloning of the meiosis-induced rec10 gene of Schizosaccharomyces pombe. Curr. Genet. 27 440–446. [DOI] [PubMed] [Google Scholar]
- Lorenz, A., J. L. Wells, D. W. Pryce, M. Novatchkova, F. Eisenhaber et al., 2004. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 117 3343–3351. [DOI] [PubMed] [Google Scholar]
- Lorenz, A., A. Estreicher, J. Kohli and J. Loidl, 2006. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma 115 330–340. [DOI] [PubMed] [Google Scholar]
- Ludin, K., J. Mata, S. Watt, E. Lehmann, J. Bahler et al., 2008. Sites of strong Rec12/Spo11 binding in the fission yeast genome are associated with meiotic recombination and with centromeres. Chromosoma 117 431–444. [DOI] [PMC free article] [PubMed]
- Lydall, D., Y. Nikolsky, D. K. Bishop and T. Weinert, 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383 840–843. [DOI] [PubMed] [Google Scholar]
- Malapeira, J., A. Moldon, E. Hidalgo, G. R. Smith, P. Nurse et al., 2005. A meiosis-specific cyclin regulated by splicing is required for proper progression through meiosis. Mol. Cell. Biol. 25 6330–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, R. E., S. J. Haring, K. E. Foreman, M. L. Pansegrau, S. M. Smith et al., 2004. The signal from the initiation of meiotic recombination to the first division of meiosis. Eukaryot. Cell 3 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Castellanos, C., M. Blanco, A. E. Rozalen, L. Perez-Hidalgo, A. I. Garcia et al., 2005. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr. Biol. 15 2056–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masukata, H., J. A. Huberman, M. G. Frattine and T. J. Kelly, 2004. DNA replication in S. pombe, pp. 73–99 in The Molecular Biology of Schizosaccharomyces pombe, edited by R. Egel. Springer-Verlag, Berlin.
- Mata, J., R. Lyne, G. Burns and J. Bahler, 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32 143–147. [DOI] [PubMed] [Google Scholar]
- Miyamoto, M., K. Tanaka and H. Okayama, 1994. res2+, a new member of the cdc10+/SWI4 family, controls the ‘start’ of mitotic and meiotic cycles in fission yeast. EMBO J. 13 1873–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, K., Y. Emura, M. Baur, J. Kohli, K. Ohta et al., 1997. The meiotic recombination hot spot created by the single-base substitution ade6–M26 results in remodeling of chromatin structure in fission yeast. Genes Dev. 11 876–886. [DOI] [PubMed] [Google Scholar]
- Mizuno, K., T. Hasemi, T. Ubukata, T. Yamada, E. Lehmann et al., 2001. Counteracting regulation of chromatin remodeling at a fission yeast cAMP response element-related recombination hotspot by stress-activated protein kinase, cAMP-dependent kinase and meiosis regulators. Genetics 159 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, M., J. Bahler, M. Sipiczki and J. Kohli, 1995. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics 141 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, M., J. Bahler, J. Kohli and Y. Hiraoka, 2001. a Live observation of fission yeast meiosis in recombination-deficient mutants: a study on achiasmate chromosome segregation. J. Cell Sci. 114 2843–2853. [DOI] [PubMed] [Google Scholar]
- Molnar, M., S. Parisi, Y. Kakihara, H. Nojima, A. Yamamoto et al., 2001. b Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, M., E. Doll, A. Yamamoto, Y. Hiraoka and J. Kohli, 2003. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J. Cell Sci. 116 1719–1731. [DOI] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194 795–823. [DOI] [PubMed] [Google Scholar]
- Murayama, Y., Y. Kurokawa, K. Mayanagi and H. Iwasaki, 2008. Formation and branch migration of Holliday junctions mediated by eukaryotic recombinases. Nature 451 1018–1021. [DOI] [PubMed] [Google Scholar]
- Nabeshima, K., Y. Kakihara, Y. Hiraoka and H. Nojima, 2001. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 20 3871–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K., 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35 673–745. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K., and A. Schleiffer, 2004. From a single double helix to paired double helices and back. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K., P. Nurse and R. S. Fraser, 1979. The effect of cell mass on the cell cycle timing and duration of S-phase in fission yeast. J. Cell Sci. 39 215–233. [DOI] [PubMed] [Google Scholar]
- Nielsen, O., 2004. Mating-type control and differentiation, pp. 281–296 in The Molecular Biology of Schizosaccharomyces pombe, edited by R. Egel. Springer-Verlag, Berlin.
- Niwa, O., M. Shimanuki and F. Miki, 2000. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19 3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse, P., 1985. Mutants of the fission yeast Schizosaccharomyces pombe which alter the shift between cell proliferation and sporulation. Mol. Gen. Genet. 198 497–502. [Google Scholar]
- Nurse, P., and Y. Bissett, 1981. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature 292 558–560. [DOI] [PubMed] [Google Scholar]
- Ogino, K., and H. Masai, 2006. Rad3-Cds1 mediates coupling of initiation of meiotic recombination with DNA replication. Mei4-dependent transcription as a potential target of meiotic checkpoint. J. Biol. Chem. 281 1338–1344. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2003. Chromosome choreography: the meiotic ballet. Science 301 785–789. [DOI] [PubMed] [Google Scholar]
- Paques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, S., M. J. McKay, M. Molnar, M. A. Thompson, P. J. van der Spek et al., 1999. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol. Cell. Biol. 19 3515–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Hidalgo, L., S. Moreno and P. A. San-Segundo, 2003. Regulation of meiotic progression by the meiosis-specific checkpoint kinase Mek1 in fission yeast. J. Cell Sci. 116 259–271. [DOI] [PubMed] [Google Scholar]
- Ponticelli, A. S., and G. R. Smith, 1989. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics 123 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce, D. W., A. Lorenz, J. B. Smirnova, J. Loidl and R. J. McFarlane, 2005. Differential activation of M26-containing meiotic recombination hot spots in Schizosaccharomyces pombe. Genetics 170 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill, B., and G. S. Roeder, 1990. Meiosis in asynaptic yeast. Genetics 126 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakem, B., 2005. Competition between the meiotic recombination hotspots ade6-M26 and ura4A of Schizosaccharomyces pombe and analysis of DNA double strand breaks at ura4A. Ph.D. Thesis, University of Berne, Berne, Switzerland.
- Sauvageau, S., A. Z. Stasiak, I. Banville, M. Ploquin, A. Stasiak et al., 2005. Fission yeast Rad51 and Dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol. Cell. Biol. 25 4377–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, M., K. Nabeshima, T. Tougan and H. Nojima, 2002. The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. EMBO J. 21 2807–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki, M., and L. Ferenczy, 1977. Protoplast fusion of Schizosaccharomyces pombe auxotrophic mutants of identical mating-type. Mol. Gen. Genet. 151 77–81. [DOI] [PubMed] [Google Scholar]
- Slater, M. L., S. O. Sharrow and J. J. Gart, 1977. Cell cycle of Saccharomyces cerevisiae in populations growing at different rates. Proc. Natl. Acad. Sci. USA 74 3850–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich, R., 2004. Meiotic DNA replication. Curr. Top. Dev. Biol. 61 29–60. [DOI] [PubMed] [Google Scholar]
- Sugiyama, A., K. Tanaka, K. Okazaki, H. Nojima and H. Okayama, 1994. A zinc finger protein controls the onset of premeiotic DNA synthesis of fission yeast in a Mei2-independent cascade. EMBO J. 13 1881–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda, A., J. Bahler and J. Kohli, 1995. Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma 104 203–214. [DOI] [PubMed] [Google Scholar]
- Tomonaga, T., K. Nagao, Y. Kawasaki, K. Furuya, A. Murakami et al., 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14 2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonami, Y., H. Murakami, K. Shirahige and M. Nakanishi, 2005. A checkpoint control linking meiotic S phase and recombination initiation in fission yeast. Proc. Natl. Acad. Sci. USA 102 5797–5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., and P. Nurse, 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400 461–464. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., Y. Lino, K. Furuhata, C. Shimoda and M. Yamamoto, 1988. The S.pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 7 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., S. Yokobayashi, M. Yamamoto and P. Nurse, 2001. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature 409 359–363. [DOI] [PubMed] [Google Scholar]
- Wells, J. L., D. W. Pryce, A. Estreicher, J. Loidl and R. J. McFarlane, 2006. Linear element-independent meiotic recombination in Schizosaccharomyces pombe. Genetics 174 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, D. H., L. H. Johnston, D. J. Fennell and G. Simchen, 1983. The timing of the S phase and other nuclear events in yeast meiosis. Exp. Cell Res. 145 209–217. [DOI] [PubMed] [Google Scholar]
- Yamada, T., K. I. Mizuno, K. Hirota, N. Kon, W. P. Wahls et al., 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 23 1792–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., R. R. West, J. R. McIntosh and Y. Hiraoka, 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Y., 2004. Initiation of meiosis, pp. 297–309 in The Molecular Biology of Schizosaccharomyces pombe, edited by R. Egel. Springer-Verlag, Berlin.
- Yoshida, S. H., H. Al-Amodi, T. Nakamura, C. J. McInerny and C. Shimoda, 2003. The Schizosaccharomyces pombe cdt2(+) gene, a target of G1-S phase-specific transcription factor complex DSC1, is required for mitotic and premeiotic DNA replication. Genetics 164 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J. A., R. W. Hyppa and G. R. Smith, 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]