Abstract
The histone deacetylase activity of Sir2p is dependent on NAD+ and inhibited by nicotinamide (NAM). As a result, Sir2p-regulated processes in Saccharomyces cerevisiae such as silencing and replicative aging are susceptible to alterations in cellular NAD+ and NAM levels. We have determined that high concentrations of NAM in the growth medium elevate the intracellular NAD+ concentration through a mechanism that is partially dependent on NPT1, an important gene in the Preiss–Handler NAD+ salvage pathway. Overexpression of the nicotinamidase, Pnc1p, prevents inhibition of Sir2p by the excess NAM while maintaining the elevated NAD+ concentration. This growth condition alters the epigenetics of rDNA silencing, such that repression of a URA3 reporter gene located at the rDNA induces growth on media that either lacks uracil or contains 5-fluoroorotic acid (5-FOA), an unusual dual phenotype that is reminiscent of telomeric silencing (TPE) of URA3. Despite the similarities to TPE, the modified rDNA silencing phenotype does not require the SIR complex. Instead, it retains key characteristics of typical rDNA silencing, including RENT and Pol I dependence, as well as a requirement for the Preiss–Handler NAD+ salvage pathway. Exogenous nicotinamide can therefore have negative or positive impacts on rDNA silencing, depending on the PNC1 expression level.
IN the budding yeast, Saccharomyces cerevisiae, there are three general locations that are silenced in the genome, the silent-mating type loci HML and HMR, the telomeres, and the ribosomal DNA (rDNA) (see Rusche et al. 2003 for review). Silencing at these locations is dependent on the silent information regulator genes, SIR1–SIR4. All four SIR genes are required for the efficient establishment, maintenance, and inheritance of silent chromatin structure at the HM loci (Pillus and Rine 1989). SIR2, SIR3, and SIR4 are critical for silencing at telomeres (Aparicio et al. 1991), but only SIR2 is required for silencing and suppression of recombination at the rDNA (Gottlieb and Esposito 1989; Bryk et al. 1997; Fritze et al. 1997; Smith and Boeke 1997). SIR2 encodes a highly conserved NAD+-dependent histone deacetylase that is predominantly localized in the nucleolus and in perinuclear foci that harbor the telomeres (Gotta et al. 1997). In the nucleolus, Sir2p is a subunit of the multiprotein deacetylase complex known as RENT. This complex also contains Net1p and Cdc14p, and functions in rDNA silencing and regulation of the exit from mitosis (Shou et al. 1999; Straight et al. 1999), as well as rDNA transcription by RNA polymerase I (Shou et al. 2001). At the telomeres and HM loci, Sir2p is a subunit of the SIR complex, which minimally consists of Sir2p and Sir4p (Ghidelli et al. 2001; Tanny et al. 2004). Sir3p can also be part of the complex, which is a heterotrimer when purified from insect cells (Cubizolles et al. 2006). The sharing of Sir2p by all forms of yeast silencing leads to competition between the various compartments for a limiting amount of Sir2p (Smith et al. 1998). rDNA silencing is especially sensitive to changes in Sir2p levels, as SIR2 overexpression dramatically strengthens silencing at this locus (Fritze et al. 1997; Smith et al. 1998), and extends replicative life span through the suppression of rDNA recombination (Gottlieb and Esposito 1989; Kaeberlein et al. 1999). Similarly, deletion of SIR4 releases Sir2p from the telomeres and HM loci, causing it to accumulate in the nucleolus and strengthen rDNA silencing (Kennedy et al. 1997; Smith et al. 1998).
The Sir2p family of protein deacetylases (collectively known as sirtuins) utilize NAD+ as a cosubstrate. For every lysine that is deacetylated, one molecule of NAD+ is hydrolyzed, yielding one molecule each of nicotinamide (NAM) and O-acetyl-ADP-ribose (AAR) (Landry et al. 2000; Tanny and Moazed 2001). The consumption of NAD+ implies there is a constant need for NAD+ production in the cell if sirtuins are to remain active. NAD+ production in yeast cells occurs through four known pathways (see Figure 1A for schematic). The de novo NAD+ synthesis pathway converts tryptophan into NAD+ through a series of steps also known as the kynurenine pathway that are catalyzed by products of the BNA genes (Kucharczyk et al. 1998). This pathway is cytosolic and does not usually contribute to rDNA silencing regulation unless nicotinic acid in the growth medium is limiting (Anderson et al. 2002; Sandmeier et al. 2002). A second pathway involves the conversion of imported nicotinamide riboside (NR) into NAD+ via the NR kinase (Nrk1p) (Bieganowski and Brenner 2004). A third, newly identified pathway involves direct conversion of NR into NAM by a set of NR hydrolases and phosphorylases (Belenky et al. 2007). In the fourth pathway, NAM produced by sirtuin-mediated protein deacetylation or breakdown of NR is converted into nicotinic acid by the nicotinamidase, Pnc1p (Ghislain et al. 2002), and then converted back into NAD+ by the Preiss–Handler pathway, primarily in the nucleus (Anderson et al. 2002; Sandmeier et al. 2002). The nicotinic acid phosphoribosyltransferase, Npt1p, is a critical step of this pathway (Rajavel et al. 1998; Smith et al. 2000), as deletion of NPT1 causes a two- to threefold reduction in the intracellular NAD+ concentration (Lin et al. 2000; Smith et al. 2000). Collectively, Pnc1p and Npt1p are often referred to as the NAD+ salvage pathway, a term that we will use throughout the article.
Figure 1.—
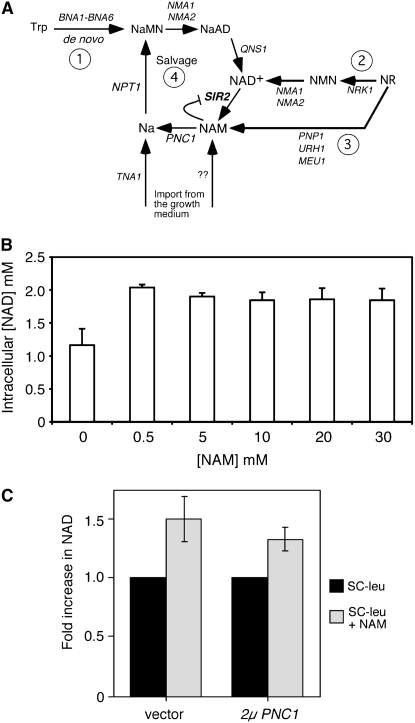
Exogenous nicotinamide raises the intracellular NAD+ concentration. (A) Schematic of known NAD+ biosynthesis pathways in S. cerevisiae. (1) The de novo NAD+ synthesis pathway begins with tryptophan (Trp) and ends with the conversion of nicotinic acid adenine dinucleotide (NaAD) into NAD+ by the NAD synthetase, QNS1. (2) The nicotinamide riboside (NR) pathway begins with the import of exogenous NR and its phosphorylation by Nrk1p into nicotinamide mononucleotide (NMN). (3) NR can also be broken down into NAM by Pnp1p, Urh1p, and Meu1p. (4) The NAD+ salvage pathway begins with NAM produced by sirtuins or imported from the growth media and merges with the de novo pathway through the production of nicotinic acid mononucleotide (NaMN) by NPT1. Na, nicotinic acid. (B) Increase in cellular NAD+ concentrations caused by various concentrations of exogenous NAM in the growth medium with a WT strain (YSB348). (C) PNC1 overexpression does not elevate the NAD+ level, even when clearing a high concentration of exogenous NAM (10 mm). The NAD+ level in the absence of nicotinamide was normalized to a value of 1.0 for each strain. Error bars represent the standard deviation from six independent experiments.
The nicotinamide produced by Sir2 and the other sirtuins is a potent noncompetitive inhibitor of their deacetylase activity (Landry et al. 2000; Bitterman et al. 2002). As a result, researchers commonly use high concentrations of exogenous NAM in the growth medium (0.5–25 mm) to inhibit the various sirtuins in their experiments (Bitterman et al. 2002; Yeung et al. 2004; Tsuchiya et al. 2006). For example, inhibition of yeast Sir2p with 5 mm NAM eliminates all three forms of silencing and shortens replicative life span (Bitterman et al. 2002). Overexpression of PNC1 suppresses these silencing defects by converting the excessive NAM into nicotinic acid (Gallo et al. 2004). However, NAM is also a key intermediate of both the NAD+ and NR salvage pathways that could potentially have positive effects on sirtuin function by influencing NAD+ production. In this study, therefore, we have investigated the effects of inhibitory NAM concentrations in the growth medium on yeast NAD+ synthesis to gain a better understanding of the relevant pathways. We find that the exogenous NAM elevates the intracellular NAD+ concentration partly through the Preiss–Handler and NR salvage pathways. Interestingly, when PNC1 is overexpressed to clear the excess NAM, the increased flux through the Preiss–Handler pathway triggers an epigenetic modification in the silencing of a mURA3 reporter gene positioned at the rDNA, such that growth occurs on both synthetic complete (SC) −ura and SC +FOA, a phenotype that is very similar to telomeric silencing of URA3. The results strongly suggest that NAM clearance by Pnc1 stabilizes the rDNA chromatin structure.
MATERIALS AND METHODS
Strains and plasmids:
Yeast media were as previously described (Smith and Boeke 1997; Sandmeier et al. 2002). Yeast extract–peptone–dextrose (YPD) and SC media (Burke et al. 2000) were supplemented with NAM at the appropriate concentrations where indicated. Counterselection against URA3 expression was carried out in SC medium containing 0.1% (w/v) 5-FOA (Toronto Research Chemicals). All yeast strains were grown at 30°. BNA1, NPT1, NRK1, PNC1, TNA1, SIR2, SIR3, and SIR4 open reading frames (ORF) were deleted and replaced with kanMX4 using a one-step PCR-mediated gene replacement protocol (Lorenz et al. 1995). All gene deletions were confirmed by PCR. The genotypes of strains used in this study are listed in Table 1. Plasmids pJOE30 and pJOE31 were constructed by PCR amplification of PNC1, including 451 bp upstream and 339 bp downstream of the ORF, from genomic DNA (Gallo et al. 2004). The PCR product was digested with XhoI and ligated into plasmid pRS424 (TRP1) or pRS425 (LEU2). Plasmids pJSS95-3 and pFR1 were constructed by PCR amplification of the yeast PNC1 open reading frame from genomic DNA or Escherichia coli pncA open reading frame from genomic DNA with HindIII tails on the oligonucleotides. The resulting PCR products were digested with HindIII and ligated into pAAH5 at the HindIII site downstream of the ADH1 promoter (Ammerer 1983). All plasmids used in this study are listed in Table 2.
TABLE 1.
Yeast strains
| Strain | Genotype | Figures |
|---|---|---|
| YSB348a | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3b | 1B, 6B, 6C |
| CGY101 | YSB348 pRS425 | 1C, 2A, 2C, 3A, 4C, 6A |
| CGY102 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(300L)∷mURA3-HIS3 pRS425 | 3A, 3B |
| CGY103 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(600L)∷mURA3-HIS3 pRS425 | 3A, 3B |
| CGY111 | YSB348 pJOE31 | 1C, 2B, 2C, 3A, 3B, 4C, 6A |
| CGY112 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(300L)∷mURA3-HIS3 pJOE31 | 3A, 3B |
| CGY113 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(600L)∷mURA3-HIS3 pJOE31 | 3A, 3B |
| CGY129 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3 sir2Δ∷kanMX4 pRS425 | 4C |
| CGY130 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3 sir2Δ∷kanMX4 pJOE31 | 4C |
| CGY132 | MATα his3Δ200 leu2Δ1 ura3-167 trp1Δ∷mURA3-HIS3 pRS425 | 3A, 3B |
| CGY133 | MATα his3Δ200 leu2Δ1 ura3-167 trp1Δ∷mURA3-HIS3 pJOE31 | 3A, 3B |
| CGY145 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3 npt1Δ∷kanMX4 | 6B, 6C |
| CGY146 | CGY145 pRS425 | 6A |
| CGY147 | CGY145 pJOE31 | 6A |
| CGY164 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3 sir4Δ∷kanMX4pRS425 | 4C |
| CGY153 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3 bna1Δ∷kanMX4 | 6B |
| CGY165 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3 sir4Δ∷kanMX4 pJOE31 | 4C |
| CGY166 | CGY153 pRS425 | 6A |
| CGY167 | CGY153 pJOE31 | 6A |
| DSY35c | MATahis3Δ200 leu2Δ0 lys2Δ202 trp1Δ63 ura3-52 ADH4∷URA3-TEL pRS425 | 4A |
| DSY37c | MATahis3Δ200 leu2Δ0 lys2Δ202 trp1Δ63 ura3-52 ADH4∷URA3-TEL pJOE31 | 4A |
| JS1153c | MATahis3Δ200 leu2Δ0 lys2Δ202 trp1Δ63 ura3-52 ADH4∷URA3-TEL sir2Δ∷kanMX4 pRS425 | 4A |
| JS1154c | MATahis3Δ200 leu2Δ0 lys2Δ202 trp1Δ63 ura3-52 ADH4∷URA3-TEL sir2Δ∷kanMX4 pJOE31 | 4A |
| JM98 | MATα his3Δ200 leu2Δ1 met15Δ0 ura3-167 RDN1(50L)∷mURA3-HIS3hst1Δ∷kanMX4 | 6D |
| JM212 | MATα his3Δ200 leu2Δ1 met15Δ0 ura3-167 RDN1(50L)∷mURA3-HIS3 nrk1Δ∷kanMX4 tna1Δ∷kanMX4 | 6B |
| JM234 | MATα his3Δ200 leu2Δ1 met15Δ0 ura3-167 RDN1(50L)∷mURA3-HIS3 nrk1Δ∷kanMX4 bna1Δ∷kanMX4 | 6B |
| JM236 | MATα his3Δ200 leu2Δ1 met15Δ0 ura3-167 RDN1(50L)∷mURA3-HIS3 nrk1Δ∷kanMX4 npt1Δ∷kanMX4 | 6B |
| JM238 | JS932 pRS425 | 6A |
| JM240 | JS932 pJOE31 | 6A |
| JM242 | JS944 pRS425 | 6A |
| JM244 | JS944 pJOE31 | 6A |
| JM254 | JM236 pRS425 | 6A |
| JM256 | JM236 pJOE31 | 6A |
| JM265d | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hmrΔA∷TRP1 pRS425 | 4B |
| JM267d | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hmrΔA∷TRP1 pJOE31 | 4B |
| JM269d | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hmrΔA∷TRP1 sir2∷HIS3pRS425 | 4B |
| JM271d | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hmrΔA∷TRP1 sir2∷HIS3pJOE31 | 4B |
| JM311 | YSB348 pFR1 | 2C |
| JM313 | YSB348 pJSS95-3 | 2C |
| JM315 | YSB348 pAAH5 | 2C |
| JS902 | MATα his3Δ200 leu2Δ1 ura3-167 pnc1Δ∷kanMX4 | 6B |
| JS932 | MATahis3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3 tna1Δ∷kanMX4 | 6B |
| JS944 | YSB348 nrk1Δ∷kanMX4 | 6B |
| JS1011 | MATα his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1(50L)∷mURA3-HIS3 | 6D |
| JS1041 | MATα his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1(50L)∷mURA3-HIS3sir2Δ∷kanMX4 | 6D |
| JS1046 | JS1041 pGLC463, pRS424 | 5 |
| JS1047 | JS1041 pGLC463, pJOE30 | 5 |
| JS1048 | JS1041 pGLC26, pRS424 | 5 |
| JS1049 | JS1041 pGLC26, pJOE30 | 5 |
| JS1050 | JS1041 pGLC65, pRS424 | 5 |
| JS1051 | JS1041 pGLC65, pJOE30 | 5 |
| JS1056 | JS1041 pGLC252, pRS424 | 5 |
| JS1057 | JS1041 pGLC252, pJOE30 | 5 |
| JS1098 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3 sir3Δ∷kanMX4 pRS425 | 4C |
| JS1099 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(50L)∷mURA3-HIS3 sir3Δ∷kanMX4 pJOE31 | 4C |
| JS1100 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(PRO+-61L)∷mURA3-HIS3 pRS425 | 2D |
| JS1101 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(PRO+-61L)∷mURA3-HIS3 pJOE31 | 2D |
| JS1102 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(proΔ-61L)∷mURA3-HIS3 pRS425 | 2D |
| JS1103 | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(proΔ-61L)∷mURA3-HIS3 pJOE31 | 2D |
| JS1129 | YSB348 pSB766 | 2B, 3B |
| JS1133 | YSB348 pRS425, pRS424 | 3C |
| JS1134 | YSB348 pSB766, pRS424 | 3C |
| JS1135 | YSB348 pRS425, pJOE30 | 3C |
| JS1136 | YSB348 pSB766, pJOE30 | 3C |
Strain described in Buck et al. (2002).
Other parental strains with mURA3-HIS3 located in unique sequence flanking the rDNA locus were described in Buck et al. (2002).
Parental strain JJSy143 was described in Gallo et al. (2004).
Parental strain YLS59 was described in Sussel and Shore (1991).
TABLE 2.
Plasmids used in the study
| Plasmid | Description | Reference |
|---|---|---|
| pRS425 | 2μ LEU2 shuttle vector | Christianson et al. (1992) |
| pRS424 | 2μ TRP1 shuttle vector | Christianson et al. (1992) |
| pJOE30 | PNC1 in pRS424 | Gallo et al. (2004) |
| pJOE31 | PNC1 in pRS424 | This study |
| pSB766 | SIR2 in pRS425 | Buck et al. (2002) |
| pGLC463 | CEN LEU2 shuttle vector | Cuperus et al. (2000) |
| pGLC26 | SIR2 in pGLC463 | Cuperus et al. (2000) |
| pGLC65 | sir2-81 in pGLC463 | Cuperus et al. (2000) |
| pGLC252 | sir2-424 in pGLC463 | Cuperus et al. (2000) |
| pAAH5 | 2μ LEU2 expression vector | Ammerer (1983) |
| pJSS95-3 | Yeast PNC1 ORF in pAAH5 | This study |
| pFR1 | E. coli pncA ORF in pAAH5 | This study |
Silencing assays:
Strains were patched onto SC medium lacking leucine, tryptophan, or both (where indicated) and allowed to grow for ∼15–20 hr. Cells were resuspended in sterile water, normalized to an OD600 of 1.0, serially diluted in fivefold increments in a 96-well plate, and then 5 μl of each dilution spotted onto the appropriate SC agar plates. To assay for telomeric silencing, cells were spotted onto SC −leucine plates to measure overall growth while selecting for a LEU2-containing plasmid, and SC −leucine +FOA agar plates to detect repression of a URA3 reporter gene positioned at the left arm telomere of chromosome VII as previously described (Gottschling et al. 1990; Smith et al. 2000). To measure rDNA silencing, strains were spotted onto SC −leu plates to measure overall growth ability, and SC −leu −ura or SC −leu +FOA plates to detect silencing of the mURA3 reporter gene positioned 50, 300, or 600 bp left of the rDNA array where indicated (Buck et al. 2002). To measure HMR silencing, strains were spotted onto SC −leu to evaluate overall growth ability, and SC −leu −trp to detect silencing of the TRP1 reporter integrated into HMR of strain background YLS59 (Sussel and Shore 1991). NAM was added to the medium where indicated. Photos of SC, SC −leu, and SC −leu −trp plates were taken after 2 days growth and photos of all −ura and FOA plates were taken after 4 days growth.
Cellular NAD+ measurements:
Determination of relative NAD+ levels in various strains was performed as previously described (Smith et al. 2000). Two hundred fifty milliliters yeast cultures in SC medium with the indicated supplements were grown to an OD600 of ∼1.0 and then harvested by centrifugation. Cell pellets were extracted for 30 min with 2.5 ml of ice-cold 1 m formic acid (saturated with butanol). Six hundred twenty-five microliters of 100% trichloroacetic acid (TCA) were added and incubated on ice for 15 min. The mixture was centrifuged at 4000× g for 20 min, and the acid soluble supernatant (containing the NAD+) was saved. The pellet was reextracted with 1.25 ml of 20% TCA and pelleted again. The supernatants were combined and used for the NAD+ measurement. Acid extract (150 μl) was added to a reaction buffer (1 ml final volume) containing 300 mm Tris-HCl, pH 9.7, 200 mm lysine-HCl, 0.2% ethanol, and 150 μg/ml alcohol dehydrogenase (Sigma). Reactions were incubated at 30° for 20 min. The absorbance was then measured at 340 nm with a Shimadzu UV-1201S spectrophotometer and the cellular NAD+ concentration calculated as previously described (Belenky et al. 2007).
RESULTS
Exogenous nicotinamide raises the intracellular NAD+ concentration:
NAM that is produced by the breakdown of NAD+ and NR, or is imported from the growth medium (Figure 1A, pathways 2 and 4), is converted to nicotinic acid in yeast cells by the nicotinamidase, Pnc1p. Since exogenous NAM is commonly used as a sirtuin inhibitor at high (0.5–25 mm) concentrations (Bitterman et al. 2002; Yeung et al. 2004; Tsuchiya et al. 2006), we were interested in determining the effects of NAM addition on the intracellular NAD+ concentration. In an earlier study, rDNA and telomeric silencing in a PNC1 strain were inhibited by 5 mm NAM in the growth medium, but not 0.5 mm (Gallo et al. 2004). As shown in Figure 1B, NAM concentrations between 0.5 and 30 mm all caused a similar 30–50% increase in the intracellular NAD+ concentration. We hypothesized that overexpressing PNC1 would increase flux through the salvage pathway at the higher NAM concentrations, leading to even higher NAD+. However, as shown in Figure 1C for 10 mm NAM, this was not the case. There was no greater increase in NAD+ levels for cells containing a high-copy PNC1 plasmid compared to cells containing an empty plasmid. Pnc1p is therefore not a limiting factor for overall cellular NAD+ production from exogenous NAM at these concentrations.
Modifying the epigenetic properties of rDNA silencing:
Having established that yeast can utilize exogenous NAM for NAD+ synthesis, we next tested whether the increased NAD+ levels affected Sir2p-mediated silencing activity. An increase in nuclear NAD+ concentration could potentially improve the histone deacetylase activity of Sir2p, leading to stronger silencing at the rDNA. To optimize the chances of detecting an increase in rDNA silencing, we utilized a reporter strain in which a modified URA3 gene (mURA3) was stably integrated within unique chromosome XII sequence, 50 bp left (50L) of the tandem array (Figure 2A). Silencing at this position is easily monitored by poor growth on SC medium lacking uracil (Buck et al. 2002) (Figure 2B, SC −leu −ura, 0 mm NAM). The level of mURA3 repression that occurs with this reporter is usually not sufficient to support growth on counterselectable SC medium containing 0.1% 5-FOA, which is toxic for Ura+ cells (Figure 2B, SC −leu +FOA, vector). Strengthening silencing through the introduction of a 2μ SIR2 plasmid suppressed colony formation even further on SC −leu −ura, and triggered robust colony growth on the FOA plate (Figure 2B, 2μ SIR2, 0 mm NAM). Therefore, a threshold of mURA3 silencing has to be reached to develop an FOA-resistant colony with this stable reporter system, making it ideal for detecting improvements in rDNA silencing, and consequently Sir2p activity.
Figure 2.—
Nicotinamide clearance alters the epigenetic properties of rDNA silencing. (A) Schematic of the centromere-proximal rDNA repeat on chromosome XII. The mURA3-HIS3 silencing reporter cassette is integrated in unique sequence, 50 bp left (50L) of the rDNA NTS1 region. The directions of Pol I and Pol II transcription are indicated by horizontal black arrows. (B) rDNA silencing assay in which the 50L∷mURA3-HIS3 silencing reporter strain (YSB348) was transformed with an empty LEU2 vector (pRS425) or a 2μ PNC1 vector (pJOE31). The concentration of NAM in the plates is indicated. (C) rDNA silencing assay showing the FOAR phenotype in the presence of 0, 10, or 20 mm NAM. YSB348 was transformed with pRS425, pJOE31, an empty ADH1-promoter vector (pAAH5), or the same ADH vector expressing PNC1 (pJSS95-3), or E. coli pncA (pFR1). (D) Requirement of modified rDNA silencing for RNA Pol I transcription. A control strain (PRO+; YSB505) and a strain deleted for the leftmost Pol I promoter (proΔ; YSB509) were transformed with the pRS425 vector or pJOE31 PNC1 vector. The mURA3-HIS3 reporter was located 61 bp left of the tandem array. An X in the schematic indicates the location of the promoter deletion. The addition of 10 mm nicotinamide is indicated as +NAM.
Enhancement of rDNA silencing (strong FOA-resistant growth) by the 2μ SIR2 plasmid was blocked by NAM concentrations of 5 or 10 mm (Figure 2B), suggesting that any positive effect of the higher NAD+ levels on rDNA silencing would require clearance of the excess NAM. Introducing the 2μ PNC1 plasmid had little effect on FOA-resistant growth with either 0 or 5 mm NAM in the media, but induced strong growth on FOA plates containing 10 mm NAM (Figure 2B, bottom row). Despite the strong growth on FOA, which was suggestive of improved silencing, the PNC1 overexpression strain also grew well on SC −leu −ura plates containing 10 mm NAM. This phenotype closely resembled telomeric silencing of URA3, where growth occurs on both FOA and −ura media because there are two classes of cells in the population: one in which URA3 is silenced and one in which it is not silenced (Gottschling et al. 1990). Ribosomal DNA silencing may, therefore, take on this telomere position effect (TPE)-like epigenetic characteristic under the PNC1 overexpression/high NAM growth condition.
Since 10 mm NAM produced the strong FOAR phenotype, but 5 mm NAM did not, we tested whether an even higher NAM concentration would improve the effect. Twenty millimolar NAM consistently produced growth on FOA that was stronger than with 10 mm (Figure 2C). However, concentrations of ≥30 mm produced nonspecific growth defects even when PNC1 was overexpressed (data not shown). Overexpression of the E. coli pncA gene, a close homolog of yeast PNC1, was sufficient to produce the strong FOAR phenotype in either 10 or 20 mm NAM (Figure 2C), suggesting that the nicotinamidase activity of the enzyme is the key trigger of silencing activity, not some other intrinsic property of yeast Pnc1p. Despite causing an equivalent increase in NAD+ concentration, clearance of 0.5 or 5 mm NAM was not sufficient to induce the FOAR silencing phenotype (Figure 2B and data not shown), suggesting that very high flux through the Preiss–Handler pathway is critical. Throughout the rest of the article we will refer to this 2μ PNC1/10 mm NAM-induced FOA-resistant phenotype as “modified rDNA silencing.”
Silencing of mURA3 positioned in the unique sequence flanking the leftmost rDNA repeat was earlier shown to require transcription by Pol I, such that deleting the Pol I promoter of the leftmost repeat (Figure 2D, schematic) prevented silencing of the downstream mURA3 reporter (Buck et al. 2002). We used this promoter-deletion strain (proΔ) to test whether modified rDNA silencing was also dependent on Pol I transcription. As shown in Figure 2D, without the addition of NAM, silencing was lost in the promoter-deletion strain as expected (SC −leu −ura, vector/proΔ). When NAM was added, FOA-resistant growth was observed only when PNC1 was overexpressed in the control PRO+ strain, but not in the proΔ strain (Figure 2D, bottom row). These results indicate that modified rDNA silencing retains the defining property of Pol I dependence.
Spreading of modified rDNA silencing:
Typical rDNA silencing is also characterized by its ability to unidirectionally spread from the rDNA toward the centromere, although without SIR2 overexpression, the spreading progresses only <600 bp. Overexpressing SIR2 extends the spreading to at least 2000 bp beyond the rDNA (Buck et al. 2002). Since modified rDNA silencing and SIR2 overexpression both induced a strong FOA-resistant phenotype when mURA3 was positioned 50 bp (50L) from the rDNA (Figure 2), it was possible that modified rDNA silencing was also spreading beyond the 50L position. We therefore tested the effect of modified rDNA silencing of mURA3 positioned 300 bp (300L) and 600 bp (600L) away from the rDNA (Figure 3A). The FOA-resistant phenotype was easily observed at the 300L position when PNC1 was overexpressed in the presence of NAM, but was extremely weak at the 600L position. This was very different from the effect of overexpressing SIR2 in the absence of NAM, which easily spread silencing to the 600L position (Figure 3B). Modified rDNA silencing, therefore, does not appear to spread from the rDNA any more efficiently than typical rDNA silencing. Importantly, the NAM/PNC1 effect on mURA3 was specific for the rDNA location because the mURA3 expression was unaffected when it was positioned at the nonsilenced TRP1 locus (Figure 3, A and B).
Figure 3.—
Spreading of modified rDNA silencing. (A) rDNA silencing in which the mURA3-HIS3 cassette was positioned 50 bp (50L), 300 bp (300L), or 600 bp (600L) away from the rDNA array or at the nonsilenced TRP1 locus on chromosome IV. Plates contained 0 or 10 mm NAM, and the strains harbored either the empty pRS425 vector or the 2μ PNC1 vector (pJOE31). (B) Effects of SIR2 overexpression on rDNA silencing at the 50L, 300L, 600L, and TRP1 locations. NAM was not added. (C) Effect of overexpressing SIR2 (pSB766) and PNC1 (pJOE30) at the same time on rDNA silencing in the absence or presence of 10 mm NAM. The mURA3-HIS3 reporter was positioned at the 50L position.
As mentioned above, growth on both FOA and −ura media is an epigenetic characteristic of URA3 telomeric silencing. The similarities with modified rDNA silencing suggested that NAM clearance by Pnc1p may have stabilized the silent rDNA chromatin state without significantly increasing the percentage of these cells in the population. We hypothesized that increasing the SIR2 dosage would stabilize the silenced phenotype even more and perhaps shift more cells in the population toward the silenced state. To test this possibility we overexpressed PNC1 and SIR2 at the same time in the presence of 10 mm NAM. As shown in Figure 3C, the addition of 2μ SIR2 produced an even stronger FOA-resistant phenotype than 2μ PNC1 alone. Importantly, it also reduced the proportion of cells that were able to grow on plates lacking uracil (SC −leu −trp −ura), which is consistent with more cells being in the silenced state.
Modified rDNA silencing does not occur at the expense of telomeric and HMR silencing:
The telomeric and rDNA silenced domains compete for a limited pool of Sir2p (Smith et al. 1998). Therefore, we reasoned that the modified rDNA silencing phenotype could involve Sir2p mobilization from telomeres or the HM loci to the rDNA, resulting in weakened telomeric or HM silencing. To test this idea, we analyzed the effect of PNC1 overexpression and 10 mm NAM on TPE and HM silencing (Figure 4, A and B). As expected, 10 mm NAM in the growth medium caused a loss of TPE when the empty vector was present, as indicated by a lack of growth on the SC −leu +FOA (10 mm NAM) plate (Figure 4A, top row). PNC1 overexpression restored normal levels of silencing on the NAM plate in a SIR2-dependent manner (Figure 4A), indicating there was no significant loss of TPE under the same condition that induced FOA resistance for rDNA silencing. Similarly, PNC1 overexpression almost completely restored silencing of a TRP1 reporter gene integrated at the HMR locus in the presence of 10 mm NAM (Figure 4B, SC −leu −trp + 10 mm NAM plate). The modified rDNA silencing phenotype in this growth condition is therefore probably not caused by a large-scale redistribution of Sir2 from telomeres or the HM loci to the rDNA.
Figure 4.—
Modified rDNA silencing does not disturb HM and telomeric silencing, but requires SIR2. (A and B) Telomeric and HM silencing are not significantly weakened or strengthened by 10 mm NAM coupled with PNC1 overexpression. (A) Fivefold serial dilutions of WT and sir2Δ strains containing pRS425 or pJOE31. The URA3 gene is silenced by an artificial telomere formed at the ADH4 locus. Silencing is indicated by growth on 5-FOA. (B) Fivefold serial dilutions of strain YLS59 (WT) and MC119 (sir2Δ) containing either pRS425 vector or pJOE31 (PNC1). The TRP1 gene is silenced at the HMR locus. Silencing is indicated by lack of growth on SC −leu −trp plates. (C) Effects of deleting SIR2, SIR3, or SIR4 on rDNA silencing in the presence of 10 mm nicotinamide. The empty vector (pRS425) or PNC1 overexpression vector (pJOE31) was transformed into rDNA reporter strains that were deleted for SIR2 (YSB408), SIR3 (CGY135), or SIR4 (CGY152). Fivefold serial dilutions were spotted.
Modified rDNA silencing is dependent on Sir2p and RENT, but not the SIR complex:
Another defining characteristic of typical rDNA silencing is dependence on Sir2p, but not the other Sir proteins (Smith and Boeke 1997). Since the modified rDNA silencing phenotype resembled telomeric silencing of URA3, it was feasible that the SIR complex was being utilized for rDNA silencing under this growth condition. To test this possibility, we deleted SIR2, SIR3, or SIR4 from the 50L∷mURA3 rDNA silencing reporter strain. As shown in Figure 4C, deletion of SIR2 derepressed silencing of mURA3 as measured by robust Ura+ growth either with or without 10 mm NAM added. As expected, deleting SIR2 also prevented strong FOA resistance when PNC1 was overexpressed in the presence of 10 mm NAM (Figure 4C, +NAM). However, deleting SIR3 or SIR4 had little effect. These mutations instead improved the repression phenotype on SC −leu −ura when NAM was cleared by PNC1 overexpression, which was previously observed with typical rDNA silencing (Smith and Boeke 1997).
To determine whether the defect in modified rDNA silencing caused by the sir2 deletion reflected a defect in RENT, we tested the effects of sir2 point mutations that inhibited either the RENT or SIR complex. Low-copy LEU2 CEN plasmids expressing either wild-type SIR2 or mutant versions of sir2 that were specifically defective in either telomeric/HM silencing (class I mutant) or rDNA silencing (class II mutant) (Cuperus et al. 2000) were transformed into a sir2Δ reporter strain. A TRP1 2μ PNC1 plasmid (pJOE30) or empty vector (pRS424) was then introduced such that the strains harbored two selectable plasmids on SC −leu −trp media. As shown in Figure 5 (−NAM, top row), the sir2Δ mutant was again defective for silencing when transformed with the empty vectors, as indicted by full growth on the SC −leu −trp −ura plate. The wild-type CEN SIR2 plasmid restored some silencing on SC −leu −trp −ura, but not enough to trigger FOA resistance. The class II mutant (sir2-81) did not strengthen silencing at all in the absence of NAM (Figure 5, −NAM), which was consistent with this type of sir2 mutant being rDNA silencing defective (Cuperus et al. 2000). The class I mutant (sir2-424) restored silencing even better than wild type (WT) (Figure 5, −NAM), most likely due to redistribution of telomeric/HM Sir2p to the nucleolus (Cuperus et al. 2000), which also occurs in sir4Δ mutants (Gotta et al. 1997; Kennedy et al. 1997; Smith et al. 1998; Cuperus et al. 2000). When 10 mm NAM was added to the plates and PNC1 was overexpressed, the WT SIR2 and sir2-424 (class I) mutant produced the FOA-resistant silencing phenotype (Figure 5, +NAM). No FOA resistance was observed with sir2-81 (class II mutant). Combined with the sir3Δ and sir4Δ phenotypes (Figure 4C), these results demonstrate that the modified form of rDNA silencing is similar to typical rDNA silencing in that it utilizes RENT, but not the SIR complex. Despite the epigenetic similarities, modified rDNA silencing retains the key rDNA-specific properties that differentiate rDNA silencing from TPE and HM silencing.
Figure 5.—
Modified rDNA silencing is dependent on the RENT, but not the SIR complex. The effects of expressing a telomere/HM silencing-defective sir2 mutation (sir2-424, class I) and rDNA silencing-defective mutation (sir2-81, class II) on rDNA silencing in the presence of 10 mm NAM and PNC1 overexpression. The strain background was JS1041, which lacks the endogenous SIR2 gene and has the 50L∷mURA3-HIS3 reporter gene.
Nicotinamide effects on rDNA silencing are mediated by the Preiss–Handler pathway:
Efficient rDNA silencing was previously shown to be dependent on the Preiss–Handler pathway, of which the Npt1 protein is partially localized in the nucleus (Anderson et al. 2002; Sandmeier et al. 2002). The FOA-resistant phenotype of modified rDNA silencing was shown in this study to be dependent on the clearance of high NAM concentrations by Pnc1, a key step of the salvage pathway (Figure 2). Therefore, we tested whether Npt1, or other salvage and biosynthesis enzymes, was required for development of the FOA-resistant phenotype. Deleting BNA1, TNA1, or NRK1 caused little effect compared to WT (Figure 6A). In contrast, deleting NPT1 completely eliminated growth on FOA in the presence of NAM, suggesting that the nicotinic acid produced by Pnc1p must pass through the Preiss–Handler pathway to induce the modified rDNA silencing phenotype.
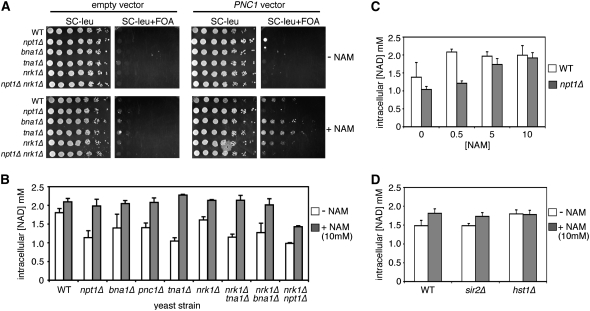
Figure 6.—
Effects of NAD+ salvage and biosynthesis mutants on modified rDNA silencing and intracellular NAD+ concentrations. (A) Modified rDNA silencing in WT (YSB348), npt1Δ (CGY145), bna1Δ (CGY153), tna1Δ (JS932), nrk1Δ (JS944), and npt1Δ nrk1Δ (JM236) mutants. The reporter gene is RDN1(50L)∷mURA3-HIS3. (B) NAD+ concentrations in the various NAD+ biosynthesis and salvage mutants when grown in the presence or absence of 10 mm NAM. (C) NPT1 is required for increasing NAD+ levels when cells are grown in low concentrations (0.5 mm) of NAM. (D) Intracellular NAD+ concentrations in WT (JS1011), sir2Δ (JS1041), and hst1Δ (JM98) strains in the presence or absence of 10 mm NAM.
Deleting NPT1 was proposed to inhibit silencing by reducing the nuclear pool of NAD+ (Anderson et al. 2002; Sandmeier et al. 2002). We therefore predicted that the absence of NPT1 or PNC1 would block the increase in NAD+ caused by the addition of 10 mm NAM. But surprisingly, these genes were not required for the elevation in NAD+ (Figure 6B). In fact, no single mutations in the NAD+ salvage/synthesis genes that we tested blocked the increase (Figure 6B). We hypothesized that Npt1p was utilizing a specific proportion of the excess NAM in the nucleus to impact silencing, but that the very high concentrations saturated the cellwide Preiss–Handler pathway. To test this hypothesis we examined whether NPT1 was required for the increase in NAD+ caused by NAM concentrations <10 mm. As shown in Figure 6C, the lack of NPT1 did block the elevation in NAD+ caused by 0.5 mm NAM, indicating that NPT1 is capable of converting a relatively small proportion of external NAM into NAD+, but that higher concentrations overwhelm the system.
It was likely that another redundant pathway(s) was responsible for the rest of the NAM at higher concentrations. Deleting NRK1 by itself did not block the increase in NAD+ (Figure 6B) and had little effect on modified rDNA silencing (Figure 6A). So we next tested whether there was any redundancy between NRK1 and the other pathways in utilizing the high NAM concentrations. Interestingly, the npt1Δ nrk1Δ double mutant partially blocked the increase in NAD+ caused by the addition of 10 mm NAM (Figure 6B), suggesting that part of the effect is mediated by NR when the Preiss–Handler pathway is blocked. The remainder of the NAD+ increase could potentially come from the inhibition of Hst1 by NAM. Deleting the yeast HST1 gene was previously shown to cause an increase in cellular NAD+ concentration through the activation of genes in the de novo NAD+ synthesis pathway (Bedalov et al. 2003). Hst1 normally functions as a repressor of these genes (Bedalov et al. 2003). As shown in Figure 6D, deletion of HST1 did increase the NAD+ concentration under this growth condition, but the addition of 10 mm NAM did not cause an additional increase. Taken together these results suggest that there are redundant pathways (both direct and indirect) in which yeast cells utilize NAM to raise the intracellular NAD+ concentration. The results also support a model for Sir2p-dependent heterochromatin formation in the yeast rDNA, in which flux through the Preiss–Handler NAD+ salvage pathway (perhaps in the nucleus) leads to activation of the nuclear sirtuin, Sir2p.
DISCUSSION
The epigenetics of rDNA silencing:
The rDNA is an unusual locus in that it is highly transcribed by RNA polymerase I, yet Pol II-transcribed genes that are endogenous to the rDNA or artificially integrated into the rDNA are silenced in a SIR2- and Pol I-dependent manner (Bryk et al. 1997, 2002; Fritze et al. 1997; Smith and Boeke 1997; Cioci et al. 2003). Silencing at the HM loci and telomeres is stably maintained and inherited through a process that requires not only SIR2, but also SIR3 and SIR4, which together encode the SIR complex (Loo and Rine 1995). Other factors involved in the maintenance include chromatin assembly factor (CAF) and PCNA (Monson et al. 1997; Enomoto and Berman 1998; Zhang et al. 2000). Silencing at these locations is characterized by the formation of two independent chromatin states, one repressive to transcription and the other not repressive. These two states can occasionally switch, especially with telomeric silencing and HM silencing when SIR1 is deleted (Pillus and Rine 1989; Gottschling et al. 1990).
It is not clear whether silencing in the rDNA involves two independent chromatin states or is the average of a mixture of varying degrees of repression (Smith and Boeke 1997). Whatever defines the silent state, it is not thought to be stably maintained from cell to cell due to the dynamic and stochastic nature of DNA replication and Pol I transcription within the tandem array, two processes that promote rDNA silencing (Smith et al. 1999; Buck et al. 2002; Cioci et al. 2003). Only about one in five rDNA repeats are replicated in any given S phase (Linskens and Huberman 1988; Fangman and Brewer 1991), and ∼50% of the rDNA genes are transcribed by Pol I at any given cell cycle (Dammann et al. 1993). Moreover, the distribution of the active genes within the tandem array is stochastic (Dammann et al. 1995). Since the Pol I transcriptional status of a single rDNA repeat can influence the silencing of an adjacent reporter gene (Buck et al. 2002), there is an ∼50% chance that a given reporter gene will be silenced, which could change at each cell cycle. As a result, when silencing of a color-indicator reporter gene such as MET15 is monitored, instead of colonies with large sectors of brown (silenced) and white (not silenced) cells, a tan color is observed, suggesting that the silent state is not stable enough to be inherited over multiple generations (Smith and Boeke 1997). A similar phenomenon is observed with silencing of a telomeric ADE2 reporter gene. In a WT strain, large sectors of silenced (red) and nonsilenced (white) cells are typically observed. But when the maintenance of silencing is impaired by deletion of the CAC1 gene, an overall pink colony color develops (Monson et al. 1997).
If a URA3 reporter is placed next to a telomere, there are cells in the population that are not silencing URA3 and grow on SC −ura and cells that are silencing URA3 that grow only on FOA plates. The result is the ability of the cell population to grow on both SC −ura and SC +FOA. Such a situation clearly does not exist in the typical rDNA silencing population. Silencing of mURA3 when positioned next to the rDNA (the 50L position) is characterized by poor growth on SC −ura plates (silencing), but no growth on SC +FOA plates. The combination of overexpressing PNC1 with high NAM concentrations in the growth medium modified the epigenetic pattern of rDNA silencing, resulting in growth on both SC −ura and FOA plates. Since this mURA3 reporter is not subjected to the high recombinational loss rates that plague reporters integrated within the rDNA array (data not shown), we propose that this growth condition sufficiently stabilizes the silent rDNA chromatin state in a subpopulation of cells to allow growth of a colony on FOA. At the same time, a large proportion of cells in the same population remained in a nonsilenced state, allowing them to grow on SC −ura plates. SIR2 overexpression also induces strong silencing of the mURA3 reporter gene, such that the cells can grow on FOA, but in this case, the population of cells that are not silencing mURA3 is greatly reduced. This is reminiscent of telomeric silencing, in which the population of cells silencing URA3 can be increased by overexpressing SIR3 (Renauld et al. 1993). SIR2 overexpression was able to do the same thing with the PNC1/NAM-induced modified rDNA silencing, shifting the cell population toward the FOA-resistant phenotype (Figure 3C). Future work will utilize this system to investigate the inheritance properties of rDNA silencing.
Stabilizing rDNA silencing possibly by increasing flux through the Preiss–Handler NAD+ salvage pathway:
The similarities between modified rDNA silencing and telomeric silencing of URA3 initially suggested that the SIR complex was functioning in the rDNA. Sir3p had previously been shown to weakly interact with the rDNA (Hoppe et al. 2002), which we have also observed (C. Gallo and M. Matecic, unpublished data). However, deleting SIR3 or SIR4 had little effect on the FOA-resistant phenotype. Additionally, only sir2 mutations that affect the RENT complex (class II mutants), and not the SIR complex (class I) were deficient in modified rDNA silencing. Other key characteristics of typical rDNA silencing were retained, including the requirement for Pol I transcription and the ability to spread outward from the rDNA into adjacent unique sequence. These results strongly suggest that the combination of PNC1 overexpression and high NAM concentrations alters the function of Sir2p within the RENT complex, although we cannot rule out the possibility that another unidentified repressive factor is also contributing.
Supplementing the yeast growth media with NAM raises the intracellular NAD+ concentration (this study), and PNC1 overexpression prevents the exogenous NAM from inhibiting Sir2p (Anderson et al. 2003a; Gallo et al. 2004). We initially hypothesized that this combination of NAM clearance and high NAD+ concentration was triggering the FOA-resistant silencing phenotype. However, we did not observe the FOA-resistant phenotype at NAM concentrations of ≤5 mm, even though the overall intracellular NAD+ levels remained elevated. Coupled with the absolute requirement of NPT1, these results instead suggest that the modified rDNA silencing phenotype may involve increased flux through the Preiss–Handler NAD+ salvage pathway, likely in the nucleus. Such a model for Sir2p activation via Npt1 and Pnc1 has also been proposed for typical rDNA silencing and the extension of replicative life span, in which increasing the copy number of NPT1 or PNC1, or calorie-restriction growth conditions strengthens silencing and extends life span without measurably increasing the overall NAD+ concentration (Anderson et al. 2002, 2003a,b). It is expected that this modified rDNA silencing phenotype will be a useful tool for investigating nuclear NAD+ production in yeast.
How does exogenous nicotinamide raise the intracellular NAD+ concentration of yeast cells?
The only known pathway in S. cerevisiae that can convert exogenous NAM into NAD+ is the Pnc1p/Npt1p-mediated NAD+ salvage pathway. This statement is supported by the finding that a qns1Δ mutant, which blocks both the de novo NAD+ synthesis and the salvage pathways (Figure 1A), is inviable when provided with exogenous NAM, but survives when given nicotinamide riboside (Bieganowski and Brenner 2004). We have observed similar results with a bna1Δ npt1Δ double mutant, which is inviable even in the presence of 10 or 20 mm NAM, but lives with 10 μm NR (data not shown). While not measurable due to its inviability, presumably there is very little NAD+ produced in the bna1Δ npt1Δ mutant, implying that the amount of NAD+ produced by exogenous NAM outside of these pathways is not sufficient to maintain cell growth. Alternatively, the NAM-derived pool of NAD+ may not be utilized efficiently by essential NAD+-dependent processes such as glycolysis and the TCA cycle.
It was therefore surprising that the addition of 10 mm NAM to the growth medium elevated the intracellular NAD+ concentration by 30–50%, even when NPT1 or PNC1 was deleted (Figure 6B). At lower NAM concentrations, NPT1 was fully required for the increase in NAD+ (Figure 6C), suggesting that the high NAM may induce one or more cryptic pathways. Yeast and most eukaryotic organisms lack a nicotinamide phosphoribosyltransferase enzyme (NAMPRT), which in mammals converts NAM into nicotinamide mononucleotide (NMN) (Revollo et al. 2004). Since mammals lack a nicotinamidase homolog of Pnc1p, the NAMPRT enzyme also likely serves to clear the inhibitory nicotinamide that would otherwise accumulate in the cell. In these organisms, the NMN is converted to NAD+ by the universally conserved nicotinamide mononucleotide adenylyltransferases (encoded by NMA1 and NMA2 in yeast) (Anderson et al. 2002). NAMPRT was reported as the rate-limiting step in NAD+ biosynthesis for mouse NIH3T3 cells because the addition of 5 mm NAM did not increase NAD+ levels, but overexpression of the transferase gene did cause an increase (Revollo et al. 2004). However, an independent study showed that adding 5 or 10 mm NAM would raise the NAD+ level of normal human fibroblasts (Kang et al. 2006). In Salmonella typhimurium, the nicotinic acid phosphoribosyl transferase PncB (Npt1p in yeast) was identified as the limiting step of the Preiss–Handler pathway (Imsande 1964). Our data is consistent with Npt1p also being a limiting factor of the pathway in yeast, except at high NAM concentrations, where other cryptic pathways are involved, including one utilizing Nrk1p. NR is not a component of standard yeast growth media, but it was recently determined that the NR salvage enzymes as a group (Nrk1p, Pnp1p, and Urh1p) contribute to NAD+ biosynthesis even in the absence of supplemented NR, suggesting that NR is not only a vitamin, but also a metabolite (Belenky et al. 2007).
Nicotinamide can also raise the NAD+ concentration through an indirect mechanism by inhibiting Hst1. However, this mechanism is unlikely to play a role in formation of the modified rDNA silencing phenotype because PNC1 overexpression prevents exogenous NAM from inhibiting Hst1 (Gallo et al. 2004). Given the importance of NAD+ as a key energy currency for the cell, it makes sense that there would be multiple pathways for the cell to respond to nicotinamide, which is both an NAD+ precursor and inhibitor of NAD+ consuming enzymes such as the sirtuins. However, there also appears to be a limit to the amount of NAD+ that the cell can synthesize in response to NAM. Despite the multiple mutants and NAM concentrations utilized in this study, the maximum NAD+ concentration we observed was always close to 2 mm and likely reflects a high degree of homeostasis for this critical cellular cofactor.
Acknowledgments
We thank David Shore for kindly providing the class I and II sir2 mutant plasmids. We also thank Joe Sandmeier for technical assistance and Charles Brenner for critical reading of the manuscript and for generously providing nicotinamide riboside. This work was supported in part by National Institutes of Health grant GM-075240 and American Heart Association Grant-in-Aid award 0555490U to J.S.S.
References
- Ammerer, G., 1983. Expression of genes in yeast using the ADC1 promoter. Methods Enzymol. 101 192–201. [DOI] [PubMed] [Google Scholar]
- Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, H. Cohen et al., 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277 18881–18890. [DOI] [PubMed] [Google Scholar]
- Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik and D. A. Sinclair, 2003. a Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, R. M., M. Latorre-Esteves, A. R. Neves, S. Lavu, O. Medvedik et al., 2003. b Yeast life-span extension by calorie restriction is independent of NAD fluctuation. Science 302 2124–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, O. M., B. L. Billington and D. E. Gottschling, 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66 1279–1287. [DOI] [PubMed] [Google Scholar]
- Bedalov, A., M. Hirao, J. Posakony, M. Nelson and J. A. Simon, 2003. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 23 7044–7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky, P., F. G. Racette, K. L. Bogan, J. M. McClure, J. S. Smith et al., 2007. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129 473–484. [DOI] [PubMed] [Google Scholar]
- Bieganowski, P., and C. Brenner, 2004. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117 495–502. [DOI] [PubMed] [Google Scholar]
- Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves and D. A. Sinclair, 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277 45099–45107. [DOI] [PubMed] [Google Scholar]
- Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel et al., 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11 255–269. [DOI] [PubMed] [Google Scholar]
- Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis et al., 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12 165–170. [DOI] [PubMed] [Google Scholar]
- Buck, S. W., J. J. Sandmeier and J. S. Smith, 2002. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111 1003–1014. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Stearns, 2000. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero and P. Hieter, 1992. Multifunctional yeast high-copy number shuttle vectors. Gene 110 119–122. [DOI] [PubMed] [Google Scholar]
- Cioci, F., L. Vu, K. Eliason, M. Oakes, I. N. Siddiqi et al., 2003. Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell 12 135–145. [DOI] [PubMed] [Google Scholar]
- Cubizolles, F., F. Martino, S. Perrod and S. M. Gasser, 2006. A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol. Cell 21 825–836. [DOI] [PubMed] [Google Scholar]
- Cuperus, G., R. Shafaatian and D. Shore, 2000. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 19 2641–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann, R., R. Lucchini, T. Koller and J. M. Sogo, 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann, R., R. Lucchini, T. Koller and J. M. Sogo, 1995. Transcription in the yeast rRNA gene locus: Distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol. 15 5294–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto, S., and J. Berman, 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 12 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman, W. L., and B. J. Brewer, 1991. Activation of replication origins within yeast chromosomes. Annu. Rev. Cell Biol. 7 375–402. [DOI] [PubMed] [Google Scholar]
- Fritze, C. E., K. Verschueren, R. Strich and R. Easton Esposito, 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo, C. M., D. L. Smith, Jr. and J. S. Smith, 2004. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 24 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidelli, S., D. Donze, N. Dhillon and R. T. Kamakaka, 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20 4522–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain, M., E. Talla and J. M. Francois, 2002. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 19 215–224. [DOI] [PubMed] [Google Scholar]
- Gotta, M., S. Strahl-Bolsinger, H. Renauld, T. Laroche, B. K. Kennedy et al., 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb, S., and R. E. Esposito, 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56 771–776. [DOI] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63 751–762. [DOI] [PubMed] [Google Scholar]
- Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie et al., 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22 4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsande, J., 1964. A cross-linked control system. I. Properties of a triphosphate-dependent nicotinic acid mononucleotide pyrophosphorylase from Bacillus subtilis. Biochim. Biophys. Acta 85 255–264. [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M., M. McVey and L. Guarente, 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H. T., H. I. Lee and E. S. Hwang, 2006. Nicotinamide extends replicative lifespan of human cells. Aging Cell 5 423–436. [DOI] [PubMed] [Google Scholar]
- Kennedy, B. K., M. Gotta, D. A. Sinclair, K. Mills, D. S. McNabb et al., 1997. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89 381–391. [DOI] [PubMed] [Google Scholar]
- Kucharczyk, R., M. Zagulski, J. Rytka and C. Herbert, 1998. The yeast gene YJR025c encodes a 3-hydroxyanthranilic acid dioxygenase and is involved in nicotinic acid biosynthesis. FEBS Lett. 424 127–130. [DOI] [PubMed] [Google Scholar]
- Landry, J., J. T. Slama and R. Sternglanz, 2000. Role of NAD+ in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278 685–690. [DOI] [PubMed] [Google Scholar]
- Lin, S.-J., P.-A. Defossez and L. Guarente, 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289 2126–2128. [DOI] [PubMed] [Google Scholar]
- Linskens, M. H., and J. A. Huberman, 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, S., and J. Rine, 1995. Silencing and heritable domains of gene expression. Annu. Rev. Cell Dev. Biol. 11 519–548. [DOI] [PubMed] [Google Scholar]
- Lorenz, M. C., R. S. Muir, E. Lim, J. McElver, S. C. Weber et al., 1995. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158 113–117. [DOI] [PubMed] [Google Scholar]
- Monson, E. K., D. de Bruin and V. A. Zakian, 1997. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl. Acad. Sci. USA 94 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus, L., and J. Rine, 1989. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59 637–647. [DOI] [PubMed] [Google Scholar]
- Rajavel, M., D. Lalo, J. W. Gross and C. Grubmeyer, 1998. Conversion of a cosubstrate to an inhibitor: phosphorylation mutants of nicotinic acid phosphoribosyltransferase. Biochemistry 37 4181–4188. [DOI] [PubMed] [Google Scholar]
- Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani et al., 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7 1133–1145. [DOI] [PubMed] [Google Scholar]
- Revollo, J. R., A. A. Grimm and S. Imai, 2004. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 279 50754–50763. [DOI] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72 481–516. [DOI] [PubMed] [Google Scholar]
- Sandmeier, J. J., I. Celic, J. D. Boeke and J. S. Smith, 2002. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD+ salvage pathway. Genetics 160 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed et al., 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97 233–244. [DOI] [PubMed] [Google Scholar]
- Shou, W., K. M. Sakamoto, J. Keener, K. W. Morimoto, E. E. Traverso et al., 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 8 45–55. [DOI] [PubMed] [Google Scholar]
- Smith, J. S., and J. D. Boeke, 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11 241–254. [DOI] [PubMed] [Google Scholar]
- Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad et al., 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. S., C. B. Brachmann, L. Pillus and J. D. Boeke, 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. S., E. Caputo and J. D. Boeke, 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies et al., 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97 245–256. [DOI] [PubMed] [Google Scholar]
- Sussel, L., and D. Shore, 1991. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc. Natl. Acad. Sci. USA 88 7749–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny, J. C., and D. Moazed, 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny, J. C., D. S. Kirkpatrick, S. A. Gerber, S. P. Gygi and D. Moazed, 2004. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell. Biol. 24 6931–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, M., N. Dang, E. O. Kerr, D. Hu, K. K. Steffen et al., 2006. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell 5 505–514. [DOI] [PubMed] [Google Scholar]
- Yeung, F., J. E. Hoberg, C. S. Ramsey, M. D. Keller, D. R. Jones et al., 2004. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., K. Shibahara and B. Stillman, 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408 221–225. [DOI] [PubMed] [Google Scholar]