Abstract
Gut granules are specialized lysosome-related organelles that act as sites of fat storage in Caenorhabditis elegans intestinal cells. We identified mutations in a gene, glo-3, that functions in the formation of embryonic gut granules. Some glo-3(−) alleles displayed a complete loss of embryonic gut granules, while other glo-3(−) alleles had reduced numbers of gut granules. A subset of glo-3 alleles led to mislocalization of gut granule contents into the intestinal lumen, consistent with a defect in intracellular trafficking. glo-3(−) embryos lacking gut granules developed into adults containing gut granules, indicating that glo-3(+) function may be differentially required during development. We find that glo-3(+) acts in parallel with or downstream of the AP-3 complex and the PGP-2 ABC transporter in gut granule biogenesis. glo-3 encodes a predicted membrane-associated protein that lacks obvious sequence homologs outside of nematodes. glo-3 expression initiates in embryonic intestinal precursors and persists almost exclusively in intestinal cells through adulthood. GLO-3∷GFP localizes to the gut granule membrane, suggesting it could play a direct role in the trafficking events at the gut granule. smg-1(−) suppression of glo-3(−) nonsense alleles indicates that the C-terminal half of GLO-3, predicted to be present in the cytoplasm, is not necessary for gut granule formation. Our studies identify GLO-3 as a novel player in the formation of lysosome-related organelles.
LYSOSOME-related organelles (LROs) comprise a functionally diverse set of cellular compartments. In mammals these organelles carry out specialized cellular functions including: storage and release of lung surfactant, secretion of blood clotting signals by α-granules, dense granules, and Weibel-Palade bodies, and pigment formation and storage by melanosomes (Raposo et al. 2007). Defects in the biogenesis of these and other LROs result in the human disease Hermansky–Pudlak syndrome (Wei 2006). While the functions of many LROs are well understood, there is comparatively much less known regarding the pathways and mechanisms that function in LRO formation.
Gut granules are newly defined LROs present within the intestinal cells of C. elegans (Hermann et al. 2005). Gut granules function as major sites for fat storage in C. elegans (Ashrafi et al. 2003; Schroeder et al. 2007). These organelles also store autofluorescent and birefringent materials (Babu 1974; Laufer et al. 1980), the identity and function of which are currently unknown. To investigate the mechanisms controlling LRO biogenesis we have identified a collection of mutants defective in gut granule biogenesis. Genes that can be mutated to give a Glo (gut granule loss) phenotype (Hermann et al. 2005) include the Rab38 homolog GLO-1, the putative GLO-1 guanine nucleotide exchange factor GLO-4, the ABC transporter PGP-2, and subunits of the AP-3 and HOPS complexes (Hermann et al. 2005; Schroeder et al. 2007). Homologs of these genes function in the membrane trafficking steps mediating the biogenesis of mammalian and Drosophila LROs (Huizing et al. 2002; Ma et al. 2004; Cheong et al. 2006; Raposo et al. 2007), indicating that the cellular pathways controlling gut granule formation are evolutionarily conserved.
Here we present our phenotypic and molecular characterization of glo-3, a novel gene that functions in Caenorhabditis elegans gut granule biogenesis. We show that glo-3 encodes a membrane-associated protein conserved in nematodes that localizes to the gut granule where it likely functions in trafficking to the compartment.
MATERIALS AND METHODS
C. elegans alleles and strains:
The wild-type strain N2 was used in our studies. All strains were grown at 22° unless otherwise noted. Strains were cultured as described (Brenner 1974). Mutant alleles are listed by linkage group and references are listed at http://wormbase.org:
LGI: apt-6(ok429), dpy-5(e61), fer-1(hc13ts), pgp-2(kx48), smg-1(r861).
LGII: rrf-3(pk1426).
LGV: dpy-11(e224), glo-4(ok623).
LGX: apt-7(tm920), egl-15(n484), glo-1(zu437), glo-3(gm125), glo-3(kx1), glo-3(kx29), glo-3(kx37), glo-3(kx38), glo-3(kx90), glo-3(kx91), glo-3(kx94), glo-3(zu446), unc-27(e159).
LG unknown: pwIs50[lmp-1∷gfp; unc-119(+)] (Treusch et al. 2004), pwIs72[vha-6p∷rab-5∷gfp; unc-119(+)] (Hermann et al. 2005), pwIs87[vha-6p∷rme-1∷gfp; unc-119(+)] (Hermann et al. 2005), pwIs170[vha-6p∷rab-7∷gfp; unc-119(+)] (Chen et al. 2006).
Extrachromosomal: kxEx9[glo-1∷gfp; rol-6(su1006)D] (Hermann et al. 2005), kxEx41[glo-3∷gfp; rol-6(su1006)D] (this work), kxEx50[glo-3p∷gfp; rol-6(su1006)D] (this work).
Genetic manipulations:
glo-3 alleles were backcrossed to N2 at least two times prior to being used for phenotypic analysis. Initial complementation tests were performed by mating glo(−)/0 males with fer-1(hc13ts); glo-3(zu446) hermaphrodites that were raised from embryogenesis through adulthood at 25°. At the nonpermissive temperature fer-1(hc13ts) animals do not produce functional sperm and are self-sterile (Ward and Miwa 1978), however mating with fer-1(+); glo(−) males results in the production of outcross progeny that were assessed for an embryonic Glo phenotype. To assess the embryonic Glo phenotype of class I/class II glo-3 alleles, transheterozygotes were constructed by mating glo-3(kx91) and glo-3(kx94) males with both fer-1(hc13ts); glo-3(zu446) and fer-1(hc13ts); glo-3(kx90) self-sterile hermaphrodites and their progeny scored. To assess the embryonic Glo phenotype of class II/class III glo-3 alleles, transheterozygotes were constructed by mating glo-3(kx1) and glo-3(kx38) males with both fer-1(hc13ts); glo-3(zu446) and fer-1(hc13ts); glo-3(kx90) self-sterile hermaphrodites and their progeny scored. The embryonic and adult Glo phenotypes exhibited by all the glo-3 alleles were found to be recessive and expressed zygotically using standard genetic techniques. Maternal modification of the Glo phenotype was scored in the embryonic progeny of glo-3(−)/+ hermaphrodites.
smg-1(r861); glo-3(−) animals were generated by mating glo-3(−) males with smg-1(r861) hermaphrodites. Dpy L1 progeny (Hermann et al. 2005) of the resulting transheterozygotes were isolated to select for glo-3(−) homozygotes. Adults in the next generation that displayed a protruding vulva were selected, as smg-1(−) homozygotes display this phenotype (Hodgkin et al. 1989). In all cases multiple double mutant lines were isolated and scored.
Double mutants of glo-3(zu446) with alleles of four other glo genes were constructed by taking unmarked glo(−) mutant males and mating them to glo-3(−) hermaphrodites containing tightly linked visible markers, including dpy-5(e61), dpy-11(e224), egl-15(n484), and unc-27(e159). Glo non-Dpy, Glo non-Egl, or Glo non-Unc F2 progeny of this cross were isolated. Single Dpy, Egl, or Unc progeny were selected from the next generation and should be glo double mutants. At least two independent double-mutant lines were isolated from each cross, and in all cases they showed the same phenotype.
RNA interference (RNAi) was carried out by feeding using bacterial clones from a C. elegans RNAi library (Geneservice, Cambridge, UK) as described (Kamath et al. 2003). For glo-3(RNAi) experiments, the clone JA:F59F5.2 was used, which targets both glo-3 and glo-3short. L1 stage larvae were placed on RNAi plates and scored as young adults.
Cloning glo-3:
glo-3 alleles were identified as described in Hermann et al. 2005. Three factor genetic crosses were used to map glo-3(zu446) to the aex-2 egl-15 interval. glo-3(kx90), glo-3(kx94), and glo-3(zu446) were complemented by nDf19, indicating that glo-3(+) is not deleted by nDf19, similar to nearby genes vab-3 and daf-12. SNP mapping with strain CB4856 placed glo-3 in the interval defined by snp_ZC504[1] and pkP6040 (mapping data are available upon request). RNAi of genes in this region identified F59F5.2/F59F5.8 as exhibiting an adult Glo phenotype similar to glo-3(−). Of the eight cosmids spanning the interval, 3/7 stable transmitting lines injected with F59F5 at 10 ng/μl and 100 ng/μl of the co-injection marker pRF4[Rol-6D] (Mello et al. 1991) rescued the embryonic Glo phenotype of glo-3(zu446). The sequence of glo-3 mutants was obtained by PCR amplifying the region spanning F59F5.2/F59F5.8 with Phusion high-fidelity DNA polymerase (New England Biolabs, Beverly, MA) from each of the mutant alleles. Amplifications and DNA sequencing (OHSU Core Facility) were performed in duplicate. Two cDNAs, yk1328a05 and yk571h2, encoded by glo-3 have been previously described. The sequence for yk571h2 is known (GenBank AV190509 and AV17878) and corresponds to F59F5.2 (Wormbase WS180). We refer to this transcript as glo-3short. We completely sequenced yk1328a05 and found that it encodes a gene encompassing F59F5.2 and F59F5.8, which corresponds to an obsolete gene prediction F59F5.gc2 (Wormbase WS180). We refer to this transcript as glo-3. We verified the expression of glo-3 and glo-3short in embryos and L4-stage animals by amplifying their corresponding cDNAs from total RNA isolated using Trizol/chloroform extraction. cDNA was generated using a d(T)20 primer with Superscript III (Invitrogen, Carlsbad, CA). In L4-stage animals the glo-3 cDNA was amplified using P454 5′ ATGTTTGGTTATGTTGTTGTTAATGAAC 3′ and P456 5′ TTATTTTAACTGTTTTAACACGCATTC 3′ and the glo-3short cDNA was amplified with P454 and the nested primer P473 5′ TTCTACGTGTACAGAAGAACAAGAATC 3′ with the glo-3short 3′-UTR specific primer P455 5′ GTAGTGTAAACCATAATCAAAACTAAC 3′. In embryos, the glo-3 cDNA was amplified with P472 5′ ACTGAATCGATTTGCACTTTTTAGCTG 3′ and P473 with the nested primer P404 5′ TCGTTAGATCACTTCAAGAAG 3′. The glo-3short cDNA was amplified from embryonic cDNA as described for L4's. A full-length glo-3 cDNA was amplified with P457 5′ GGGGACAACTTTGTACAAAAAAGTTGTGTTTGGTTATGTTGTTGTTAATGAAC 3′ and P459 5′ GGGGACAACTTTGTACAAGAAAGTGTATTTTAACTGTTTTAACACGCATTC 3′ with Phusion DNA polymerase and Gateway (Invitrogen) cloned using a BP reaction into pDONR221. The glo-3 cDNA sequence in the resulting pDONR221 plasmid was identical to yk1328a05 (GenBank FJ009048). The full-length glo-3short cDNA sequence was amplified with P457 and P458 5′ GGGGACAACTTTGTACAAGAAAGTGTAGCATTTTGGGCTGTATGACAATTTTC 3′ with Phusion DNA polymerase and Gateway cloned using a BP reaction into pDONR221. The resulting pDONR221-glo-3 and pDONR221-glo-3short plasmids were sequenced to ensure a lack of PCR-generated mutations.
Generation of reporters:
The transcriptional reporter glo-3p∷gfp was constructed using PCR fusion (Hobert 2002). A 1.6-kb sequence spanning from the predicted translational start of glo-3 5′ to the nearest upstream gene F59F5.1 was amplified using P411 5′ TCACTTTGCCTTCTGTGCGAGTTG 3′ and P412 5′ CAGTGAAAAGTTCTTCTCCTTTACTCATTTTGAACGAGTTTACCTGGAA 3′ from the cosmid F59F5. gfp-nls and flanking unc-54 3′-UTR was amplified from pPD95.67 (Addgene, Cambridge, MA) using P266 5′ AAGGGCCCGTACGGCCGACTAGTAGG 3′ and P269 5′ ATGAGTAAAGGAGAAGAACTTTTCACTG 3′. P267 5′ GGAAACAGTTATGTTTGGTATATTGGG 3′and P413 5′ TCCAGATTAAAACGTCACAAGCAACCG 3′ were used in a PCR reaction that fused the glo-3 promoter and gfp-nls sequences as described (Hobert 2002). The glo-3p∷gfp-nls reporter was co-injected into wild type at 6 ng/μl with pRF4[Rol-6D] at 100 ng/μl. kxEx50[glo-3p∷gfp; rol-6(su1006)D] was used for the analysis presented here, however five other independently derived lines showed similar GFP expression patterns.
The rescuing translational reporter GLO-3∷GFP made using the glo-3 genomic sequence was constructed using PCR fusion (Hobert 2002) by amplifying the genomic region containing the glo-3 promoter and coding sequence from the cosmid F59F5 using P411 and P422 5′ AGTCGACCTGCAGGCATGCAAGCTTTTTAACTGTTTTAACACGCATTC 3′. The gfp coding sequence and flanking unc-54 3′-UTR was amplified from pPD95.75 (Addgene) using P399 5′ AGCTTGCATGCCTGCAGGTCGACT 3′ and P266. The two PCR products were fused using P413 and P267. The resulting glo-3p∷glo-3∷gfp product was co-injected into glo-3(zu446) at 4.5 ng/μl with pRF4[Rol-6D] at 100 ng/μl. glo-3(zu446); kxEx41[glo-3∷gfp; rol-6(su1006)D] was used in the analysis presented here, and six of six other independently derived lines showed a similar subcellular distribution of GFP. Seven of seven lines were Glo(+), showing wild-type patterns of birefringent material in embryos and autofluorescence in adults.
A second GLO-3∷GFP rescuing translational reporter was made using the glo-3 cDNA sequence, which was constructed by Gateway cloning glo-3(cDNA) from pDONR221-glo-3 using an LR reaction into the vha-6p∷GTWY(Asp718)∷gfp vector (Chen et al. 2006). The resulting plasmid has a C-terminal GFP fusion to GLO-3 and is regulated by the vha-6 promoter. PCR fusion was used to place the glo-3(cDNA)∷gfp sequence under the control of the glo-3 promoter by amplifying the glo-3 promoter and coding sequence from the cosmid F59F5 using P411 and P503 5′ GTTCATTAACAACAACATAACCAAACAT 3′. The glo-3(cDNA)∷gfp coding sequence and flanking unc-54 3′-UTR was amplified from vha-6p∷glo-3(cDNA)∷gfp using P454 and P500. The two PCR products were fused using P413 and P271 5′ GACTAGTTTTCCTTCCTCCTCTATAT 3′. The resulting glo-3p∷glo-3(cDNA)∷gfp product was co-injected into glo-3(kx90) at 4.5 ng/μl with pRF4[Rol-6D] at 100 ng/μl. Thirteen of 13 lines were Glo(+), showing wild-type patterns of birefringent material in embryos and autofluorescence in adults.
The translational reporter GLO-3short∷GFP was constructed using PCR fusion by amplifying the genomic region containing the glo-3 promoter and glo-3short coding sequence from the cosmid F59F5 using P411 and P440 5′ AGTCGACCTGCAGGCATGCAAGCTGCATTTTGGGCTGTATGACAATTTTCG 3′. P440 contains a silent mutation that removes the 5′ splice site at the end of exon 4 without altering the amino acid sequence of GLO-3short. This ensures that the short isoform is produced. The gfp coding sequence and flanking unc-54 3′-UTR was amplified from pPD95.75 (Addgene) using P399 and P266. The two PCR products were fused using P413 and P267. The resulting glo-3p∷glo-3short∷gfp product was co-injected into wild type at 5 ng/μl with pRF4[Rol-6D] at 100 ng/μl. Eight of 8 independently derived lines showed a similar subcellular distribution of GFP.
The translational reporter GFP∷GLO-3short was constructed using the glo-3short cDNA sequence by Gateway cloning glo-3short using an LR reaction into the vha-6p∷GTWY(EcoRI)∷gfp vector (Chen et al. 2006). The resulting plasmid has an N-terminal GFP fusion to GLO-3short and is regulated by the vha-6 promoter. PCR fusion was used to place the gfp∷glo-3short sequence under the control of the glo-3 promoter by amplifying the glo-3 promoter and coding sequence from the cosmid F59F5 using P411 and P412. The gfp∷glo-3short coding sequence and flanking unc-54 3′-UTR was amplified from vha-6p∷gfp∷glo-3short using P269 and P500. The two PCR products were fused using P413 and P271 5′ GACTAGTTTTCCTTCCTCCTCTATAT 3′. The resulting glo-3p∷gfp∷glo-3short(cDNA) product was co-injected into wild type at 4.5 ng/μl with pRF4[Rol-6D] at 100 ng/μl. Thirteen of 13 lines showed a similar subcellular distribution of GFP.
The translational reporter glo-3p∷GLO-3short was constructed using the vha-6p∷gfp∷glo-3short vector. PCR fusion was used to place the glo-3short sequence under the control of the glo-3 promoter by amplifying the glo-3 promoter and coding sequence from the cosmid F59F5 using P411 and P503. The glo-3short coding sequence and flanking unc-54 3′-UTR were amplified from vha-6p∷gfp∷glo-3short(cDNA) using P454 and P500. The two PCR products were fused using P413 and P271. The resulting glo-3p∷glo-3short(cDNA) product was co-injected into glo-3(kx90) at 4.5 ng/μl with pRF4[Rol-6D] at 100 ng/μl. Seven of seven lines showed embryonic and adult Glo phenotypes.
Microscopy:
A Zeiss Axioskop II plus microscope (Thornwood, NY) equipped with DIC, polarization, and fluorescence optics was used for all light microscopy. Images were captured using a Diagnostic Instruments Insight Spot QE 4.1 digital camera with Spot Basic software (Sterling Heights, MI). Images were placed into separate channels in Photoshop 7.0 (Adobe, San Jose, CA) and analyzed for colocalization. Adults and larvae were immobilized prior to analysis by being mounted in 1× M9 containing 10 mm levamisole. Embryos that had developed beyond the 1.5-fold stage were visualized immediately after becoming hypoxic and immobilized due to the addition of excess Escherichia coli when the embryos were mounted. Intestinal autofluorescence in living animals was detected using the Zeiss 09 (Ex:BP450-490; Em:LP515), Zeiss 15 (Ex:BP586/12; Em:LP590), or custom (Ex:BP480/20; Em:BP530/20) fluorescence filters. Acidic compartments within intestinal cells were stained with Lysotracker Red (Molecular Probes, Eugene, OR) or acridine orange (Sigma, St. Louis) as described (Hermann et al. 2005). Fluid-phase endocytosis into adult intestinal cells was analyzed by feeding animals TRITC-dextran (Sigma) or TRITC-BSA (Sigma) using the procedures described by Hermann et al. 2005. Fat stores in living animals were stained with Nile Red (Molecular Probes) or BODIPY (493/503) (Molecular Probes) (Schroeder et al. 2007). Zeiss 15 and a custom (Ex:BP480/20; Em:BP530/20) fluorescence filter were used to analyze the colocalization of autofluorescence and vital staining in adult intestinal cells. Fat stores in fixed adults were stained with Sudan Black B (Sigma) as described (Ogg and Ruvkun 1998). Standard polarization optics were used to detect birefringent material in the embryonic intestine.
For immunostaining with affinity-purified rabbit FUS-1 (Kontani et al. 2005), RAB-5 (Audhya et al. 2007), PGP-2 (C-term) (Schroeder et al. 2007), VPS-27 (Roudier et al. 2005), and mouse 3E6 GFP antisera (Qbiogene), embryos (Leung et al. 1999) and adults (Currie et al. 2007) were fixed and stained as described. These protocols result in the loss of birefringent and autofluorescent gut granule contents.
RESULTS
glo-3 alleles define three distinct gut granule phenotypes:
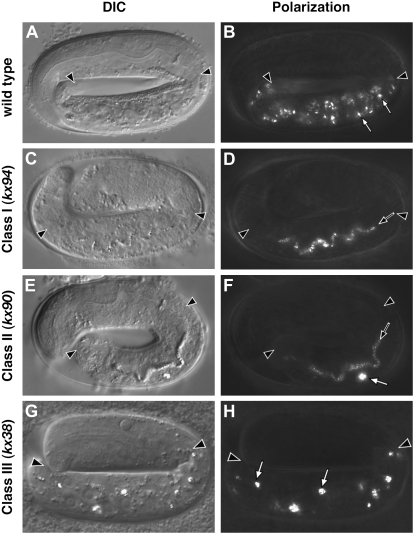
glo-3 was identified in screens for C. elegans mutants defective in the formation and/or morphology of birefringent and autofluorescent gut granules (Hermann et al. 2005). We previously described glo-3(zu446) embryos as displaying a Glo phenotype [for a description of C. elegans embryogenesis see (Sulston et al. 1983)]. glo-3(zu446) is characterized by the mislocalization of birefringent material into the intestinal lumen, often accompanied by the presence of one to five enlarged birefringent organelles in intestinal cells. glo-3(zu446) adults contain reduced numbers of autofluorescent compartments when compared to wild type (Hermann et al. 2005), and of the glo mutants characterized to date they display the highest number. Four other glo-3 alleles (gm125, kx29, kx37, and kx90) were phenotypically indistinguishable from glo-3(zu446) (Hermann et al. 2005).
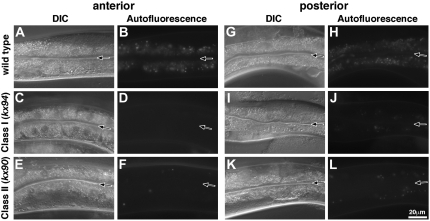
Continued genetic analysis of mutants in our collection of glo mutants led to the identification of four alleles of glo-3 which were phenotypically distinct from glo-3(zu446). glo-3(kx91) and glo-3(kx94) embryos mislocalized birefringent material into the embryonic intestinal lumen, however they completely lacked the one to five enlarged birefringent organelles often seen in glo-3(zu446) (Figure 1, D and F, Table 1). As adults, glo-3(kx91) and glo-3(kx94) typically lacked autofluorescent gut granules in anterior intestinal cells unlike glo-3(zu446), which typically contained higher numbers (Figure 2D and Table 2). glo-3(zu446), glo-3(kx91), and glo-3(kx94) alleles contained similar numbers of autofluorescent compartments in posterior adult intestinal cells (Figure 2J and Table 2). On the basis of the different effects these alleles have on gut granule formation (Tables 1 and 2), we define glo-3(kx91) and glo-3(kx94) as glo-3 class I alleles and glo-3(gm125), glo-3(kx29), glo-3(kx37), glo-3(kx90), and glo-3(zu446) as defining glo-3 class II alleles.
Figure 1.—
Mutations in glo-3 result in three different embryonic Glo phenotypes. (A and B) Birefringent gut granules (open arrows in B) were present within the intestinal cells of wild-type pretzel-stage embryos. (C and D) glo-3(−) alleles that exhibit a class I phenotype mislocalized birefringent material in the intestinal lumen (solid arrow in D) and did not contain birefringent material within embryonic intestinal cells. (E and F) glo-3(−) alleles that exhibit a class II phenotype mislocalized birefringent material in the intestinal lumen (solid arrow in F) and often contained a few enlarged birefringent granules (open arrow in E) within embryonic intestinal cells. (G and H) glo-3(−) alleles that exhibit a class III phenotype typically contained a reduced number of enlarged birefringent granules within embryonic intestinal cells (open arrows in H) and did not contain birefringent material within the intestinal lumen. Intestinal cells are located between the solid arrowheads in A–H. C. elegans embryos are ∼50 μm in length.
TABLE 1.
Birefringent gut granules in glo-3(−) embryos
| Genotype | % of embryos that lacked birefringence in intestinal cells (% of embryos that also mislocalized birefringent material into the intestinal lumen) | % of embryos with 1–10 enlarged birefringent granules in intestinal cells (% of embryos that also mislocalized birefringent material into the intestinal lumen) | % of embryos with 11–30 enlarged birefringent granules in intestinal cells (% of embryos that also mislocalized birefringent material into the intestinal lumen) | n |
|---|---|---|---|---|
| Wild typea | 0 | 0 | 0 | 52 |
| Class I alleles | 0 | |||
| glo-3(kx91) | 100 (47) | 0 | 0 | 77 |
| glo-3(kx94) | 100 (56) | 0 | 0 | 107 |
| Class II alleles | 0 | |||
| glo-3(gm125) | 41 (40) | 59 (23) | 0 | 83 |
| glo-3(kx29) | 41 (29) | 59 (23) | 0 | 73 |
| glo-3(kx37) | 68 (38) | 32 (22) | 0 | 69 |
| glo-3(kx90) | 47 (26) | 53 (23) | 0 | 115 |
| glo-3(zu446) | 54 (38) | 46 (15) | 0 | 125 |
| Class III alleles | ||||
| glo-3(kx1) | 0 | 12 (0) | 88 (0) | 91 |
| glo-3(kx38) | 0 | 25 (5) | 75 (0) | 92 |
| smg-1(−) suppression | ||||
| smg-1(r861)b | 0 | 0 | 2 (0) | 54 |
| Class I alleles | ||||
| glo-3(kx91); smg-1(r861) | 100 (94) | 0 | 0 | 85 |
| glo-3(kx94); smg-1(r861) | 100 (63) | 0 | 0 | 68 |
| Class II alleles | ||||
| glo-3(gm125); smg-1(r861) | 0 | 0 | 100 (0) | 26 |
| glo-3(kx29); smg-1(r861) | 0 | 0 | 100 (0) | 41 |
| glo-3(kx37); smg-1(r861) | 0 | 3 (100) | 97 (0) | 59 |
| glo-3(kx90); smg-1(r861) | 0 | 0 | 100 (0) | 24 |
| glo-3(zu446); smg-1(r861) | 0 | 68 (6) | 32 (0) | 34 |
| Class III alleles | ||||
| glo-3(kx1); smg-1(r861) | 0 | 0 | 100 (0) | 27 |
| glo-3(kx38); smg-1(r861) | 0 | 0 | 100 (0) | 28 |
| Parental genotypec | ||||
| Class I alleles | ||||
| glo-3(kx91)/+ | 15 (9) | 19 (11) | 0 | 186 |
| glo-3(kx94)/+ | 9 (4) | 15 (9) | 0 | 110 |
| Class II alleles | ||||
| glo-3(kx90)/+ | 10 (6) | 12 (6) | 0 | 177 |
| glo-3(zu446)/+ | 11 (4) | 14 (3) | 0 | 109 |
All strains were grown at 22°. Twofold and later-stage embryos were analyzed using polarization microscopy and scored for the presence, morphology, and localization of birefringent material in the intestine. n, number of embryos scored.
Wild-type embryos contained >100 birefringent granules in intestinal cells and lacked birefringent material in the intestinal lumen.
Ninety-eight percent of smg-1(r861) embryos contained >100 birefringent granules in intestinal cells and lacked birefringent material in the intestinal lumen.
The progeny of heterozygous glo-3(−)/+ hermaphrodites were scored. Twenty-five percent are predicted to be glo-3(−)/glo-3(−) and exhibit a Glo phenotype.
Figure 2.—
Mutations in glo-3 result in two distinct adult Glo phenotypes. Numerous autofluorescent gut granules visualized with a standard FITC filter were present within anterior (A and B) and posterior (G and H) intestinal cells of wild-type animals. glo-3(−) alleles that exhibit a class I phenotype lacked autofluorescent gut granules in anterior intestinal cells (C and D) and displayed substantially reduced numbers of autofluorescent organelles in posterior intestinal cells (I and J). glo-3(−) alleles that exhibit a class II phenotype contained substantially reduced numbers of autofluorescent organelles in both anterior (E and F) and posterior intestinal cells (K and L). The intestinal lumen is marked with a solid arrow in A–L.
TABLE 2.
Autofluorescent gut granules in glo-3(−) adults
| % of animals with the specified no. of autofluorescent granules in anterior intestinal cells
|
% of animals with the specified no. of autofluorescent granules in posterior intestinal cells
|
Phenotype (anterior/posterior) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Very low | Low | Medium | High | Very low | Low | Medium | High | n | |
| Wild type | 0 | 0 | 0 | 100 | 0 | 0 | 1 | 99 | High/high | 89 |
| glo-3(RNAi)a | 0 | 100 | 0 | 0 | 0 | 85 | 5 | 0 | Low/low | 40 |
| Class I alleles | ||||||||||
| glo-3(kx91) | 100 | 0 | 0 | 0 | 16 | 84 | 0 | 0 | Very low/low | 109 |
| glo-3(kx94) | 96 | 4 | 0 | 0 | 8 | 88 | 4 | 0 | Very low/low | 77 |
| Class II alleles | ||||||||||
| glo-3(gm125) | 1 | 96 | 3 | 0 | 0 | 100 | 0 | 0 | Low/low | 77 |
| glo-3(kx29) | 23 | 77 | 0 | 0 | 0 | 99 | 1 | 0 | Low/low | 86 |
| glo-3(kx37) | 52 | 48 | 0 | 0 | 0 | 91 | 9 | 0 | Low/low | 67 |
| glo-3(kx90) | 23 | 77 | 0 | 0 | 0 | 96 | 4 | 0 | Low/low | 74 |
| glo-3(zu446) | 33 | 67 | 0 | 0 | 1 | 86 | 13 | 0 | Low/low | 95 |
| Class III alleles | ||||||||||
| glo-3(kx1) | 2 | 83 | 12 | 3 | 0 | 50 | 45 | 5 | Low/low | 58 |
| glo-3(kx38) | 0 | 100 | 0 | 0 | 0 | 79 | 21 | 0 | Low/low | 71 |
All strains were grown at 22°. Individual young adults were analyzed using fluorescence microscopy with a fluorescein isothiocyanate (FITC) filter and were scored for the number of autofluorescent gut granules within the anterior and posterior areas of the intestine. Young animals were scored as very low when 0–5 autofluorescent gut granules were present, as low when 6–100 autofluorescent gut granules were present, as medium when 101–200 autofluorescent gut granules were present, and as high when >200 autofluorescent gut granules were present. n, number of animals scored.
RNAi was carried out in the rrf-3(pk1426) RNAi-sensitive background (Simmer et al. 2003). Similar results were seen in the N2 strain.
Two alleles of glo-3 were identified that rarely mislocalized birefringent material into the embryonic intestinal lumen. Instead, glo-3(kx1) and glo-3(kx38) embryos contained 5–30 enlarged birefringent organelles, which is reduced compared to wild type but substantially more than class II alleles (Figure 1, F and H; Table 1). In adult stages, glo-3(kx1), glo-3(kx38), and class II animals contained similar numbers of autofluorescent compartments (Table 2). Due to the unique embryonic phenotype displayed by glo-3(kx1) and glo-3(kx38) we define them as glo-3 class III alleles.
Class I alleles likely cause the strongest glo-3 loss-of-function phenotype:
We investigated which class of glo-3 alleles exhibited the stronger phenotype. Unfortunately, glo-3 is not deleted by nDf19 (our unpublished observations), the only reported deletion spanning the glo-3 genomic region, preventing an analysis of glo-3(−) phenotypic changes when individual alleles are placed over a deficiency. We therefore took a number of different approaches to investigate the relative severity of glo-3(−) alleles.
Often when two alleles that differentially affect gene function are placed in trans, the phenotype observed is that of the weaker allele (the allele that perturbs function the least). In four different glo-3 class I/glo-3 class II transheterozygous combinations we found that the class II embryonic phenotype was expressed (supplemental Table 1). We found in four different glo-3 class II/glo-3 class III transheterozygous combinations that the class III embryonic phenotype was expressed (supplemental Table 1). These data suggest that the relative strengths of glo-3 alleles from strongest to weakest are class I > class II > class III.
We observed a maternal effect modification of the class I glo-3 embryonic phenotype that further supports the idea that glo-3 class I alleles exhibit a stronger phenotype than glo-3 class II alleles. All of the glo-3 alleles identified to date are strict recessive, zygotic effect for the Glo phenotype (Table 1 and data not shown). However, glo-3(−)/glo-3(−) class I progeny of glo-3(+)/glo-3(−) parents displayed a class II embryonic phenotype (Table 1). In contrast, glo-3(−)/glo-3(−) class I progeny of homozygous glo-3(−) class I parents display the class I phenotypes (Table 1). The phenotype of glo-3(−)/glo-3(−) class II mutants produced by glo-3(+)/glo-3(−) parents are unchanged relative to being produced by homozygous class II parents (Table 1). These results indicate that a wild-type maternal copy of glo-3(+) modifies the phenotype of class I alleles, making it more similar to the class II phenotype. One explanation of this observation is that class I alleles have less glo-3(+) activity than class II alleles, however with a contribution of wild-type glo-3(+) from the maternal parent glo-3(class I)/glo-3(class I) embryos have increased glo-3(+) activity and phenotypically resemble class II mutants.
Finally, the Glo phenotypes exhibited by the three classes of glo-3 alleles suggest that class I alleles most compromise glo-3 function. Class I mutants never contain birefringent gut granule contents within their intestinal cells (Table 1). In contrast, class II and class III mutants retain organelles with birefringent material, albeit at reduced numbers when compared to wild type (Figure 1). Class I mutant adults typically lack or have very few organelles with autofluorescent gut granule contents in anterior intestinal cells. Both class II and class III mutant adults display higher numbers of autofluorescent organelles (Table 2). Together, these data strongly suggest that glo-3 class I alleles are more severe in their effect on gut granule biogenesis than either class II or class III alleles. We therefore present our phenotypic analysis of the class I allele glo-3(kx94). We have also carried out a detailed phenotypic analysis of the autofluorescent organelles class II alleles glo-3(kx90) and glo-3(zu446) at the adult stage and not seen substantially different results than we present for the class I allele glo-3(kx94).
glo-3(−) adults contain reduced numbers of gut granules:
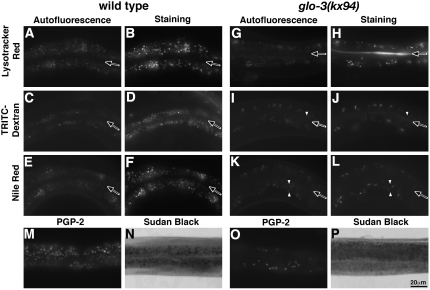
The intestinal cells of wild-type adults contain hundreds of autofluorescent gut granules that are characterized by: (1) their acidification, (2) their function as terminal endocytic compartments, and (3) the presence of the gut granule membrane-associated ABC transporter PGP-2, and (4) the presence of fat (Clokey and Jacobson 1986; Schroeder et al. 2007). To determine whether the reduction of autofluorescent intestinal compartments is reflective of a decrease in the number of gut granules, we examined glo-3(kx94) adults for organelles with gut granule characteristics. glo-3(kx94) adults contained substantially reduced numbers of acidified organelles marked by Lysotracker Red (Figure 3, A, B, G, and H) and acridine orange (supplemental Figure 1, C, D, I, and J). Similarly, reduced numbers of organelles in glo-3(kx94) adults were labeled with TRITC-dextran (Figure 3, C, D, I, and J) and TRITC-BSA (supplemental Figure 1, E, F, K, and L) that was endocytosed across the apical surface. Staining of fixed animals showed that reduced numbers of PGP-2-containing compartments were present within glo-3(kx94) adults (Figure 3, M and O). These results indicate that glo-3(kx94) adults contain reduced numbers of gut granules, indicative of defects in gut granule formation and/or stability.
Figure 3.—
glo-3(−) adults contain reduced numbers of gut granules. (A–F) In living animals, wild-type gut granules contained autofluorescent material (A, C, and E) that brightly stained with markers for acidity (B), terminal endocytic activity (D), and fat (F). In fixed wild-type adults, gut granules were marked by the PGP-2 protein (M). glo-3(−) adults contained dramatically reduced numbers of autofluorescent gut granules, the majority of which were acidified (H), terminal endocytic compartments (J), that contained fat (L). Nile Red staining of glo-3(−) adults was quite weak and its signal has been increased three to four times that in wild type to aid in its visualization. (O) glo-3(−) adults contained reduced numbers of PGP-2-stained organelles. A few autofluorescent compartments in glo-3(−) adults did not accumulate TRITC-dextran or Nile Red (open arrowheads in J and L). (N and P) Similar numbers of Sudan Black-stained organelles were present in wild-type and glo-3(−) adults. In A–L the intestinal lumen is marked with a solid arrow. Posterior intestinal cells are shown A–P.
We used three different labels, Nile Red, BODIPY 493/503, and Sudan Black, to mark fat-containing compartments in glo-3(kx94) adults. glo-3(kx94) animals fed Nile Red contained reduced numbers of stained compartments (Figure 3, F, K, and L). However, these compartments were only weakly stained when compared to wild type (data not shown). A similar loss of Nile Red staining was reported for glo-3(RNAi) in a genomewide screen for genes regulating C. elegans body fat levels (Ashrafi et al. 2003). BODIPY 493/503-fed glo-3(kx94) animals completely lacked staining (supplemental Figure 1, A, B, G, and H). Our prior studies of a subset of glo mutants demonstrated that the loss of staining by vital stains such as Nile Red and BODIPY 493/503 did not correlate with a decrease in the overall fat content at the level of the organism (Schroeder et al. 2007). To investigate whether this might be the case in glo-3(−) animals, we fixed and stained glo-3(kx94) adults with Sudan Black, a sensitive marker for fat in C. elegans (Kimura et al. 1997). glo-3(kx94) and wild-type animals displayed similar levels of Sudan Black staining (Figure 3, N and P). Consistent with our biochemical analysis of fat content in glo mutants that lack Nile Red staining (Schroeder et al. 2007), we did not observe a difference in Sudan Black staining between wild-type, apt-7(tm920), glo-1(zu437), and pgp-2(kx48) adults (supplemental Figure 2). These data show that Nile Red and BODPIY 493/503 only stain a subset of fat-containing organelles, namely gut granules, in C. elegans and strongly suggest that body fat levels are not dramatically altered in glo-3(−) adults.
The loss of gut granule-associated BODIPY 493/503 and reduced Nile Red staining in glo-3(−) animals raises the question of whether the autofluorescent gut granules present in glo-3(−) are properly formed. To address this issue, we analyzed whether the autofluorescent organelles in glo-3(kx94) adults contained gut granule-associated markers. Nearly all of the autofluorescent gut granules in glo-3(kx94) were marked by Lysotracker Red and TRITC-BSA (supplemental Table 2); 62 and 69% of autofluorescent gut granules were stained by Nile Red and TRITC-dextran, respectively (supplemental Table 2). Due to experimental limitations we were unable to analyze the colocalization of autofluorescent material with acridine orange or anti-PGP-2 staining. However, similar numbers of autofluorescent, acridine orange-stained, and PGP-2-containing organelles were seen in glo-3(kx94) adults (Table 2 and supplemental Table 3). The formation of autofluorescent compartments in glo-3(−) adults required the activity of glo-1 and glo-4 (Table 3), two genes that function in gut granule biogenesis (Hermann et al. 2005). Together these data indicate that the autofluorescent organelles in glo-3(−) adults, with the exception of stored fat, appear to be indistinguishable from wild-type gut granules. The reduction/loss of vital staining for gut granule-associated fat could result from a bona fide defect in gut granule-associated fat accumulation or from alterations in trafficking to, or retention of Nile Red and BODIPY 493/503 in gut granules.
TABLE 3.
glo-3(−) double mutants
| % of animals with the specified no. of autofluorescent granules in intestinal cells
|
|||||
|---|---|---|---|---|---|
| Genotype | None | Low | Medium | High | n |
| Wild type | 0 | 0 | 0 | 100 | 24 |
| glo-3(zu446)a | 0 | 95 | 5 | 0 | 85 |
| glo-1(zu437) | 100 | 0 | 0 | 0 | 41 |
| glo-4(ok623)b | 100 | 0 | 0 | 0 | 32 |
| pgp-2(kx48)c | 0 | 3 | 97 | 0 | 35 |
| apt-6(ok429) | 16 | 84 | 0 | 0 | 19 |
| glo-1(zu437) glo-3(zu446) | 100 | 0 | 0 | 0 | 49 |
| glo-4(ok623); glo-3(zu446) | 100 | 0 | 0 | 0 | 95 |
| pgp-2(kx48); glo-3(zu446) | 100 | 0 | 0 | 0 | 54 |
| apt-6(ok429); glo-3(zu446) | 91 | 9 | 0 | 0 | 31 |
All strains were grown at 22°. Individual L4/young adults were analyzed using fluorescence microscopy with a fluorescein isothiocyanate (FITC) filter and were scored for the number of autofluorescent gut granules within the intestine. Animals were scored as none when autofluorescent gut granules were lacking, as low when 1–100 autofluorescent gut granules were present, as medium when 101–200 autofluorescent gut granules were present, and as high when >200 autofluorescent gut granules were present.
The linked markers egl-15(n484) or unc-27(e159) did not alter the Glo phenotype of glo-3(zu446). n, number of animals scored.
The linked marker dpy-11(e224) did not alter the Glo phenotype of glo-4(ok623).
The linked marker dpy-5(e61) did not alter the Glo phenotype of pgp-2(kx48).
glo-3(−) embryos lack gut granules:
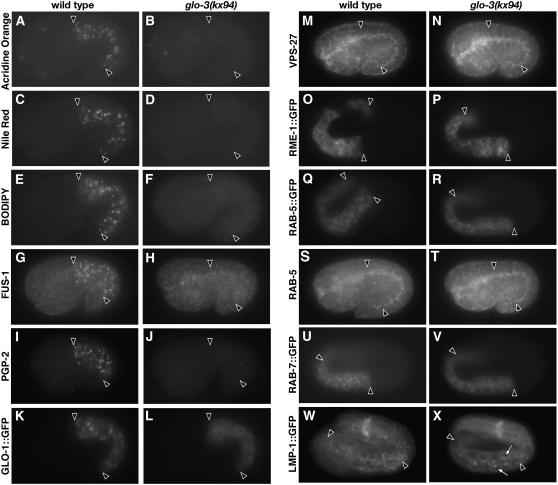
glo-3(kx94) embryos lack birefringent compartments in their intestinal cells and mislocalize birefringent material into the intestinal lumen (Figure 1, C and D). These phenotypes are consistent with glo-3(kx94) embryos being defective in gut granule biogenesis and lacking gut granules (Hermann et al. 2005). We investigated whether this was the case by staining with dyes and antibodies that label gut granules in wild-type embryos. Embryonic gut granules are acidified, fat-storing compartments that contain subunits of the V-ATPase, the Rab GTPase GLO-1, and PGP-2 (Hermann et al. 2005; Schroeder et al. 2007). Unlike wild type, glo-3(kx94) embryonic intestinal cells lacked acridine orange stained, acidified compartments (Figure 4, A and B). Consistent with the lack of acidification, FUS-1, an integral membrane-associated subunit of the V-ATPase (Kontani et al. 2005), was not detected in the intestinal cells of glo-3(kx94) embryos (Figure 4, G and H). Wild-type embryonic gut granules are stained by Nile Red and BODIPY 493/503 (Schroeder et al. 2007). Neither of these dyes stained compartments within intestinal cells of glo-3(kx94) embryos (Figure 4, D and F). In wild-type embryos, GLO-1∷GFP localizes to gut granules (Hermann et al. 2005). GLO-1∷GFP did not localize to any distinct organelles in the embryonic intestine of glo-3(kx94) (Figure 4L). We conclude that gut granules are lacking in glo-3(kx94) embryos and that embryonic gut granule biogenesis requires the function of glo-3.
Figure 4.—
glo-3(−) embryos lack gut granules but appear to contain other endosomal compartments. Acridine orange stained, acidified compartments present in wild-type 1.5-fold-stage embryos (A) were lacking in glo-3(−) embryos (B) Wild-type embryos displayed fat-containing, Nile Red and BODIPY 493/503-stained compartments (C and E) that were lacking in glo-3(−) embryos (D and F). Wild-type embryos contained anti-FUS-1 and anti-PGP-2 antibody marked compartments (G and I), which were not present in glo-3(−) embryos (H and J). GLO-1∷GFP marked compartments present in wild-type (K) were lacking in glo-3(−) embryos (L). Antibodies that recognize VPS-27 and RAB-5 stained similar organelles in wild-type (M and S) and glo-3(−) (N and T) embryos. Anti-GFP antibodies used to stain embryos expressing RME-1, RAB-5, RAB-7, and LMP-1 fusion proteins showed similar staining patterns in wild-type (O, Q, U, and W) and glo-3(−) embryos (P, R, V, and X), with the exception that enlarged LMP-1∷GFP marked organelles were present in glo-3(−) (open arrows in X). Intestinal cells are located between the solid arrowheads.
We examined whether glo-3(kx94) embryos had defects in forming other endosomal organelles. The recycling endosome-associated protein RME-1∷GFP (Grant et al. 2001), and early endosome-associated proteins RAB-5 (Audhya et al. 2007), RAB-5∷GFP (Hermann et al. 2005), and VPS-27 (Roudier et al. 2005) had similar localizations and morphologies in wild-type and glo-3(kx94) embryonic intestinal cells (Figure 4, M–T). The localization of late endosome-associated proteins RAB-7∷GFP and LMP-1∷GFP (Chen et al. 2006) was unchanged in glo-3(kx94) embryonic intestinal cells (Figure 4, U and V). However, as seen in two other glo mutants, glo-1(−) and pgp-2(−) (Hermann et al. 2005; Schroeder et al. 2007), glo-3(kx94) embryos contained slightly enlarged LMP-1∷GFP compartments (Figure 4, W and X). It is possible that LMP-1∷GFP compartments become enlarged due to the mistrafficking of gut granule contents when there are defects in gut granule biogenesis (Currie et al. 2007). Our results show that many aspects of the endosomal system are properly organized in glo-3(kx94) embryonic intestinal cells, despite defects in gut granule formation.
glo-3 encodes a novel predicted membrane-associated protein:
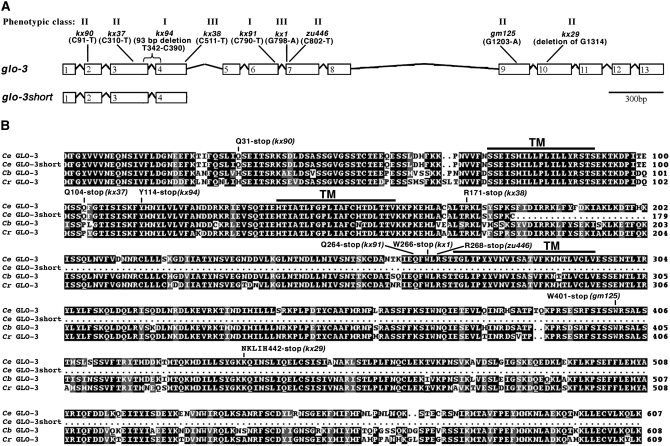
We mapped glo-3 to a 220-kb interval defined by SNPs snp_ZC504[1] and pkP6040. An RNAi-based screen of genes in the interval identified RNAi clone JA:F59F5.2 as inducing an adult Glo phenotype very similar to glo-3 class II and class III alleles (Table 2). JA:F59F5.2 targets two predicted genes, F59F5.2 and F59F5.8. Two previously isolated cDNAs, yk1328a05 and yk571h2, suggested that F59F5.2 and F59F5.8 are a single gene that is alternatively spliced to produce two transcripts. We independently isolated F59F5.2/.8 cDNAs from both adult and embryonic stages, supporting the conclusion that F59F5.2 and F59F5.8 are a single gene (see materials and methods). We refer to the long F59F5.2/.8 transcript as glo-3 and the F59F5.2 transcript as glo-3short (Figure 5A). glo-3short is generated by a lack of splicing at the 5′ end of intron 4. The Glo phenotypes of glo-3(−) were completely rescued by the cosmid F59F5 and by both glo-3(genomic)∷gfp and glo-3(cDNA)∷gfp expressed under the control of the glo-3 promoter. DNA sequencing showed that all nine glo-3 alleles had a mutation in the coding sequence of F59F5.2/.8 (Figure 5A).
Figure 5.—
Predicted structure of the glo-3 gene and encoded protein. (A) The structure of the glo-3 gene and the location and phenotypic class of mutations are shown. (B) The predicted protein sequences and alignment of GLO-3 from C. elegans (Ce), C. briggsae (Cb), and C. remanei (Cr). The location of predicted membrane-spanning domains (TM) identified using TMpred (Hofmann and Stoffel 1993) are shown.
glo-3 encodes a predicted protein that is conserved in nematodes, but lacks obvious sequence homologs in any other phyla (Figure 5B). The nematodes containing glo-3, including C. elegans, C. briggsae, and C. remanei, are known to be highly genetically divergent from one another (Kiontke et al. 2004). GLO-3 is predicted to possess three membrane spanning domains and GLO-3short is predicted to possess two of these (Figure 5B). The likely topology of GLO-3 positions its 312-amino-acid C terminus in the cytoplasm. Extensive database searches did not reveal any other functional or structural motifs in the GLO-3 sequence.
GLO-3 is expressed in the intestine and associates with the gut granule membrane:
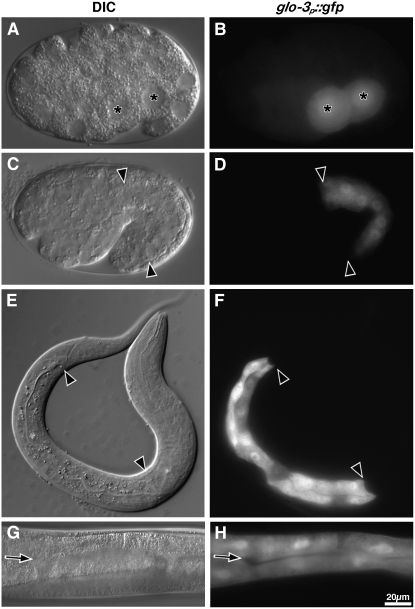
Genes that mediate embryonic gut granule biogenesis begin to be expressed early in the developmental program of the intestine, typically during gastrulation of the two intestinal precursors (Hermann et al. 2005; Schroeder et al. 2007). To determine whether this is the case for glo-3, we placed gfp under the transcriptional control of a 1.6-kb glo-3 promoter fragment containing all of the sequences between the translational start sites of glo-3 and the upstream gene F59F5.1. glo-3p∷gfp expression was first detected at gastrulation in the two intestinal precursors (Figure 6, A and B). glo-3p∷gfp continued to be expressed in the intestine through embryogenesis (Figure 6, C and D), larval stages (Figure 6, E and F), and into adulthood (Figure 6, G and H). Weak expression of glo-3p∷gfp was sometimes seen in a pair of cells in the head with neuronal morphology and the intestinal valve cell (data not shown), which represented the only nonintestinal expression of glo-3p∷gfp.
Figure 6.—
glo-3 is expressed in embryonic and adult intestinal cells. Strains carrying an extrachromosomal transgene in which the glo-3 promoter drives the transcription of gfp (glo-3p∷gfp) resulted in the expression of gfp in intestinal precursors at the E2 (A and B) and 1.5-fold stages (C and D). gfp was also expressed in the intestinal cells of L1 larvae (E and F) and adults (G and H). In A and B, the asterisks mark the nuclei of the two gastrulating intestinal precursors. In C–F, intestinal cells are located between the solid arrowheads. The solid arrows mark the intestinal lumen in G and H.
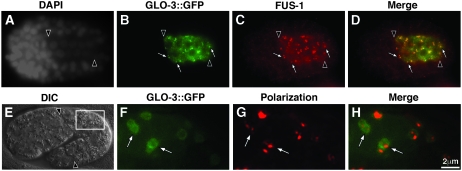
We analyzed the localization of GLO-3 with a carboxy terminally tagged GLO-3∷GFP fusion expressed under the control of the glo-3 promoter. The GLO-3∷GFP fusion fully rescued the embryonic and adult Glo phenotypes of glo-3(zu446) indicating that GLO-3∷GFP is functional and likely localized properly. GLO-3∷GFP had a punctate distribution within the intestinal primordium reminiscent of gut granules (Figure 7B). We compared the localization of GLO-3∷GFP to the gut granule localized FUS-1 protein in fixed embryos and found that GLO-3∷GFP was associated with gut granules from bean stage (Figure 7, A–D) through hatching (data not shown). The punctate distribution of GLO-3∷GFP was lacking in glo-1(−) embryos (data not shown), which do not generate gut granules (Hermann et al. 2005). Furthermore, apt-7(−) and pgp-2(−) embryos, which contain reduced numbers of gut granules (Schroeder et al. 2007), lacked or displayed only a few GLO-3∷GFP puncta, respectively (data not shown). In living embryos, GLO-3∷GFP apparently localized to the limiting membrane of gut granules containing birefringent material (Figure 7, E–H). The association of GLO-3∷GFP with the gut granule membrane is consistent with the prediction of three transmembrane segments within GLO-3. Moreover, the probable localization of GFP at the gut granule membrane in living embryos suggests that the carboxy terminus of GLO-3 is localized in the cytoplasm, as GFP fluorescence is quenched by low pH (Kneen et al. 1998), a well-defined characteristic of gut granules (Clokey and Jacobson 1986; Hermann et al. 2005).
Figure 7.—
GLO-3∷GFP is localized to the gut granule membrane. (A–D) A bean-stage glo-3(zu446) embryo expressing GLO-3∷GFP from the glo-3 promoter showing colocalization of anti-GFP and anti-FUS-1 antibody staining at the same intestinal organelles (white arrows). (F–H) High magnification of the intestine of a GLO-3∷GFP expressing 1.75-fold-stage embryo (white box in E). Birefringent material (pseudocolored red in G and H) is present within GLO-3∷GFP-containing vesicles (white arrows in F–H). Intestinal cells are located between the black arrowheads in A–E.
To analyze the localization of GLO-3short we expressed GLO-3short carboxy (GLO3short∷GFP) and amino terminal (GFP∷GLO-3∷short) GFP fusions under the control of the glo-3 promoter. The GLO-3short GFP fusions did not localize to the gut granule membrane and instead were localized to the nuclear envelope or cytoplasm (data not shown). However, the GLO-3short GFP fusions and an untagged glo-3short cDNA expressed under control of the glo-3 promoter did not rescue the Glo phenotype of glo-3(−) embryos or induce a dominant Glo phenotype when expressed in wild type. On the basis of these phenotypic data it is unclear whether the distribution we documented represents the bona fide localization of GLO-3short within intestinal cells.
smg suppression of glo-3 alleles:
All of the glo-3 alleles we have characterized contain premature stop codons, none of which are predicted to alter consensus splice sites (Blumenthal and Steward 1997). Due to the locations of the stop codons, the nonsense mediated RNA decay (NMD) pathway, encoded by the smg genes in C. elegans (Mango 2001; Maquat 2004), should target and degrade these glo-3(−) mRNAs, resulting in a similar Glo phenotype. It is therefore surprising that the nonsense alleles fall into three distinct phenotypic classes (Table 1 and Figure 5A). One possible explanation for the different phenotypes could be due to differential NMD of the glo-3(−) alleles, so that some alleles produce partially functional truncated GLO-3 proteins.
To test if the NMD pathway targets glo-3 alleles and investigate whether the production of truncated GLO-3 might contribute to phenotypic differences between the alleles, we placed all nine glo-3 alleles in the smg-1(r861) background. smg-1 encodes a protein kinase essential for NMD in C. elegans (Grimson et al. 2004). smg-1(−) partially suppressed the embryonic Glo phenotype of all five class II glo-3(−) alleles (Table 1). When placed into the smg-1(−) background, class II alleles no longer mislocalized birefringent material into the intestinal lumen and displayed substantially increased numbers of birefringent gut granules so that they phenotypically resembled class III alleles (supplemental Figure 3 and Table 1), indicating that these alleles are targets for NMD. While smg-1(−) suppression did not result in wild-type numbers of gut granules, birefringent gut granules in smg-1(r861); glo-3(zu446), smg-1(r861); glo-3(gm125), and smg-1(r861); glo-3(kx29) embryos were acidified, showing that they are likely properly formed compartments (data not shown). Notably, class I alleles, kx91 and kx94, were not suppressed by smg-1(−) (Table 1), indicating that the truncated GLO-3 proteins produced by these alleles are not functional in gut granule biogenesis. The Glo phenotypes of both class III alleles were not substantially altered by smg-1(−) (Table 1), suggesting that they are not efficient targets of the SMG pathway, and produce truncated GLO-3 proteins with partial activity in the smg-1(+) background.
glo-3 likely functions in parallel or downstream of the AP-3 complex and PGP-2:
Our prior studies have shown that AP-3 and PGP-2 function independently to promote the formation of gut granules in C. elegans (Schroeder et al. 2007). The AP-3 adapter complex in C. elegans is composed of four proteins, including the β3 subunit encoded by apt-6 (Boehm and Bonifacino 2001). pgp-2 encodes an ABC transporter, localized to the gut granule membrane, most similar to members of the ABCB subfamily of proteins that includes human P-glycoprotein, which mediates tumor multidrug resistance (Borst and Elferink 2002; Schroeder et al. 2007). Single AP-3 and pgp-2(−) null mutants display reduced numbers of gut granules in adults and embryos (Hermann et al. 2005; Schroeder et al. 2007), indicating that pathways for gut granule biogenesis are functioning in these mutants. On the basis of the severe Glo phenotype exhibited by glo-3(−), it is unlikely that glo-3(+) functions upstream of AP-3 or pgp-2(+). To determine whether glo-3(+) promotes gut granule formation in AP-3 or pgp-2(−) mutants, we introduced glo-3(zu446) into apt-6(−) and pgp-2(−) genetic backgrounds. We found that pgp-2(kx48); glo-3(zu446) double mutants completely lacked autofluorescent organelles (Table 3). apt-6(ok429); glo-3(zu446) animals typically lacked and rarely contained very low numbers of autofluorescent compartments (Table 3). Both double mutants lacked birefringent compartments in embryos (data not shown). The phenotypes of the double mutants indicate that GLO-3 acts in parallel or downstream of AP-3 and PGP-2 in gut granule biogenesis.
DISCUSSION
glo-3 alleles:
The nine glo-3 alleles identified to date are nonsense alleles that fall into an allelic series with graded severity of embryonic and adult gut granule biogenesis phenotypes. Our phenotypic and genetic analyses strongly suggest that the relative mutant strengths are class I > class II > class III (Tables 1, 2, and supplemental Table 1). Surprisingly, the most 5′ allele kx90, which is predicted to severely truncate GLO-3, results in a class II phenotype. The most 3′ alleles, gm125 and kx29, predicted to encode the longest GLO-3 mutant proteins also result in a class II phenotype. Class I alleles are flanked by class II alleles. In fact, three alleles, kx91, kx1, and zu446, located within a 12-nucleotide region, each display a distinct embryonic Glo phenotype (Figure 5). The complex relationships between genotype and phenotype suggest that alternative transcriptional start sites, alternative mRNA splicing, or nonsense mediated mRNA decay leading to the production of varied amounts of truncated GLO-3, may differentially affect glo-3 alleles. These results, while not altering our conclusions as to the relative strength of glo-3 alleles, prevent a definitive statement as to whether any of the alleles represent a glo-3 null. Our conclusions regarding glo-3 activity are based upon analysis of a class I mutant, which we believe contains a strong loss-of-function glo-3 allele.
All five class II alleles are partially suppressed by mutations in smg-1, indicating that the nonsense mediated mRNA decay pathway targets their transcripts. Loss of this pathway is predicted to stabilize mRNAs with premature stop codons (Mango 2001). The formation of gut granules in smg-1(−); glo-3(zu446) embryos indicates that the C-terminal half of GLO-3, which includes the third transmembrane domain and likely a large cytoplasmic domain, are not required for partial activity in gut granule biogenesis (Table 1). More surprisingly, glo-3(kx90) and glo-3(kx37) were partially suppressed by smg-1(−) (Table 1). These alleles are predicted to produce extremely truncated forms of GLO-3 (Figure 5). Rather than these truncated GLO-3 proteins being functional, we favor the possibility that low levels of alternative splicing or alternative translation initiation bypass the early stop codons and promote the formation of amino terminally truncated GLO-3 with partial activity. Our analysis of glo-3(−); smg-1(−) mutants strongly suggests that carboxy and possibly amino terminal domains of GLO-3 are not essential for its function in gut granule biogenesis; instead the region of GLO-3 containing transmembrane domains 2 and 3 appears to be critical for its activity.
glo-3 function:
The glo-3 locus encodes two transcripts, glo-3 and glo-3short, which share the same predicted translational start site but differ at their 3′ ends due to alternative splicing and polyadenylation (Figure 5). The proteins encoded by these transcripts are identical for the first 178 amino acids; however, glo-3 encodes a protein with an additional 427 amino acids at its carboxy terminus (Figure 5). We suggest that glo-3short does not significantly function in gut granule biogenesis. Five glo-3 alleles disrupting gut granule biogenesis specifically affect glo-3 and not glo-3short (Figure 5). The glo-3short cDNA expressed under control of the glo-3 promoter did not rescue the Glo phenotypes of glo-3(kx90) (data not shown) indicating that expression of glo-3short is not sufficient to support gut granule biogenesis when glo-3 function is compromised. Moreover, amino and carboxy terminal-tagged forms of GLO-3short were not localized to the gut granule membrane (data not shown).
In contrast to glo-3short, glo-3 plays an essential role in the formation of embryonic gut granules. Our examination of six different markers that label embryonic gut granules failed to show any organelles with gut granule characteristics in glo-3(kx94) (Figure 4). glo-3(+) is likely to specifically function in gut granule formation as other endolysosomal organelles appeared to be properly formed in glo-3(kx94) embryos (Figure 4). Class I and class II alleles disrupting glo-3 function result in the extracellular mislocalization of birefringent material into the embryonic intestinal lumen (Figure 1 and Table 1). Mutations affecting the function of factors with known roles in protein trafficking, such as GLO-1/Rab-38 and AP-3, result in the same phenotype (Hermann et al. 2005), suggesting that GLO-3 functions in trafficking to the embryonic gut granule. GLO-3 is likely positioned to play a direct role in the processes that regulate the formation, maturation, and/or stability of gut granules as a rescuing GLO-3∷GFP fusion is localized to the gut granule membrane (Figure 7).
glo-3 clearly functions in the formation of adult gut granules, as they are lacking in anterior intestinal cells of class I alleles (Figure 2 and Table 2). Interestingly, glo-3(kx94) adults contained a significant number of autofluorescent organelles in posterior intestinal cells (Figure 2 and Table 2). These organelles appear to be gut granules, as they are acidified, terminal endocytic compartments (Figure 3 and supplemental Table 2), both defining gut granule characteristics (Clokey and Jacobson 1986), and glo-1 and glo-4, two genes necessary for gut granule biogenesis (Hermann et al. 2005), are required for the formation of these compartments in glo-3(−) adults (Table 3). While the cellular basis of the anterior–posterior difference in the Glo phenotype is not known, there is clear precedent for differential gene expression along this axis in C. elegans intestinal cells (Schroeder and Mcghee 1998; Fukushige et al. 2005). We suggest that an additional factor or process that can functionally substitute for glo-3 is expressed in posterior intestinal cells of larvae/adults.
The mechanisms that control gut granule formation in embryos and larvae/adults may be functionally distinct, as glo-3(−) class I embryos that lacked observable gut granules developed into adults that contained gut granules (Figures 1 and 2). The gut granules present in glo-3(−) adults might result from a glo-3 independent pathway that functions exclusively in larvae/adults. However, it is also possible that both class I glo-3 alleles we analyzed retain partial activity, which is sufficient to specifically promote gut granule formation in posterior intestinal cells of larvae/adults and not embryos.
The developmental pathway controlling the formation of birefringent gut granules in C. elegans embryos has long been known to be tightly coupled to endoderm fate specification (Laufer et al. 1980). A pathway involving Wnt signaling promotes the expression of two GATA transcription factors END-1/END-3 that induce gut cell fate and gut granule formation (Maduro 2006). These transcription factors promote the expression of genes at the E2 stage, precisely when glo-3 expression initiates (Figure 6), suggesting that glo-3 may be a direct transcriptional target of END-1/END-3. glo-3 joins glo-1, pgp-2, and mrp-4, genes with intestine restricted E2 expression profiles that mediate the formation and differentiation of gut granules (Hermann et al. 2005; Currie et al. 2007; Schroeder et al. 2007). As such, gut granules provide an attractive and unique system to study the developmental pathways that control the cellular mechanisms that regulate the initiation of organelle formation and specialization.
Currently, we can only speculate as to the function of GLO-3 in gut granule formation. GLO-3 likely acts independently of the AP-3 complex and PGP-2 in the biogenesis of gut granules (Table 3). The lack of observable gut granules in glo-3(−) class I embryos is consistent with a role in membrane trafficking. However, GLO-3 is not homologous to known proteins involved in vesicle formation, movement, docking, and fusion. Alternatively, GLO-3 might function in the selection of cargo trafficked to the gut granule. C. elegans lacks obvious homologs of mannose-6-phosphate (Drickamer and Dodd 1999) and sortilin receptors (our unpublished results), the two best-characterized receptors for lysosomal cargo (Ghosh et al. 2003; Ni et al. 2006), indicating that the mechanisms in C. elegans selecting soluble lysosomal contents utilizes unidentified and possibly novel proteins. Recently, LIMP-2 a multipass lysosomal-associated protein was shown to act as a new type of sorting receptor for lysosomal cargo in mammalian cells (Reczek et al. 2007). While GLO-3 and LIMP-2 lack sequence homology, they have similar domain structures, raising the possibility that GLO-3 might function in the recognition of cargo trafficked to the gut granule. Testing this hypothesis awaits the identification of soluble gut granule-associated proteins.
Conservation of proteins controlling lysosome biogenesis:
The proteins that mediate lysosome biogenesis are highly conserved in eukaryotes. The retromer, ESCRT, and HOPS protein complexes first identified as being necessary for trafficking to the yeast vacuole, a lysosomal equivalent, are well conserved and present in a broad range of metazoan taxa (Bowers and Stevens 2005). Similarly conserved are the vesicle coats, adapter complexes, phosphatidylinositol modifying enzymes, Rabs, and SNAREs that regulate cargo selection, vesicle budding, transport, and fusion within the endolysosomal system (Mullins and Bonifacino 2001). The formation of LROs requires many of the same highly conserved proteins required for lysosome biogenesis, including the AP-1, AP-3, and HOPS complexes (Raposo et al. 2007).
Studies of LRO biogenesis have led to the identification of proteins that function in organelle formation that are not as broadly conserved. Rab38/GLO-1, orthologs of which are not found in yeast (Pereira-Leal and Seabra 2001), are required for LRO biogenesis in C. elegans (Hermann et al. 2005), Drosophila (Ma et al. 2004), and mammals (Loftus et al. 2002; Wasmeier et al. 2006). Eight proteins composing the BLOC-1 and BLOC-2 complexes that function in LRO biogenesis in mammals (Raposo et al. 2007), are found only in metazoans (Dell'angelica 2004). Nearly all of the subunits of the BLOC-1 and BLOC-2 complexes are conserved in Drosophila, where at least one subunit is necessary for the biogenesis of a LRO (Falcon-Perez et al. 2007; Syrzycka et al. 2007). Notably, few BLOC-1 or BLOC-2 subunits are obviously conserved in C. elegans (De Voer et al. 2008).
GLO-3 is unique among factors functioning in LRO biogenesis, being present in only one group of organisms. Extensive searches failed to identify any proteins outside nematodes with sequence similarity to GLO-3. It is possible that proteins functionally homologous to GLO-3 have significantly diverged so as not to be detectable. Alternatively, nematodes may have evolved a novel mechanism involving GLO-3 that mediates the assembly of lysosome-related organelles. Future studies aimed at identifying the molecular function of GLO-3 should resolve this issue.
Acknowledgments
We gratefully acknowledge members of the Hermann, Binford, Lycan, and Reiness laboratories for advice. We thank Anjon Audhya, Barth Grant, Yuji Kohara, Kenji Kontani, and Renaud Legouis for gifts of strains, antisera, and plasmids. Some nematode strains were provided by the Caenorhabditis Genetics Center, the C. elegans Knockout Consortium, and the National Bioresource Project for C. elegans. We thank Karen Kelly for genetic characterization of class I glo-3 alleles. This work was supported by grants from the National Science Foundation (MCB-0314332 and MCB-0716280), the John S. Rogers Summer Research Program, and the M. J. Murdock Charitable Trust.
References
- Ashrafi, K., F. Y. Chang, J. L. Watts, A. G. Fraser, R. S. Kamath et al., 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421 268–272. [DOI] [PubMed] [Google Scholar]
- Audhya, A., A. Desai and K. Oegema, 2007. A role for Rab5 in structuring the endoplasmic reticulum. J. Cell Biol. 178 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu, P., 1974. Biochemical genetics of Caenorhabditis elegans. Mol. Gen. Genet. 135 39–44. [Google Scholar]
- Blumenthal, T., and K. Steward, 1997. RNA processing and gene structure, pp. 117–145 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- Boehm, M., and J. S. Bonifacino, 2001. Adaptins: the final recount. Mol. Biol. Cell 12 2907–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst, P., and R. O. Elferink, 2002. Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 71 537–592. [DOI] [PubMed] [Google Scholar]
- Bowers, K., and T. H. Stevens, 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1744 438–454. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. C., P. J. Schweinsberg, S. Vashist, D. P. Mareiniss, E. J. Lambie et al., 2006. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol. Biol. Cell 17 1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, N., M. Madesh, L. W. Gonzales, M. Zhao, K. Yu et al., 2006. Functional and trafficking defects in ATP binding cassette A3 mutants associated with respiratory distress syndrome. J. Biol. Chem. 281 9791–9800. [DOI] [PubMed] [Google Scholar]
- Clokey, G. V., and L. A. Jacobson, 1986. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech. Ageing Dev. 35 79–94. [DOI] [PubMed] [Google Scholar]
- Currie, E., B. King, A. L. Lawrenson, L. K. Schroeder, A. M. Kershner et al., 2007. Role of the Caenorhabditis elegans multidrug resistance gene, mrp-4, in gut granule differentiation. Genetics 177 1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Voer, G., D. Peters and P. E. Taschner, 2008. Caenorhabditis elegans as a model for lysosomal storage disorders. Biochim. Biophys. Acta 1782 433–446. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E. C., 2004. The building BLOC(k)s of lysosomes and related organelles. Curr. Opin. Cell Biol. 16 458–464. [DOI] [PubMed] [Google Scholar]
- Drickamer, K., and R. B. Dodd, 1999. C-type lectin-like domains in Caenorhabditis elegans: predictions from the complete genome sequence. Glycobiology 9 1357–1369. [DOI] [PubMed] [Google Scholar]
- Falcon-Perez, J. M., R. Romero-Calderon, E. S. Brooks, D. E. Krantz and E. C. Dell'Angelica, 2007. The Drosophila pigmentation gene pink (p) encodes a homologue of human Hermansky-Pudlak syndrome 5 (HPS5). Traffic 8 154–168. [DOI] [PubMed] [Google Scholar]
- Fukushige, T., B. Goszczynski, J. Yan and J. D. McGhee, 2005. Transcriptional control and patterning of the pho-1 gene, an essential acid phosphatase expressed in the C. elegans intestine. Dev. Biol. 279 446–461. [DOI] [PubMed] [Google Scholar]
- Ghosh, P., N. M. Dahms and S. Kornfeld, 2003. Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell. Biol. 4 202–212. [DOI] [PubMed] [Google Scholar]
- Grant, B., Y. Zhang, M.-C. Paupard, S. X. Lin, D. Hall et al., 2001. Evidence that RME-1, a conserved C. elegans EH domain protein, functions in endocytic recycling. Nat. Cell Biol. 3 573–579. [DOI] [PubMed] [Google Scholar]
- Grimson, A., S. O'Connor, C. L. Newman and P. Anderson, 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell. Biol. 24 7483–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, G. J., L. K. Schroeder, C. A. Hieb, A. M. Kershner, B. M. Rabbitts et al., 2005. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell 16 3273–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques 32 728–730. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., A. Papp, R. Pulak, V. Ambros and P. Anderson, 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K., and W. Stoffel, 1993. TMBASE: a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374 166. [Google Scholar]
- Huizing, M., R. E. Boissy and W. A. Gahl, 2002. Hermansky-Pudlak syndrome: vesicle formation from yeast to man. Pigment Cell Res. 15 405–419. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231–237. [DOI] [PubMed] [Google Scholar]
- Kimura, K. D., H. A. Tissenbaum, Y. Liu and G. Ruvkun, 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 942–946. [DOI] [PubMed] [Google Scholar]
- Kiontke, K., N. P. Gavin, Y. Raynes, C. Roehrig, F. Piano et al., 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneen, M., J. Farinas, Y. Li and A. S. Verkman, 1998. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys. J. 74 1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontani, K., I. P. G. Moskowitz and J. H. Rothman, 2005. Repression of cell-cell fusion by components of the C. elegans vacuolar ATPase complex. Dev. Cell 8 787–794. [DOI] [PubMed] [Google Scholar]
- Laufer, J. S., P. Bazzicalupo and W. B. Wood, 1980. Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell 19 569–577. [DOI] [PubMed] [Google Scholar]
- Leung, B., G. J. Hermann and J. R. Priess, 1999. Organogenesis of the Caenorhabditis elegans intestine. Dev. Biol. 216 114–134. [DOI] [PubMed] [Google Scholar]
- Loftus, S. K., D. M. Larson, L. L. Baxter, A. Antonellis, Y. Chen et al., 2002. Mutation of melanosome protein RAB38 in chocolate mice. Proc. Natl Acad. Sci. USA 99 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., H. Plesken, J. E. Treisman, I. Edelman-Novemsky and M. Ren, 2004. Lightoid and Claret: A rab GTPase and its putative guanine nucleotide exchange factor in biogenesis of Drosophila eye pigment granules. Proc. Natl. Acad. Sci. USA 101 11652–11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro, M. F., 2006. Endomesoderm specification in Caenorhabditis elegans and other nematodes. BioEssays 28 1010–1022. [DOI] [PubMed] [Google Scholar]
- Mango, S. E., 2001. Stop making nonSense: the C. elegans smg genes. Trends Genet. 17 646–653. [DOI] [PubMed] [Google Scholar]
- Maquat, L. E., 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5 89–99. [DOI] [PubMed] [Google Scholar]
- Mello, C. C., J. M. Kramer, D. Stinchcomb and V. Ambros, 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, C., and J. S. Bonifacino, 2001. The molecular machinery for lysosome biogenesis. BioEssays 23 333–343. [DOI] [PubMed] [Google Scholar]
- Ni, X., M. Canuel and C. R. Morales, 2006. The sorting and trafficking of lysosomal proteins. Histol. Histopathol. 21 899–913. [DOI] [PubMed] [Google Scholar]
- Ogg, S., and G. Ruvkun, 1998. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell 2 887–893. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J. B., and M. C. Seabra, 2001. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313 889–901. [DOI] [PubMed] [Google Scholar]
- Raposo, G., M. S. Marks and D. F. Cutler, 2007. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr. Opin. Cell Biol. 19: 394–401. [DOI] [PMC free article] [PubMed]
- Reczek, D., M. Schwake, J. Schroder, H. Hughes, J. Blanz et al., 2007. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell 131 770–783. [DOI] [PubMed] [Google Scholar]
- Roudier, N., C. Lefebvre and R. Legouis, 2005. CeVPS-27 is an endosomal protein required for the molting and the endocytic trafficking of the low-density lipoprotein receptor-related protein 1 in Caenorhabditis elegans. Traffic 6 695–705. [DOI] [PubMed] [Google Scholar]
- Schroeder, D. F., and J. D. McGhee, 1998. Anterior-posterior patterning within the Caenorhabditis elegans endoderm. Development 125 4877–4887. [DOI] [PubMed] [Google Scholar]
- Schroeder, L. K., S. Kremer, M. J. Kramer, E. Currie, E. Kwan et al., 2007. Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle. Mol. Biol. Cell 18 995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F., C. Moorman, A. M. van der Linden, E. Kuijk, P. V. van den Berghe et al., 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1 E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J. E., E. Schierenberg, J. G. White and J. N. Thomson, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100 64–119. [DOI] [PubMed] [Google Scholar]
- Syrzycka, M., L. A. McEachern, J. Kinneard, K. Prabhu, K. Fitzpatrick et al., 2007. The pink gene encodes the Drosophila orthologue of the human Hermansky-Pudlak syndrome 5 (HPS5) gene. Genome 50 548–556. [DOI] [PubMed] [Google Scholar]
- Treusch, S., S. Knuth, S. A. Slaugenhaupt, E. Goldin, B. D. Grant et al., 2004. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA 13 4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, S., and J. Miwa, 1978. Characterization of temperature-sensitive, fertilization-defective mutants of the nematode Caenorhabditis elegans. Genetics 88 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmeier, C., M. Romao, L. Plowright, D. C. Bennett, G. Raposo et al., 2006. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J. Cell Biol. 175 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, M. L., 2006. Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res. 19 19–42. [DOI] [PubMed] [Google Scholar]