Abstract
Critically shortened telomeres can be subjected to DNA repair events that generate end-to-end chromosome fusions. The resulting dicentric chromosomes can enter breakage–fusion–bridge cycles, thereby impeding elucidation of the structures of the initial fusion events and a mechanistic understanding of their genesis. Current models for the molecular basis of fusion of critically shortened, uncapped telomeres rely on PCR assays that typically capture fusion breakpoints created by direct ligation of chromosome ends. Here we use independent approaches that rely on distinctive features of Caenorhabditis elegans to study the frequency of direct end-to-end chromosome fusion in telomerase mutants: (1) holocentric chromosomes that allow for genetic isolation of stable end-to-end fusions and (2) unique subtelomeric sequences that allow for thorough PCR analysis of samples of genomic DNA harboring multiple end-to-end fusions. Surprisingly, only a minority of end-to-end fusion events resulted from direct end joining with no additional genome rearrangements. We also demonstrate that deficiency for the C. elegans Ku DNA repair heterodimer does not affect telomere length or cause synthetic effects in the absence of telomerase.
THE natural ends of eukaryotic linear chromosomes present a special challenge for genome integrity: they must be protected from exonucleolytic degradation and must avoid repair by pathways that respond to DNA double-strand breaks. Furthermore, terminal DNA sequences shorten progressively due to the inability of DNA polymerases to replicate the ends of chromosomes and due to processing events that generate 3′ overhangs at chromosome termini (Watson 1972; Olovnikov 1973; Lansdorp 2005). Protection of chromosome ends is provided by telomeres: DNA-protein complexes typically consisting of short, guanine-rich tandem DNA repeats (TTAGGG in humans) and specific proteins that bind telomeric DNA to promote the integrity and proper function of telomeres (de Lange 2005). Telomere shortening is offset by telomerase, which adds telomeric DNA repeats to chromosome ends using an RNA template and the telomerase reverse transcriptase (Greider and Blackburn 1985, 1987). When cells are deficient for telomerase, their telomeres shorten progressively and eventually become dysfunctional, which can lead to end-to-end chromosome fusion and genomic instability (Harley et al. 1990; Hastie et al. 1990). In addition to telomere length, telomere structure is an important factor in capping, as perturbing telomere binding proteins by altering the telomeric DNA sequence or by expressing dominant-negative telomere binding proteins can lead to end-to-end fusion of telomeres of normal length (McEachern and Blackburn 1995; Kirk et al. 1997; Prescott and Blackburn 1997; Van Steensel et al. 1998; Ferreira and Cooper 2001; Underwood et al. 2004; Miller et al. 2005; Pardo and Marcand 2005).
Given that nonhomologous end joining (NHEJ) ligates DNA sequences that lack homology, this DNA repair pathway may mediate the fusion of uncapped telomeres. Cytological studies have revealed that while a core component of the canonical NHEJ machinery is required to fuse acutely uncapped telomeres of normal lengths, disruption of NHEJ does not significantly impair fusion of critically shortened, uncapped telomeres (Smogorzewska et al. 2002; Maser et al. 2007). Other studies have relied on PCR to capture transient fusion events that arise as a consequence of telomere erosion and have revealed DNA sequences for some end-to-end fusion breakpoints (Hackett et al. 2001; Hemann et al. 2001; Mieczkowski et al. 2003; Heacock et al. 2004; Cheung et al. 2006; Capper et al. 2007). These PCR-based studies indicated that NHEJ or microhomology-mediated end joining (MMEJ) are the major pathways that fuse uncapped telomeres. However, PCR of genomic template DNA containing end-to-end fusions can bias the fusion events that are recovered. For example, due to limits of product size and molecular structures that are amenable to PCR, large insertions or inverted repeats may have been missed in such assays. In addition, fusion breakpoints that involve exonucleolytic attack of subtelomeric DNA beyond the PCR primer target sites would be missed. Furthermore, PCR primers used in end-to-end fusion experiments typically target only a subset of chromosome ends. Although PCR-based assays have revealed that end-to-end fusion can occur as a consequence of direct ligation of uncapped chromosome ends, the frequency of direct ligation events in comparison to the full spectrum of telomeric fusion breakpoint structures has not been quantified in any system.
Mutation of trt-1, the Caenorhabditis elegans telomerase reverse transcriptase, results in telomere shortening over successive generations, end-to-end chromosome fusions, and progressive sterility (Cheung et al. 2006; Meier et al. 2006). Here, we address the frequency of direct fusion events that arise in trt-1 mutants. This frequency can be quantified in a relatively unbiased manner in C. elegans, because its chromosomes are holocentric and thus end-to-end chromosome fusions can be genetically isolated and maintained as stable lines. Our analysis of an unbiased sample of fusion events suggests that direct ligation may not be the primary mechanism of repair of critically shortened uncapped telomeres. We also investigated whether one of the core components of NHEJ contributes to telomere length homeostasis: the Ku heterodimer, an NHEJ component whose functions in telomere biology are well studied, if somewhat plastic and controversial (Fisher and Zakian 2005; Riha et al. 2006; Slijepcevic 2006).
MATERIALS AND METHODS
Strains:
All experiments were carried out at 20° under standard culture conditions. The following strains were used in this study: Bristol N2 wild type, bli-3(e767) I, trt-1(ok410) I, unc-29(e193) I, unc-54(r293) I, sqt-2(sc3) II, unc-52(e444) II, mrt-2(e2663) III, lig-4(tm750) III, lig-4(ok416) III, cku-80(tm1203) III, cku-80(ok861) III, cku-70(tm1524) III, pot-1(tm1620) III, unc-45(e286) III, unc-64(e246) III, dpy-9(e12) IV, unc-17(e245) dpy-20(e1282) IV, unc-60(e723) V, unc-51(e369) V, unc-1(e719) X, and unc-3(e151) X. Double mutants for trt-1 with cku-80, cku-70, and lig-4 were constructed by crossing NHEJ mutant males with trt-1, unc-29 and selecting for Unc F2 whose F3 embryos all displayed an egg radiation sensitive phenotype characteristic of NHEJ mutants (i.e., slow growth, vulval defects, and movement defects upon irradiation of late-stage embryos) (Clejan et al. 2006).
Isolation of end-to-end chromosome fusions:
trt-1(ok410), unc-29 or trt-1(ok410), unc-29; lig-4(tm750) strains were propagated for multiple generations until brood size had dropped as a consequence of end-to-end fusions. Hermaphrodites from these strains were crossed with wild-type males, and non-Unc F1 L4 hermaphrodites were singled (i.e., transferred to fresh plates, one hermaphrodite per plate). At larval stage L4, hermaphrodites are virgins and singling them ensured that all progeny were self-progeny. F1 that gave rise to a dominant high incidence of males (Him) phenotype in the F2 revealed the presence of an X-autosome fusion (Ahmed and Hodgkin 2000). F2 males from Him F1 were crossed with unc-1 or unc-3 hermaphrodites to map the X-linked fusion breakpoint to the left or right end of the X chromosome. For each fusion, a total of three outcrosses with unc-1 or unc-3 were performed before strains homozygous for X-autosome fusions were isolated. The unc-29-linked trt-1 mutation was removed during these outcrosses.
Complementation and linkage analysis of fusion breakpoints:
To determine which autosome ends were involved in the end-to-end chromosome fusions, complementation and linkage analyses were performed. For complementation, fusion strain hermaphrodites were crossed with wild-type males. The resulting male progeny, all carrying an X-autosome fusion, were crossed with a hermaphrodite from a different fusion strain. Four F1 hermaphrodites were singled and the F2 progeny were scored for the dominant Him phenotype. A Him phenotype indicated complementation and the presence of X-autosome fusions with different breakpoint orientations, whereas a non-Him phenotype indicated that the fusions belonged to the same complementation group. For linkage analysis, the following genetic marker strains were used: bli-3 I, unc-54 I, sqt-2 II, unc-52 II, unc-45 III, unc-64 III, dpy-9 IV, unc-17 dpy-20 IV, unc-60 V, unc-51 V, unc-1 X, and unc-3 X. Males carrying an X-autosome fusion were crossed with genetic marker hermaphrodites, F1 hermaphrodites were singled, 20 F2 progeny that appeared wild type for the genetic marker mutation were singled, and F3 progeny were scored for the segregation of the marker phenotype and the dominant Him phenotype, which is tightly linked to each fusion breakpoint. Strains homozygous for a fusion would be non-Him and the lack of marker siblings would indicate linkage. Confirmation that non-Him F2 progeny were homozygous for a fusion was obtained by crossing such strains with wild-type males and scoring for the Him phenotype in F1 cross progeny.
Molecular analysis of fusion breakpoints:
Genomic DNA was prepared using a Puregene DNA isolation kit (Gentra). For each fusion strain, all 12 chromosome ends were checked for terminal deletion by PCR using the following subtelomeric primers within the last 1 kb of the telomere:
IL: TCGTCAGCCTTGTTATGTCAACC, GCCTAAGCCTAAAAGAATATGGTAG
IR: TAAGCCTAAGACCAATACCGCAAC, CATTAGGACTGACAGATTGAAAGC
IIL: CATCGCACTTTGAGGACTTTTCC, GCCTAAGCCTAAAATAGTGACTCTG
IIR: ACGCTGTCATCCGAAGCATTGG, GCCTAAGCCTAAAAGCCGCAGC
IIIL: AGTCAGATGGAGGCACGAGTTG, AAAAATAAAATCGGGCTTTTCGACC
IIIR: TGCATTTGTTTTTCCACTTCTGCG, GGCTTTTCAGATAAAAAAATTGTTTTG
IVL: CAATCAATTTTCGGATTTTTTTTCCC, AAGCCTAAGCCTAAGAAGAGACC
IVR: TTGAAAACTCTGTTTTTTGACGGAG, CCATTTCTTGTTTTTCTTTCAATAGC
IVR: TCCCATAACCCAAGCCAGTTGC, GCCTAAGCCTAAGCCAGAGAGT
VL: TTTTGAGTTTTTCATTGAGTCGCTG, CATGTCTCTGTACCGACGATATTC
VR: CAATGTATTTTCAATGATTAAAAGCGG, GCCTAAGCCTAAGCAAATCCCC
XL: TTTTCGGAGCTGCAACTTTGTG, CTAAGCCTAAGCCTAATCTGTGC
XR: GCTCTGCTGAATCGACATTTTGC, AATTCTCATTATTCGATAGTAAACCC.
PCR assay to detect end-to-end chromosome fusions in late-generation strains deficient for telomerase:
Genomic template DNA from strains harboring end-to-end fusions were used in PCR reactions targeting chromosome ends in pairwise combinations to detect fusion between various pairs of chromosome ends. Optimal PCR conditions were determined by testing primers in pairs that targeted one chromosome end, and primers giving strong, reproducible results were selected to target pairs of chromosome ends.
Telomere-length analysis and physical analysis of fusion breakpoint:
For telomere-length analysis, DNA was digested with HinFI and separated on a 0.6% agarose gel at 1.5 V/cm. Southern blotting was carried out with a digoxigenin-dUTP-labeled probe following the manufacturer's instructions (Roche). The probe was made by PCR using primers Tel2 and T7long to amplify telomeric repeats from the plasmid cTel55X, as described (Ahmed and Hodgkin 2000). For analysis of fusion breakpoints, DNA was digested with the indicated enzymes and separated on a 0.6% agarose gel at 3.5 V/cm. Digoxigenin-dUTP-labeled probes for Southern blotting were made using N2 wild-type genomic DNA late and the following primers:
ypT27
XR probe: TTGTCAATCTAACCGAACTTATGC, ATGGCGTCACACTTTTCAGG
VL probe: CACTGTGCCATATGGATTCG, TAGCTCTTTTCGAGGCATGG
ypT21
XR probe: TTGATTGAGTAAGGGCTATTTGG, CTGGAAAAATGTGGCAAAGC
IVR probe: TTGCACGGGAAATTTTTATTG, GCACTTCCTTGTAATGCAACC.
RESULTS
Telomere length in NHEJ mutants:
In yeast and plants, the Ku heterodimer is required to maintain telomeres of normal length (Boulton and Jackson 1996a,b; Baumann and Cech 2000; Riha et al. 2002; Carter et al. 2007). Conflicting data exist in mammals regarding the involvement of Ku in telomere-length regulation; however, spontaneous chromosome fusions are observed consistently in mouse Ku mutants (though not in yeast, plants, or chickens) (Bailey et al. 1999; Difilippantonio et al. 2000; Samper et al. 2000; d'Adda di Fagagna et al. 2001; Gilley et al. 2001; Goytisolo et al. 2001; Maser et al. 2007). To address the role of the Ku heterodimer at telomeres in C. elegans, strains harboring deletions in cku-70(tm1524), cku-80(tm1203 or ok861), or lig-4(ok416 or tm750), each of which confers a strong NHEJ double-strand break (DSB) repair defect (Clejan et al. 2006), were outcrossed multiple times vs. wild type. Homozygous mutant lines were established and then examined for chromosome fusions or changes in telomere length. Propagation of these strains for 30 generations did not result in sterility, as occurs for telomerase mutants, or in modest drops in brood size (data not shown). Furthermore, the NHEJ mutants failed to exhibit even low levels of dominant Him or embryonic lethal phenotypes indicative of chromosome missegregation, which occurs during meiosis when chromosome fusions are heterozygous (data not shown) (Herman et al. 1982). To assess telomere length, genomic DNA was isolated from cku-70, cku-80, or lig-4 mutant strains and subjected to Southern analysis with a C. elegans (TTAGGC)n telomere repeat probe. For each allele, multiple independently outcrossed homozygous mutant lines were examined. Telomere length for cku-70, cku-80, and lig-4 mutants was similar to that of wild type, with telomeres ranging from 2 to 7 kb (Figure 1A and data not shown). While one or two telomeres of cku-80(tm1203) mutant strains appear longer than those of wild type, this modest effect was not observed for an independent null allele of cku-80(ok861) (Figure 1A). Further, telomere length varies among wild-type strains and fluctuates within a single wild-type strain, increasing or decreasing over successive generations (data not shown) (Ahmed and Hodgkin 2000; Ahmed et al. 2001). In contrast, pot-1(tm1620) is an example of a mutation that elicits a significant increase in telomere length by generation F8 (Figure 1A). Thus, mutation of C. elegans NHEJ proteins does not affect telomere length or result in spontaneous end-to-end fusions.
Figure 1.—
Telomere-length homeostasis in C. elegans NHEJ mutants. (A) Telomere length of cku-80 mutant strains. Genomic DNA was isolated from strains that were homozygous for their given mutations for eight generations. (B) Time to sterility for trt-1 single and trt-1; NHEJ double mutant strains. Arrows indicate median. (C) Telomere erosion in trt-1 single and trt-1; cku-80 double mutant strains.
Impact of Ku on telomere shortening in telomerase mutants:
Mutation of Ku in a telomerase deficient background results in synthetic lethality or rapid telomere shortening in yeast and plants, respectively. (Nugent et al. 1998; Baumann and Cech 2000; Riha and Shippen 2003; Heacock et al. 2004). In mice, disruption of Ku alone causes sterility (Barnes et al. 1998; Vogel et al. 1999; Samper et al. 2000), therefore synthetic effects of Ku with telomerase were examined in cells derived from mouse strains that had been homozygous for telomerase deficiency for one or more generations but had been homozygous for Ku deficiency for only a single generation; no synthetic phenotype was observed (Espejel et al. 2002). To test whether there is a functional interaction of telomerase with NHEJ components in C. elegans, we constructed double mutants defective for trt-1, the telomerase reverse transcriptase, and deletion alleles of three NHEJ components that correspond to Ku70, Ku80, or DNA ligase IV. Upon propagation, trt-1; cku-70, trt-1; cku-80, or trt-1; lig-4 double mutants reached sterility at similar generations as trt-1 alone, where the median generation at sterility was 17, 18, 21, and 18, respectively (Figure 1B, arrows). Consistent with these observations, Southern analysis using a telomeric probe revealed that deficiency for cku-80 or lig-4 did not affect the rate of telomere shortening in a trt-1 background (Figure 1C and data not shown). Rates of telomere shortening were 106 ± 27 bp/generation for trt-1, 119 ± 15 bp/generation for trt-1; cku-80, and 117 ± 46 bp/generation for trt-1; lig-4 (n = 2 strains each), which were not significantly different from one another (P > 0.5 in all cases). Thus, mutation of Ku or DNA ligase IV does not cause synthetic effects in the absence of telomerase in C. elegans.
Isolation of end-to-end chromosome fusions:
Molecular analysis of unstable end-to-end fusions from yeast, plants, C. elegans, mice, and humans has typically relied upon PCR reactions that utilize primers adjacent to chromosome ends to amplify fusions from genomic template DNA that contains unknown quantities of end-to-end fusions (Hackett et al. 2001; Hemann et al. 2001; Mieczkowski et al. 2003; Heacock et al. 2004; Cheung et al. 2006; Capper et al. 2007). Although these studies address pathways that process dysfunctional telomeres, the approaches used are biased to recover direct fusions, likely mediated by NHEJ or MMEJ. To test whether direct ligation was the major repair event at critically shortened telomeres, 19 X-autosome end-to-end fusions were isolated genetically from independent strains that were homozygous for the telomerase reverse transcriptase deletion mutation trt-1(ok410) (Meier et al. 2006). Given that dysfunctional telomeres may be substrates of canonical NHEJ, an additional 19 X-autosome end-to-end fusions were isolated from trt-1(ok410); lig-4 double mutant strains that were deficient for the LIG-4 ATP-dependent ligase that is vital for NHEJ-mediated double-strand break repair in C. elegans (Clejan et al. 2006; Robert and Bessereau 2007). The trt-1 telomerase mutation was separated from each of the 38 fusions that were isolated, and each end-to-end fusion was outcrossed three times vs. a genetic background with wild-type telomeres. All isolated chromosome fusions were homozygous viable, indicating that chromosome fusion in the context of telomerase deficiency typically occurs prior to resection and disruption of essential genes near uncapped chromosome ends (data not shown). Note that nonessential genes are predicted to occur within 5 kb of the telomere repeat array at every chromosome end, such that many predicted genes were deleted at some fusion breakpoints. As a preliminary step toward molecular analysis of the fusion breakpoints, linkage analysis was performed. The breakpoints of each end-to-end fusion were mapped to an end of the X chromosome and to an end of an autosome using genetic marker mutations (Figure 2A). Complementation tests confirmed the fusion breakpoint map positions, where nondisjunction was not observed for transheterozygotes carrying end-to-end fusions with the same X and autosome fusion breakpoints (data not shown). At least one fusion event was recovered for all 12 chromosome ends (Figure 2A).
Figure 2.—
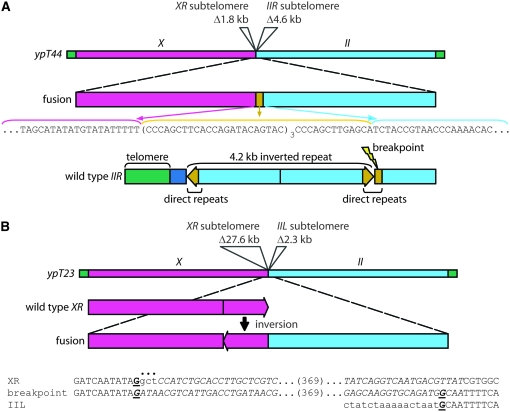
Characterization of outcrossed end-to-end chromosome fusions. (A) Fusion breakpoints of 38 X-autosome end-to-end chromosome fusions, as determined by linkage analysis. Different chromosomes are depicted as rectangles in various colors with green squares representing telomeres at every chromosome end. Genetic names of each independent fusion are to the right of their fusion orientation. End-to-end fusions that were amenable to PCR are indicated in boldface type and underlined. Although the fusion breakpoint of ypT23 was spanned by PCR, it involved a small inversion event and therefore was not a direct fusion breakpoint. (Figure 3B) (B) Terminal deletion analysis to map fusion breakpoints molecularly. PCR results are shown for one chromosome end involved in a fusion. Arrows indicate primers. In this example, one fusion strain was tested for terminal deletion at XR with primers targeting 5 kb of subtelomeric DNA. N2, wild type control; f, X-autosome fusion strain. (C) Distribution of extent of telomere deletion at chromosome ends. (D) Frequency of fusion breakpoint configurations. Observed frequencies (%), expected frequencies (Exp), and P-values are shown.
Terminal deletion analysis:
Genomic DNA was prepared from each fusion strain and PCR was performed targeting subtelomeric DNA within 1 kb of telomeric tracts at every chromosome end. If a primer pair generated a PCR product using template genomic DNA from N2 wild-type but not from a fusion strain, then the targeted region may have been deleted in the fusion strain. Deletions occurred only at chromosome ends that involved a fusion breakpoint (Tables 1 and 2), confirming that outcrossing of each end-to-end fusion had successfully eliminated most unlinked telomeric aberrations that might have been segregating in the telomerase-deficient backgrounds.
TABLE 1.
Terminal deletions at X-autosome fusion breakpoints spanned by PCR
| Deletion (bp)
|
||
|---|---|---|
| Strain | X chromosome | Autosome |
| ypT2 | 2406 | 5137 |
| ypT28 | 10230 | 4364 |
| ypT44 | 1836 | 4598 |
| ypT40 | 994 | 1261 or 8488a |
| ypT41 | 954 | 27627 |
| ypT47 | 216 | 314 |
| ypT23 | 27601 | 2321 |
On the basis of BLAST analysis of the fusion breakpoint sequence, the autosomal deletion breakpoint for ypT40 occurred at one of two possible sites within an rDNA repeat on chromosome IR (Wicky et al. 1996).
TABLE 2.
Terminal deletions at X-autosome fusion breakpoints
| Strain | Origin | X deletion (kb) | Autosome deletion (kb) |
|---|---|---|---|
| ypT29 | trt-1(ok410) | XR 0.0a | IL 0.0 |
| ypT22 | trt-1(ok410) | XR 104.7–105.3 | IIL 1.1–1.3 |
| ypT24 | trt-1(ok410) | XR 0.0 | IIL 13.2–13.4 |
| ypT25 | trt-1(ok410) | XR 11.0–11.4 | IIL 0.0–0.4 |
| ypT16 | trt-1(e2727) | XL 0.0 | IIR 4.0–4.5 |
| ypT3 | trt-1(e2727) | XL 0.0 | IIR 3.6–8.3 |
| ypT17 | trt-1(ok410) | XL 0.0 | IIR 0.0–2.1 kb |
| ypT32 | trt-1(ok410); lig-4 | XL 0.0 | IIR 10.7–11.1 |
| ypT33 | trt-1(ok410); lig-4 | XL 0.0 | IIR 0.0–2.1 kb |
| ypT42 | trt-1(ok410); lig-4 | XR 0.2–0.5 | IIR 8.3–8.8 |
| ypT43 | trt-1(ok410); lig-4 | XR 0.2–0.5 | IIR 10.7–11.1 |
| ypT52 | trt-1(ok410); lig-4 | XR 20.4–20.9 | IIR 0.0 |
| ypT20 | trt-1(ok410) | XL 0.0 | IIIL 0.6–5.6 |
| ypT4 | trt-1(ok410) | XL 0.0 | IIIL 0.9–1.2 |
| ypT34 | trt-1(ok410); lig-4 | XL 0.0–0.3 | IIIR 0.0 |
| ypT18 | trt-1(ok410) | XL 0.0–0.3 | IVL 0.0 |
| ypT26 | trt-1(ok410) | XR 10.4–11.0 | IVL >47.6 |
| ypT45 | trt-1(ok410); lig-4 | XR 30.7–31.0 | IVL 0.0 |
| ypT35 | trt-1(ok410); lig-4 | XL 0.0 | IVR 0.0 |
| ypT36 | trt-1(ok410); lig-4 | XL 0.0 | IVR 0.0 |
| ypT37 | trt-1(ok410); lig-4 | XL 0.0 | IVR 0.0 |
| ypT38 | trt-1(ok410); lig-4 | XL 0.0 | IVR 0.0 |
| ypT21 | trt-1(ok410) | XR 0.8 | IVR 0.0 |
| ypT31 | trt-1(ok410) | XR 23.9–30.7 | IVR 1.4–1.8 |
| ypT7 | trt-1(e2727) | XR 4.2–4.4 | VL 0.0 |
| ypT27 | trt-1(ok410) | XR 0.0 | VL 4.2–4.4 |
| ypT30 | trt-1(ok410) | XR 36.1–36.7 | VL 0.0 |
| ypT46 | trt-1(ok410); lig-4 | XR 42.7–43.2 | VL 1.3–1.7 |
| ypT50 | trt-1(ok410); lig-4 | XR 0.1 | VL 0.0 |
| ypT48 | trt-1(ok410); lig-4 | XR 0.0 | VR 0.8–2.7 |
| ypT49 | trt-1(ok410); lig-4 | XR 2.7–3.0 | VR 0.8–1.0 |
0.0 indicates that subtelomeric DNA sequence was intact.
Breakpoints of those fusions harboring terminal deletions were mapped at 1-kb resolution and then at 5-kb resolution for chromosome ends with >5 kb of subtelomeric DNA deleted. In the example in Figure 2B, between 2 and 3 kb of subtelomeric DNA was deleted in the fusion strain, as products were generated for primer pairs 1 and 2, but not for primer pairs 3–5. Once an intact region of DNA was found, additional primers were utilized to map the deletion breakpoint at ∼500-bp resolution.
Both ends of chromosome IV presented difficulty with precise terminal deletion analysis. The subtelomeric sequence abutting the right end of chromosome IV (IVR) has a tract of DNA composed of 49 copies of a 25-bp direct repeat (Wicky et al. 1996), which makes it difficult to PCR through (M. Lowden, unpublished data). Thus, terminal deletion analysis at IVR relied on primers upstream of the subtelomeric repeats, 1.4 kb from the start of the telomere repeat DNA. The subtelomeric repeats were deleted at one of six IVR fusion breakpoints (Table 2). Abutting IVL is a 23.5-kb inverted repeat (Wilson 1999). While the spacer between the two copies of the inverted repeat was intact for two of three IVL fusion breakpoints, this structure limited further terminal deletion analysis.
On the basis of terminal deletion analysis, three types of fusion breakpoints occurred: telomere–telomere (3%), telomere–subtelomere (44%), and subtelomere–subtelomere (53%). Telomere–telomere fusions were infrequent, on the basis of the hypothesis that the three configurations were equally likely to occur (P < 2 × 10−5) (Figure 2D). Excluding seven IVL or IVR fusion breakpoints for which no deletion could be detected, 27/69 chromosome ends that had been involved in a fusion event contained telomere repeats. Of 47 chromosome ends with subtelomeric DNA deletions, excluding ypT40 where the fusion breakpoint could have occurred at two possible sites, 14 were <1 kb, 17 were 1–5 kb, and 16 were 9–105 kb (Figure 2C).
All fusion strains containing terminal deletions appeared wild type when homozygous, consistent with the observation that, with one exception, no known genes with visible or lethal phenotypes were disrupted by the terminal deletions (http://www.wormbase.org). The exception is ypT41, which carries a 27.6-kb deletion at IIL that removes sqt-2 (SQuaT), which encodes a collagen protein (Kusch and Edgar 1986). The two alleles of sqt-2 that have been characterized, sc3 and sc108, each carry a missense mutation that results in C. elegans mutants with short fat bodies that are helically twisted so that they roll when they move (Kusch and Edgar 1986). For all C. elegans collagen mutants characterized, a defect in body morphology is caused by point mutations in collagen genes, but no phenotype occurs for null mutations. Thus, it is not surprising that ypT41 has no visible phenotype, despite harboring a deletion of sqt-2.
PCR of mapped fusion breakpoints:
After refining the locations of the deletion breakpoints of end-to-end fusions to within ∼500 bp, primers facing each fused chromosome end were utilized to amplify fusion breakpoints. At least two sets of validated primers were tested for each fusion breakpoint. For fusions involving IVR, the two IVR primers used were as follows: one upstream of the subtelomeric repeats (which may have worked if the subtelomeric repeats were deleted) and one overlapping the subtelomeric and telomeric repeats (which may have worked if IVR subtelomeric DNA were intact). No products were amplified for the IVR fusions. Fusions involving IVL were not tested. On the basis of PCR analysis, it is unclear whether the fusions of IVR or IVL were direct. For the remaining 29 fusions whose breakpoints had been clearly mapped, 7 yielded PCR products that spanned the fusion breakpoint (Figure 2A, boldface type, underlined, and ypT23). For each fusion breakpoint that was amenable to PCR, terminal deletions occurred at both chromosome ends (Table 1). The lack of telomere repeats at direct fusion breakpoints suggests that telomere repeat DNA or telomere binding proteins may repress end joining in C. elegans, as supported by in vitro studies of the mammalian telomere binding proteins RAP1 and TRF2 (Bae and Baumann 2007).
Sequence analysis demonstrated that all but one of the fusion breakpoints amplified by PCR were simple direct ligations (Table 3 and Figure 3B). For ypT2 and ypT28, isolated from trt-1, there was no homology between the X chromosome and the autosome at the fusion breakpoint, suggesting that these fusions may have been mediated by canonical NHEJ (Lieber 1999). Of four end-to-end fusions isolated in the absence of lig-4, three (ypT40, ypT41, and ytT47) displayed microhomology at the fusion breakpoints, consistent with observations that MMEJ is a significant end-joining pathway in the absence of the canonical NHEJ machinery (Moore and Haber 1996; Feldmann et al. 2000; Ma et al. 2003; Yu and Gabriel 2003; Heacock et al. 2004). The fourth trt-1; lig-4 fusion breakpoint, ypT44, lacked microhomology and occurred in the context of a 4.2-kb inverted repeat normally present at IIR, which is flanked by two segments of short tandem DNA repeats, whose repeat sequence is identical (Figure 3A). The IIR breakpoint occurs within one set of tandem repeats such that the entire inverted repeat is lost and only approximately four tandem repeats remain. In yeast and mammalian cells, direct and inverted repeats cause genomic instability and are hotspots for mitotic recombination between chromosomes, which can lead to almost complete deletion of the inverted repeat (Lobachev et al. 1998; Waldman et al. 1999; Lobachev et al. 2000). Analogously, the site of the ypT44 fusion breakpoint may reflect an unusual MMEJ- and lig-4-independent mechanism of DSB repair. Finally, the fusion breakpoint of ypT23, which had occurred in a strain containing wild-type lig-4, had a 410-bp inversion that occurred precisely at its XR fusion breakpoint (Figure 3B). The inversion is flanked on either side by 1 bp of microhomology, suggestive of MMEJ, though the mechanism by which this inversion occurred is unclear.
TABLE 3.
Fusion breakpoints defined in this study
| Strain | Origin | Fusion sequence | |
|---|---|---|---|
| ypT2 | trt-1(ok410) | XR | ACGAGCTTTAtcactaggta |
| Breakpoint | ACGAGCTTTACATTCACACA | ||
| IIL | ttcccacaacCATTCACACA | ||
| ypT28 | trt-1(ok410) | XR | GTTTCAAATCcatggaagcc |
| Breakpoint | GTTTCAAATCATTCAGTTGT | ||
| VL | atgaggtaatATTCAGTTGT | ||
| Not isolateda | trt-1(ok410).a generation F10 | IIR | TTTGTCAAAAatccaatttc |
| Breakpoint | TTTGTCAAAAGTACCGACGA | ||
| VL | tcatgtctctGTACCGACGA | ||
| ypT44 | trt-1(ok410; lig-4 | XR | GTATATTTTTtcagtacatg |
| Breakpoint | GTATATTTTTCCCAGCTTCA | ||
| IIR | gatacagaacCCCAGCTTCA | ||
| ypT40 | trt-1(ok410); lig-4 | XR | CATTTTGCTAattttttaaa |
| Breakpoint | CATTTTGCTACCTTGTTACG | ||
| IR | cctacaGCTACCTTGTTACG | ||
| ypT41 | trt-1(ok410); lig-4 | XR | ACCCAAGCTTtgccacattt |
| Breakpoint | ACCCAAGCTTCATGTTGGAA | ||
| IIL | tttgacaCTTCATGTTGGAA | ||
| ypT47 | trt-1(ok410); lig-4 | XR | GTGCACGGAGtcgagaaacc |
| Breakpoint | GTGCACGGAGGTTTTTAAAG | ||
| VR | tttacaGGAGGTTTTTAAAG |
Strains carrying outcrossed homozygous X-autosome fusions are indicated as yp strains. Sequence present at breakpoints is in uppercase, deleted sequence is in lowercase, and microhomology is underlined and in boldface type.
Non-outcrossed fusion recovered by PCR using template genomic DNA from a telomerase mutant harboring end-to-end chromosome fusions.
Figure 3.—
Fusion breakpoint structures of two direct fusions. (A) The drawing depicts the fusion breakpoint for ypT44 where sequencing revealed several subtelomeric direct repeats (yellow) remain intact. Below, the structure of inverted repeats and direct repeats at wild-type IIR, as well as the position of the fusion breakpoint for ypT44, are shown. (B) An inversion at the fusion breakpoint of ypT23 is shown. A 3-bp deletion occurred at the inversion, denoted by a dots above the breakpoint sequence. Boldface type and underlining indicate microhomology at the breakpoints.
In summary, despite careful genetic and molecular mapping of 29 fusion breakpoints, only 7 were amplified, 6 of which were direct ligations and one of which contained a small inversion. Thus, our results indicate that direct ligation may not be the major pathway for the genesis of end-to-end fusions that occur as a consequence of telomere attrition.
PCR of unmapped fusion breakpoints:
To confirm our observations that the frequency of direct fusion was low, we utilized an approach similar to previous PCR-based studies performed in other organisms. Genomic template DNA from mid- to late-generation C. elegans telomere replication mutant strains that had accumulated end-to-end fusions was analyzed using a PCR-based strategy that examines almost all possible permutations that could result from end-to-end fusion of C. elegans chromosomes. The haploid number of chromosomes in C. elegans is six. Although some regions of subtelomeric DNA are repeated at internal genomic sites, the subtelomeric sequences at each chromosome end differ from one another and possess unique primer targets (http://www.wormbase.org, Wicky et al. 1996). Fifty pairwise combinations of subtelomeric primers were utilized to encompass every possible fusion event between different chromosomes, excluding IVR (see above). To test whether PCR could amplify fusion breakpoints from late-generation telomere replication defective strains that may have accumulated multiple fusions, controls were performed with pooled genomic DNA from wild-type and from six isolated X-autosome fusion strains where direct ligation of chromosome ends had occurred (Figure 2) (such that DNA from each fusion strain was diluted 1:9). Under these conditions of nonhomogeneous template genomic DNA, mapped fusion breakpoints were amplified successfully and robustly (data not shown). Additional controls were performed to test primers targeting 11 chromosome ends (excluding IVR), individually and in pairs, to ensure that all the primers worked effectively and specifically under the same reaction conditions. Once appropriate primers and conditions were selected, the following telomere replication mutants were analyzed for fusion breakpoints: trt-1 and mrt-2 single mutants, and four double mutants: trt-1; mrt-2, trt-1; cku-80(ok861), trt-1; lig-4(ok416), and trt-1; lig-4(tm750) (n = 28 mid- to late-generation telomere replication defective strains). mrt-2 encodes a subunit of the 9-1-1 DNA damage response complex that is required for telomerase activity in vivo (Ahmed and Hodgkin 2000; Meier et al. 2006). Strains were subjected to a population bottleneck of six individuals every two generations and passaged until every animal contained multiple fusions, some of which might have shared a common ancestry prior to preparation of genomic DNA for PCR analysis. In an exhaustive attempt to amplify fusion breakpoints, each chromosome end was targeted by one primer within 2 kb of the telomeric tract, which mimics other PCR-based end-to-end fusion assays where primers are usually targeted within 1 kb of telomere repeat tracts. A total of 50 primer pairs were tested on 28 telomere replication mutant strains. Although weak bands were occasionally observed, none were reproducible, and no fusion breakpoints were amplified (Figure 4A).
Figure 4.—
PCR amplification of end-to-end fusions from mid- to late-generation telomerase deficient strains. PCR products were analyzed by separation on an ethidium bromide-stained agarose gel. Three primer pairs were utilized on 16 strains, including the wild-type control, as indicated. M, 1-kb ladder (Invitrogen). (A) PCR targeting IIR and VL. All of the PCR reactions containing genomic DNA from telomerase-deficient strains displayed nonspecific, weak bands, or smears. (B) PCR targeting IIL and VL. Arrowheads indicate PCR products for one strain, trt-1(ok410).a, for all three primer pairs. While all three reactions contained the same primer targeting IIR, each reaction contained a different primer targeting VL at sites 1371 bp, 1651 bp, and 1768 bp from the start of the telomere repeats for primers VL3, VL29, and VL31, respectively.
In agreement with our poor success at amplifying unmapped fusion breakpoints, deletions of 2 kb typically occurred at one or both chromosome ends for genetically isolated end-to-end fusions where direct ligation had occurred (Table 1). In addition, most of the isolated X-autosome fusion strains harbored fusion events involving chromosomes II and V (Figure 2A). Accordingly, PCR reactions using template DNA from each of the individual mid- to late-generation telomere replication strains were performed using 24 primer pairs 1–5 kb away from the telomere tracts of chromosomes II and V. Three reactions amplified a fusion breakpoint for one trt-1 strain, whose PCR product sizes were consistent with the amplification of the same fusion breakpoint (Figure 4B). Sequencing of those PCR products revealed a fusion breakpoint that contained deletions of 2229 and 112 bp at IIR and VL, respectively, and displayed no microhomology (Table 3). The lack of microhomology indicates that this fusion event probably arose as a consequence of canonical NHEJ, in agreement with the two breakpoints from genetically isolated fusions derived from trt-1 strains with wild-type lig-4 (ypT2 and ypT28). Thus, we recovered a single autosome–autosome fusion event from genomic DNA of 28 strains that each harbored multiple end-to-end fusions.
Taken together, analysis of genetically isolated end-to-end fusions whose breakpoints were precisely mapped, as well as PCR-based analysis of non-outcrossed, unmapped fusions of mid- to late-generation telomere replication mutant strains, revealed that end-to-end fusions typically do not occur as a consequence of simple end joining. Under our conditions, the maximum size of the PCR products that could be amplified from C. elegans genomic DNA was 11 kb. Thus, the remaining fusion breakpoints may involve genome duplications >11 kb in length. An alternative possibility is that all fusion breakpoints occurred as a consequence of direct ligation of uncapped chromosome ends, but that PCR failed at these fusion breakpoints due to the presence of DNA sequences that are not amenable to PCR. To distinguish between these possibilities, Southern blotting was performed.
Physical analysis of fusion breakpoints:
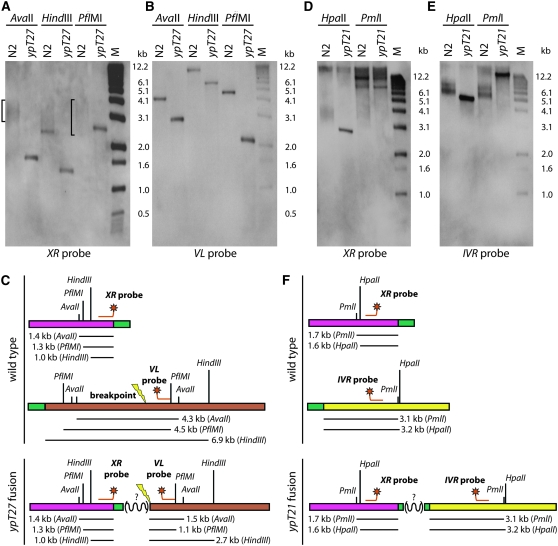
By using genetically isolated end-to-end fusions, large quantities of DNA from many animals homozygous for a single fusion event can be obtained, which allows for physical analysis by Southern blotting. To examine the physical structure of fusion breakpoints that were refractory to PCR, genomic DNA isolated from fusion strains ypT21 and ypT27 was subjected to Southern analysis using probes designed to hybridize to either side of their mapped fusion breakpoints. The genomic DNA for each strain was digested with restriction enzymes to produce restriction fragments of various sizes at each fusion breakpoint. If the fusion events occurred as a consequence of direct ligation, then flanking probes targeting both fused chromosome ends ought to detect a single fusion breakpoint restriction fragment. Alternatively, detection of restriction fragments of different sizes would indicate that an insertion containing additional restriction sites had occurred at the fusion breakpoint. For strain ypT27, which carries a XR–VL fusion, probes targeting XR and VL hybridized to different restriction fragments: AvaII, 1.7-kb and 3.2-kb fragments, respectively; HindIII, 1.3-kb and 5.2-kb fragments, respectively; and PflMI, 2.9-kb and 2.4-kb fragments, respectively (Figure 5, A and B). For ypT21, an XR–IVR fusion, genomic DNA digested with HpaII, the XR and IVR probes hybridized to 2.4-kb and 3.5-kb fragments, respectively (Figure 5, D–F). For ypT21 digested with PmlI, the results were ambiguous because the XR probe hybridized to multiple fragments and showed the same pattern as wild type. Nevertheless, the HpaII data clearly support the conclusion that there is an insertion containing additional restriction sites at the fusion breakpoint of ypT21. The probes used to analyze the fusion breakpoints for ypT27 or ypT21 detected the predicted restriction fragments of wild-type genomic DNA, confirming that the probes were targeting the correct restriction fragments (Figure 5). Thus, physical analysis supports the interpretation that fusion breakpoints that are refractory to PCR do not occur as a consequence of direct ligation, but instead represent complex DNA repair events that result in duplication of one or more segments of the genome at a fusion breakpoint.
Figure 5.—
The physical structure of two fusion breakpoints that were refractory to PCR, as investigated by Southern analysis. Wild-type and mutant DNA digests using the indicated enzymes are shown. ypT27 fusion breakpoint was probed with (A) XR and (B) VL subtelomeric probes. For wild type, the signals agreed with the predicted sizes and appearances: >1.4 kb, >1.3 kb, and >1.0 kb (XR) and 4.3 kb, 4.5 kb, and >7.2 kb (VL) for AvaII, HindIII, or PflMI, respectively. The smeary signals associated with terminal restriction fragments (containing telomeric DNA) are delimited by a bracket to the left of a given lane. (C) The restriction fragments predicted to be detected by each probe for wild type and ypT27 are shown. ypT21 was probed with (D) XR and (E) IVR subtelomeric probes. Signals for wild type agreed with predictions: >1.6 kb and >1.7 kb (XR) or >3.2 kb and >3.1 kb (IVR) for HpaII and PmlI, respectively. (F) The restriction fragments predicted to be detected by each probe for wild type and ypT21 are shown.
DISCUSSION
In this study, we have demonstrated that disruption of C. elegans Ku does not affect telomere length or cause spontaneous telomere uncapping and end-to-end chromosome fusion, phenotypes that have been observed in other systems, but are either controversial within a species or not conserved (Figure 1 and data not shown). Furthermore, in a telomerase-deficient background, disruption of Ku did not increase the rate of telomere shortening or cause a synthetic lethal or accelerated senescence phenotype (Figure 1). The severe effects of Ku deficiency in yeast or plant telomerase mutants contrast sharply with the lack of such effects in C. elegans or mice, suggesting that the role of Ku at telomeres in multicellular animals has been significantly altered, possibly in the context of functional redundancy (Boulton and Jackson 1996a,b; Baumann and Cech 2000; Riha et al. 2002; Carter et al. 2007).
Here, we have established that C. elegans telomere replication mutants can accumulate end-to-end fusions in the absence of DNA ligase IV. Our findings are in strong agreement with studies that show DNA ligase IV is not required for end-to-end fusion of critically shortened telomeres in telomerase mutants in Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis thaliana, or mice (Baumann and Cech 2000; Hackett et al. 2001; Heacock et al. 2004; Maser et al. 2007). In contrast, end-to-end fusion of uncapped telomeres of normal length in S. cerevisiae, S. pombe, Kluyveromyces lactis, and mice depends on DNA ligase IV (Ferreira and Cooper 2001; Smogorzewska et al. 2002; Mieczkowski et al. 2003; Pardo and Marcand 2005; Carter et al. 2007). Thus, processing and fusion of acutely uncapped long telomeres can rely on a specific DNA repair pathway (canonical NHEJ), whereas fusion of telomeres that shorten progressively in the absence of telomerase may be a more promiscuous process that can occur via several DNA repair pathways (Riha et al. 2006).
Molecular analyses of dysfunctional telomeres in yeast, plants, worms, mice, and humans have revealed that end-to-end fusion can be mediated by direct ligation of uncapped telomeres (Hackett et al. 2001; Hemann et al. 2001; Mieczkowski et al. 2003; Heacock et al. 2004; Cheung et al. 2006; Capper et al. 2007). However, the frequency of directly ligated fusion breakpoints could not be determined in these studies, which would miss (1) direct fusion events that involve extensive terminal deletion or (2) complex fusion breakpoints with insertions of large segments of DNA. The holocentric chromosomes of C. elegans result in stable end-to-end fusions where an unbiased collection of initial fusion events can be outcrossed and recovered for quantitative analysis in a manner that is not possible in most model systems. We found that direct fusion events at critically shortened telomeres occur infrequently (Figure 2A and Table 3). In a parallel approach resembling PCR-based experiments conducted in other systems, PCR primers at 11 of 12 chromosome ends failed to reveal significant levels of direct end-to-end fusions in genomic DNA from 28 strains harboring end-to-end fusions. Thus, independent approaches indicate that direct ligation does not explain the majority of DNA repair events at critically shortened telomeres.
Although other molecular studies have not measured the frequency of direct fusions, some were able to detect a greater absolute number of direct fusions than we have observed. One possible explanation for this discrepancy is that some studies in monocentric organisms have relied on Southern analysis to detect fusion PCR products (Heacock et al. 2004; Capper et al. 2007) or have cloned and sequenced the products of entire PCR reactions (Heacock et al. 2004; Cheung et al. 2006) to recover direct fusions that may not have been apparent on gels stained with ethidium bromide (Figure 4A). In addition, direct fusion events may be more frequent in specific cell types (e.g., somatic cells) or organisms, and we might have missed cell-type specific fusions using our PCR-based approach, which was designed to detect stable fusions that were transmitted via germ cells prior to preparation of genomic template DNA for PCR. In this context, HR is the primary DSB repair pathway in C. elegans germ cells, whereas NHEJ is a major DSB repair pathway in noncycling somatic cells (Clejan et al. 2006). Nevertheless, we were able to recover some NHEJ-mediated direct fusion breakpoints, consistent with independent reports that NHEJ-mediated DSB repair can occur in C. elegans germ cells (Morton et al. 2006; Robert and Bessereau 2007). We speculate that developmental regulation of DNA repair pathways may modulate end-to-end fusion during tumorigenesis in mammals.
How do the majority of end-to-end chromosome fusions arise? Southern analysis of genetically isolated end-to-end fusions revealed that probes flanking fusion breakpoints that were refractory to PCR detected restriction fragments of different sizes, whereas a single fusion breakpoint restriction fragment would have been detected for a direct ligation event (Figure 5). Thus, many initial fusion breakpoints that arise when telomerase is deficient may contain insertions of large segments of genomic DNA. In human cells, end-to-end chromosome fusion results in dicentric chromosomes that often break during anaphase and subsequently form new fusions, which impedes analysis of their fusion breakpoints (Bailey and Murnane 2006). Either these initial fusion events or the ensuing genomic instability associated with breakage–fusion–bridge cycles (McClintock 1941) may promote tumorigenesis (Murnane and Sabatier 2004). Our results suggest that a significant fraction of end-to-end chromosome fusion breakpoints that arise when telomerase is deficient may represent complex recombination events.
Studies in other systems are consistent with our observations. Molecular and genetic analyses of telomerase-deficient yeast mutants have revealed that dysfunctional telomeres can copy large segments of DNA from an intact chromosome end via break-induced replication (Hackett et al. 2001). End-to-end fusion and recombination between sister chromatids occurs at dysfunctional telomeres in mammalian cells and can generate duplications at chromosome ends, as shown in cytological studies (Bailey and Murnane 2006). Furthermore, a complex fusion breakpoint involving a small duplication of one chromosome end was recovered by PCR in the context of telomerase deficiency in A. thaliana, an observation that agrees with cytogenetic measurements that revealed fusion of homologous chromosomes or sister chromatids (Siroky et al. 2003; Heacock et al. 2004). Sister chromatid recombination events have also been observed in C. elegans using template genomic DNA from trt-1 mutants and a PCR primer that targets a single chromosome end (Cheung et al. 2006). Together, the latter observations suggest that alternatives to direct end-to-end fusion exist. Thus, the low frequency of direct fusion events observed for end-to-end chromosome fusions genetically isolated from C. elegans, as well as in genomic DNA from late-generation telomerase mutants, may be broadly relevant.
We conclude that the majority of end-to-end fusions that occur as a consequence of telomere erosion may be fundamentally different in nature from the direct fusion events that can be detected by PCR-based analysis of heterogeneous mixtures of genomic DNA harboring end-to-end fusions.
Acknowledgments
We thank I. Clejan, Y. Liu, and V. Alldredge for technical assistance. We thank the C. elegans Gene Knockout Consortium, the National Bioresource Project for the Experimental Animal C. elegans (Shohei Mitani), and the C. elegans Genetics Center for providing strains, and members of the Ahmed lab for discussion, technical assistance, and critical reading of the manuscript. M.L. was supported by National Institutes of Health Predoctoral Fellowship Award for Minority Students GM-072150, National Institutes of Health Research Supplement for Underrepresented Minorities GM-066228-02S1, and National Science Foundation grant HRD0450099. T.L. was supported by the Smallwood Foundation Summer Undergraduate Research Fellowship through the University of North Carolina Office of Undergraduate Research. T.L., B.M., J.H., M.L., and S.A. were supported by National Institutes of Health grant GM-066228.
References
- Ahmed, S., A. Alpi, M. O. Hengartner and A. Gartner, 2001. C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr. Biol. 11 1934–1944. [DOI] [PubMed] [Google Scholar]
- Ahmed, S., and J. Hodgkin, 2000. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403 159–164. [DOI] [PubMed] [Google Scholar]
- Bae, N. S., and P. Baumann, 2007. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell 26 323–334. [DOI] [PubMed] [Google Scholar]
- Bailey, S. M., J. Meyne, D. J. Chen, A. Kurimasa, G. C. Li et al., 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA 96 14899–14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, S. M., and J. P. Murnane, 2006. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 34 2408–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, D. E., G. Stamp, I. Rosewell, A. Denzel and T. Lindahl, 1998. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8 1395–1398. [DOI] [PubMed] [Google Scholar]
- Baumann, P., and T. R. Cech, 2000. Protection of telomeres by the Ku protein in fission yeast. Mol. Biol. Cell 11 3265–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1996. a Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1996. b Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Capper, R., B. Britt-Compton, M. Tankimanova, J. Rowson, B. Letsolo et al., 2007. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 21 2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, S. D., S. Iyer, J. Xu, M. J. McEachern and S. U. Astrom, 2007. The role of nonhomologous end-joining components in telomere metabolism in Kluyveromyces lactis. Genetics. 175 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, I., M. Schertzer, A. Rose and P. M. Lansdorp, 2006. High incidence of rapid telomere loss in telomerase-deficient Caenorhabditis elegans. Nucleic Acids Res. 34 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clejan, I., J. Boerckel and S. Ahmed, 2006. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics 173 1301–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna, F., M. P. Hande, W. M. Tong, D. Roth, P. M. Lansdorp et al., 2001. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11 1192–1196. [DOI] [PubMed] [Google Scholar]
- de Lange, T., 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19 2100–2110. [DOI] [PubMed] [Google Scholar]
- Difilippantonio, M. J., J. Zhu, H. T. Chen, E. Meffre, M. C. Nussenzweig et al., 2000. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel, S., S. Franco, S. Rodriguez-Perales, S. D. Bouffler, J. C. Cigudosa et al., 2002. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 21 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, E., V. Schmiemann, W. Goedecke, S. Reichenberger and P. Pfeiffer, 2000. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 28 2585–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, M. G., and J. P. Cooper, 2001. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell 7 55–63. [DOI] [PubMed] [Google Scholar]
- Fisher, T. S., and V. A. Zakian, 2005. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair 4 1215–1226. [DOI] [PubMed] [Google Scholar]
- Gilley, D., H. Tanaka, M. P. Hande, A. Kurimasa, G. C. Li et al., 2001. DNA-PKcs is critical for telomere capping. Proc. Natl. Acad. Sci. USA 98 15084–15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytisolo, F. A., E. Samper, S. Edmonson, G. E. Taccioli and M. A. Blasco, 2001. The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol. 21 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider, C. W., and E. H. Blackburn, 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43 405–413. [DOI] [PubMed] [Google Scholar]
- Greider, C. W., and E. H. Blackburn, 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51 887–898. [DOI] [PubMed] [Google Scholar]
- Hackett, J. A., D. M. Feldser and C. W. Greider, 2001. Telomere dysfunction increases mutation rate and genomic instability. Cell 106 275–286. [DOI] [PubMed] [Google Scholar]
- Harley, C. B., A. B. Futcher and C. W. Greider, 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345 458–460. [DOI] [PubMed] [Google Scholar]
- Hastie, N. D., M. Dempster, M. G. Dunlop, A. M. Thompson, D. K. Green et al., 1990. Telomere reduction in human colorectal carcinoma and with ageing. Nature 346 866–868. [DOI] [PubMed] [Google Scholar]
- Heacock, M., E. Spangler, K. Riha, J. Puizina and D. E. Shippen, 2004. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 23 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann, M. T., M. A. Strong, L. Y. Hao and C. W. Greider, 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107 67–77. [DOI] [PubMed] [Google Scholar]
- Herman, R. K., C. K. Kari and P. S. Hartman, 1982. Dominant X-chromosome nondisjunction mutants of Caenorhabditis elegans. Genetics 102 379–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, K. E., B. P. Harmon, I. K. Reichardt, J. W. Sedat and E. H. Blackburn, 1997. Block in anaphase chromosome separation caused by a telomerase template mutation. Science 275 1478–1481. [DOI] [PubMed] [Google Scholar]
- Kusch, M., and R. S. Edgar, 1986. Genetic studies of unusual loci that affect body shape of the nematode Caenorhabditis elegans and may code for cuticle structural proteins. Genetics 113 621–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp, P. M., 2005. Major cutbacks at chromosome ends. Trends Biochem. Sci. 30 388–395. [DOI] [PubMed] [Google Scholar]
- Lieber, M. R., 1999. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells 4 77–85. [DOI] [PubMed] [Google Scholar]
- Lobachev, K. S., B. M. Shor, H. T. Tran, W. Taylor, J. D. Keen et al., 1998. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148 1507–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev, K. S., J. E. Stenger, O. G. Kozyreva, J. Jurka, D. A. Gordenin et al., 2000. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 19 3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. L., E. M. Kim, J. E. Haber and S. E. Lee, 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser, R. S., K. K. Wong, E. Sahin, H. Xia, M. Naylor et al., 2007. DNA-PKcs is not required for dysfunctional telomere fusion and checkpoint response in the telomerase deficient mouse. Mol. Cell. Biol. 27 2253–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern, M. J., and E. H. Blackburn, 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376 403–409. [DOI] [PubMed] [Google Scholar]
- Meier, B., I. Clejan, Y. Liu, M. Lowden, A. Gartner et al., 2006. trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase. PLoS Genet. 2 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski, P. A., J. O. Mieczkowska, M. Dominska and T. D. Petes, 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc. Natl. Acad. Sci. USA 100 10854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. M., M. G. Ferreira and J. P. Cooper, 2005. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 24 3128–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. K., and J. E. Haber, 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, J., M. W. Davis, E. M. Jorgensen and D. Carroll, 2006. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc. Natl. Acad. Sci. USA 103 16370–16375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane, J. P., and L. Sabatier, 2004. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. BioEssays 26 1164–1174. [DOI] [PubMed] [Google Scholar]
- Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger et al., 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8 657–660. [DOI] [PubMed] [Google Scholar]
- Olovnikov, A. M., 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41 181–190. [DOI] [PubMed] [Google Scholar]
- Pardo, B., and S. Marcand, 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 24 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, J., and E. H. Blackburn, 1997. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 11 528–540. [DOI] [PubMed] [Google Scholar]
- Riha, K., and D. E. Shippen, 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K., J. M. Watson, J. Parkey and D. E. Shippen, 2002. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha, K., M. L. Heacock and D. E. Shippen, 2006. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu. Rev. Genet. 40 237–277. [DOI] [PubMed] [Google Scholar]
- Robert, V., and J. L. Bessereau, 2007. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 26 170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper, E., F. A. Goytisolo, P. Slijepcevic, P. P. van Buul and M. A. Blasco, 2000. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 1 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siroky, J., J. Zluvova, K. Riha, D. E. Shippen and B. Vyskot, 2003. Rearrangements of ribosomal DNA clusters in late generation telomerase-deficient Arabidopsis. Chromosoma 112 116–123. [DOI] [PubMed] [Google Scholar]
- Slijepcevic, P., 2006. The role of DNA damage response proteins at telomeres: an “integrative” model. DNA Repair 5 1299–1306. [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A., J. Karlseder, H. Holtgreve-Grez, A. Jauch and T. de Lange, 2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 12 1635–1644. [DOI] [PubMed] [Google Scholar]
- Underwood, D. H., C. Carroll and M. J. McEachern, 2004. Genetic dissection of the Kluyveromyces lactis telomere and evidence for telomere capping defects in TER1 mutants with long telomeres. Eukaryot. Cell 3 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel, B., A. Smogorzewska and T. de Lange, 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92 401–413. [DOI] [PubMed] [Google Scholar]
- Vogel, H., D. S. Lim, G. Karsenty, M. Finegold and P. Hasty, 1999. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. USA 96 10770–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman, A. S., H. Tran, E. C. Goldsmith and M. A. Resnick, 1999. Long inverted repeats are an at-risk motif for recombination in mammalian cells. Genetics 153 1873–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, J. D., 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239 197–201. [DOI] [PubMed] [Google Scholar]
- Wicky, C., A. M. Villeneuve, N. Lauper, L. Codourey, H. Tobler et al., 1996. Telomeric repeats (TTAGGC)n are sufficient for chromosome capping function in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 93 8983–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R. K., 1999. How the worm was won. The C. elegans genome sequencing project. Trends Genet. 15 51–58. [DOI] [PubMed] [Google Scholar]
- Yu, X., and A. Gabriel, 2003. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics 163 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]