Abstract
The CLAVATA1 (CLV1) receptor kinase regulates stem cell specification at shoot and flower meristems of Arabidopsis. Most clv1 alleles are dominant negative, and clv1 null alleles are weak in phenotype, suggesting additional receptors functioning in parallel. We have identified two such parallel receptors, BAM1 and BAM2. We show that the weak nature of the phenotype of clv1 null alleles is dependent on BAM activity, with bam clv mutants exhibiting severe defects in stem cell specification. Furthermore, BAM activity in the meristem depends on CLV2, which is required in part for CLV1 function. In addition, clv1 mutants enhance many of the Bam− organ phenotypes, indicating that, contrary to current understanding, CLV1 function is not specific to the meristem. CLV3 encodes a small, secreted peptide that acts as the ligand for CLV1. Mutations in clv3 lead to increased stem cell accumulation. Surprisingly, bam1 and bam2 mutants suppress the phenotype of clv3 mutants. We speculate that in addition to redundant function in the meristem center, BAM1 and BAM2 act to sequester CLV3-like ligands in the meristem flanks.
THE vast majority of tissues and organs of the adult plant are formed postembryonically. To achieve continuous organ formation plants rely on the activity of stem cells located at meristems (Byrne et al. 2003; Carles and Fletcher 2003; Gross-Hardt and Laux 2003). The shoot apical meristem, which produces the aerial portion of the plant, is composed of a central pool of stem cells surrounded by descendant cells that are directed toward differentiation. Division of stem cells results in daughter cells that maintain the central stem cell population as well as daughter cells that are localized more peripherally, where they perceive positional cues that drive their differentiation.
Stem cell specification and organogenesis at the Arabidopsis shoot apical meristem is regulated by a receptor-kinase signaling system that includes the CLAVATA1 (CLV1), CLV2, and CLV3 gene products. CLV1 encodes a receptor-kinase protein with 21 leucine-rich repeats (LRRs) in its predicted extracellular domain, a single pass transmembrane domain, and a cytoplasmic serine/threonine kinase domain (Clark et al. 1997). CLV2 encodes a protein structurally similar to CLV1, however CLV2 has a very small predicted cytoplasmic domain with no known signaling motifs (Jeong et al. 1999). CLV1 accumulation is at least in part dependent on the presence of CLV2 (Jeong et al. 1999). Furthermore, CLV1 and CLV2 are hypothesized to form a heterodimer on the basis of migration in both gel chromatography and nonreducing protein gel blots (Trotochaud et al. 1999).
Several lines of evidence suggest that CLV3 is the ligand for CLV1. CLV3 is both secreted and appears to be subjected to proteolytic maturation, releasing a conserved polypetide, termed the CLV3/ESR-related (CLE) domain, from the C terminus that is capable of altering plant development when added exogenously (Fiers et al. 2005; Ito et al. 2006; Kondo et al. 2006; Ni and Clark 2006). Null mutations in CLV3 are largely epistatic to mutations in CLV1 and CLV2, while CLV3 overexpression phenotypes are dependent on CLV1, suggesting that CLV3 functions upstream of CLV1 (Clark et al. 1995; Brand et al. 2000). In addition, CLV3 is required for the formation of a large molecular mass complex containing CLV1 (Trotochaud et al. 1999). CLV1 acts to limit the diffusion of CLV3 protein, suggesting that CLV1 binds to CLV3, sequestering it (Lenhard and Laux 2003). Recent work has shown that the CLE peptide is capable of binding to the CLV1 extracellular domain (Ogawa et al. 2008). Taken together these data suggest a model in which CLV1 and CLV2 form a receptor complex that is capable of activating specific signal transduction pathways upon perception of the CLE peptide derived from CLV3.
CLV1, CLV2, and CLV3 function in a common genetic pathway to regulate the expression domain of the stem cell-promoting transcription factor, WUSCHEL (WUS) (Mayer et al. 1998; Brand et al. 2000; Schoof et al. 2000). Mutations in CLV1, CLV2, or CLV3 result in an increased WUS expression domain and a larger meristem with extra stem cells (Clark et al. 1993, 1995; Kayes and Clark 1998; Brand et al. 2000; Schoof et al. 2000), while mutations in WUS result in reduction or elimination of the meristem (Laux et al. 1996). The related phosphatases POLTERGEIST (POL) and PLL1 are signaling intermediates acting downstream of CLV1, CLV2, and CLV3. POL/PLL1 act to maintain WUS expression within the meristem, while CLV signaling represses POL/PLL1 activity (Yu et al. 2000, 2003; Song and Clark 2005; Song et al. 2006). The remaining genetic and biochemical mechanisms that link CLV1 activation and WUS expression are largely unknown.
Several lines of evidence suggest the presence of additional receptors that function in parallel with CLV1. Most of the clv1 mutant alleles identified in mutagenic screens are dominant-negative missense alleles that exhibit intermediate-to-strong phenotypes, while clv1 null alleles exhibit quite weak phenotypes by comparison (Diévart et al. 2003). In addition, clv3 null alleles exhibit strong meristem phenotypes and are epistatic to both null and dominant-negative alleles of clv1, demonstrating that CLV3 retains some function in the absence of CLV1 (Clark et al. 1995). Candidates for CLV1-redundant receptors can be found among the related BAM receptor kinases (BAM1, BAM2, and BAM3) (DeYoung et al. 2006). CLV1 and the BAM genes form a monophyletic group of Arabidopsis genes encoding related receptor kinases, although the duplication giving rise to the BAM and CLV1 clades predates the split between monocots and dicots (DeYoung et al. 2006).
Despite the related sequences of the these receptors, several functional differences exist between CLV1 and BAM receptors (DeYoung et al. 2006). While CLV1 promotes stem cell differentiation, BAM receptors are required for stem cell maintenance. In bam1 bam2 double mutants, a reduction in meristem size is observed, while in bam1 bam2 bam3 triple mutants a frequent incidence of premature shoot and flower meristem termination is observed. A second key difference lies in the specificity of the receptors. clv1 mutants only exhibit phenotypes at the shoot and flower meristem, while all three BAM receptors regulate numerous aspects of Arabidopsis development, from leaf vascular patterning to the proper development of all floral organs, especially the anthers and ovules (Clark et al. 1993; DeYoung et al. 2006; Hord et al. 2006). Despite these differences, CLV1 and BAM receptors appear to have retained significant similarities in protein function, because broadly expressed CLV1 can fully rescue bam mutants while meristem-expressed BAM receptors can partially rescue clv1 mutants (DeYoung et al. 2006). Thus, on a genetic level, the differences between CLV1 and BAM function appear to be largely driven by expression differences. Consistent with this, BAM1 and BAM2 are expressed throughout the plant and exhibit relatively high levels of expression on the meristem flanks, while CLV1 is expressed primarily in the meristem center (Clark 1997; DeYoung et al. 2006).
To understand the relationships between CLV1, CLV2, and BAM receptors, especially within the meristem where they play apparently complimentary roles, we sought to assess the genetic interactions of BAM1 and BAM2 with mutant alleles of CLV1, CLV2, and CLV3. We show that bam and clv mutants display complex gene- and allele-specific interactions. bam mutants suppress clv3 phenotypes, have no effect on clv2 phenotypes, and enhance clv1 null phenotypes. Our evidence suggests that BAM proteins function similarly to CLV1 on a biochemical level and, in fact, function in parallel with CLV1 in the meristem center, but carry out separate roles elsewhere within the plant. We propose that one of these roles is to sequester CLE peptide ligands on the flanks of the meristem, creating a buffer around the meristem that prevents these ligands from upsetting the delicate balance necessary for stem cell maintenance.
MATERIALS AND METHODS
Plant growth (Yu et al. 2003), SEM analysis (Diévart et al. 2003), and phloroglucinol staining (Prigge et al. 2005) were performed as previously described unless otherwise noted.
Genetic analysis:
clv1-1 bam1-1, clv1-4 bam1-1, clv1-7 bam1-1, clv1-11 bam1-1, clv2-1 bam1-1, clv3-2 bam1-1, clv1-1 bam2-1, clv1-4 bam2-1, clv1-7 bam2-1, clv1-11 bam2-1, clv2-1 bam2-1, clv3-2 bam2-1, clv1-7 bam1-3, clv1-11 bam1-3, clv1-7 bam2-3, and clv1-11 bam2-3 double-mutant combinations were generated by crossing plants homozygous for the bam1-1, bam2-1, bam1-3, or bam2-3 allele to plants homozygous for the respective clv1, clv2, or clv3 mutant allele. Individual F2 progeny were screened on the basis of phenotype to identify individuals homozygous for the respective clv1, clv2, or clv3 mutant alleles and, in the cases of bam1-1 and bam1-3, to identify lines homozygous for the er-1 allele. No segregation distortion was observed that might suggest clv homozygous mutants were not segregating in a 3:1 ratio. These plants were subsequently screened using a PCR strategy to identify individuals homozygous for bam1-1, bam2-1, bam1-3, or bam2-3, respectively, as previously described (DeYoung et al. 2006). In some cases, progeny from F2 plants heterozygous for bam1-1, bam2-1, bam1-3, or bam2-3 were screened in the F3 generation to identify lines homozygous for bam1-1, bam2-1, bam1-3, or bam2-3, respectively.
clv1-1 bam1-1 bam2-1, clv1-4 bam1-1 bam2-1, clv1-7 bam1-1 bam2-1, clv1-11 bam1-1 bam2-1, clv2-1 bam1-1 bzm2-1, and clv3-2 bam1-1 bam2-1 triple-mutant combinations were generated by crossing clv bam1 double-mutant plants to the respective clv bam2 mutant plants. For example, clv1-1 bam1-1 bam2-1 plants were generated by crossing clv1-1 bam1-1 plants to clv1-1 bam2-1 and screening the F2 progeny of this cross to identify plants homozygous for both bam1-1 and bam2-1. Because the bam1-1 bam2-1 allele combination causes infertility, the bam1 bam2 clv1-1 plants used for subsequent analysis were identified from segregating progeny of sibling plants heterozygous at one BAM locus and homozygous mutant at the other, as above.
Floral phenotype measurement:
The mean number of carpels per flower was determined by averaging the number of carpels per flower from the first 10 flowers of each plant.
Image analysis:
Photographic images were captured with a Nikon Coolpix 995 digital camera. Brightness and contrast of some photographic and SEM images were adjusted with Adobe PHOTOSHOP CS.
Accession numbers:
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession nos. Q9SYQ8 (AT1G75820) for CLV1, AAF02655 (AT1G65380) for CLV2, AAD27620 (AT2G27250) for CLV3, NM_125967 (AT5G65700) for BAM1, and NM_114827 (AT3G49670) for BAM2. All bam alleles used in this study have been provided to the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu/pcmb/Facilities/abrc/abrchome.htm).
RESULTS
clv3 phenotypes are suppressed by bam1 bam2 mutations:
Our previous genetic studies have suggested that BAM and CLV1 proteins have retained significant biochemical redundancy, although the developmental role of BAM1 and BAM2 is apparently antagonistic to that of CLV1 in the meristem (DeYoung et al. 2006). We therefore undertook experiments to characterize the role of BAM receptors in the absence of specific CLV pathway components in an attempt to clarify the role of the BAM receptors within meristem development. In particular, it is critical to investigate the relationship(s) between BAM and CLV3, because the expression of these genes in adjacent cell populations raises the possibility that CLV3 acts as a ligand for the BAM receptors on the flanks of the meristem.
To this end, bam1-1 and bam2-1 single and double mutants were generated in the clv3-2 null mutant background. To quantify the relative effect of these mutations on the flower meristem, we took advantage of previous observations that floral organ number, especially carpel number, reflects the relative size of the flower meristem and is a sensitive method of detecting change in flower meristem size (Clark et al. 1993; Song and Clark 2005). When carpel number per flower was assessed in bam1 clv3 and bam2 clv3 plants, we observed a modest, but statistically significant reduction when compared to clv3 single-mutant plants (Table 1). bam1 mutations lead to a larger absolute reduction in carpel number in the clv3-2 mutant background than bam2. bam1 bam2 clv3 triple mutants exhibited more obvious suppression of the Clv− phenotype, with ∼4.3 carpels per flower compared to clv3-2 single mutants, which have ∼6.7 carpels per flower (Table 1). The suppression of clv3 by bam mutations is consistent with effects of multiple bam mutants to reduce meristem size (DeYoung et al. 2006).
TABLE 1.
Phenotypic interactions of bam1 and bam2 mutations with clv3 and clv2 mutations
| Genotype | Mean carpels/flower | SEa | nb | Pc |
|---|---|---|---|---|
| Lerd | 2.00 | 0.000 | 100 | |
| bam2-1d | 2.00 | 0.000 | 100 | |
| bam2-3d | 2.00 | 0.000 | 100 | |
| bam1-1d | 2.00 | 0.000 | 100 | |
| bam1-3d | 2.00 | 0.000 | 100 | |
| bam1-1 bam2-1d | 2.00 | 0.000 | 100 | |
| bam1-3 bam2-3d | 2.00 | 0.000 | 100 | |
| clv3-2 | 6.66 | 0.081 | 100 | |
| clv3-2 bam2-1 | 5.96 | 0.057 | 300 | <0.001 |
| clv3-2 bam1-1 | 5.82 | 0.061 | 300 | <0.001 |
| clv3-2 bam1-1 bam2-1 | 4.32 | 0.066 | 140 | <0.001 |
| clv2-1 | 3.98 | 0.083 | 100 | |
| clv2-1 bam2-1 | 3.65 | 0.044 | 200 | <0.001 |
| clv2-1 bam1-1 | 3.34 | 0.057 | 200 | <0.001 |
| clv2-1 bam1-1 bam2-1 | 3.89 | 0.065 | 100 | 0.394 |
Standard error.
Number of flowers evaluated.
P-value calculated using Student's t-test comparing clv bam1, clv bam2, or clv bam1 bam2 to the respective clv single mutant.
Data from DeYoung et al. (2006).
clv2 phenotypes are unaltered by bam1 bam2:
CLV2 function is necessary for normal CLV1 function (Jeong et al. 1999). Because CLV2 expression can be detected broadly within the meristem, as well as outside of the meristem, and because clv2 mutants exhibit several phenotypes during organ development, we have hypothesized that CLV2 interacts with multiple receptor-like kinases in addition to CLV1 (Kayes and Clark 1998; Jeong et al. 1999). Considering these observations, CLV2 could conceivably act as a partner for the BAM receptors.
To test this idea, bam1-1 and bam2-1 single and double mutants were combined with the clv2-1 null mutation. While bam1 clv2 and bam2 clv2 exhibited a modest but statistically significant suppression in carpel number compared to clv2 single mutants, the mean carpel number of bam1 bam clv2 triple-mutant flowers was not significantly different from that of clv2-1 alone (Table 1). Thus, clv2 is unaltered bam1 bam2 double mutants, in direct contrast to clv3. This raises the possibility that CLV2 may be necessary for both CLV1 and BAM function, which would imply that the Clv2− phenotype is the combined result of the loss of both the meristem-inhibiting CLV1 pathway and the meristem-promoting BAM pathway.
Synergistic meristem interactions of bam1, bam2, and clv1 null alleles:
To determine the net effect of removing both BAM and CLV1 receptors from the meristem, bam1-1 and bam2-1 mutations were combined with clv1-7 or clv1-11 mutations. clv1-11 is a null allele containing a T-DNA insertion in the coding sequence for the LRR domain, while clv1-7 contains a nonsense mutation that results in truncation of a large portion of the kinase domain and appears phenotypically similar to clv1 null alleles (Diévart et al. 2003). Unexpectedly, bam1 and bam2 alleles significantly enhanced the flower meristem defects in both the clv1-11 and clv1-7 backgrounds (Table 2). To test if these results could be attributed to linked mutations in the bam1-1 or bam2-1 background, we repeated these experiments with the bam1-3 and bam2-3 alleles. Again we observed a significant enhancement of the meristem defect (Table 2). For all combinations, bam1 alleles resulted in a stronger enhancement than bam2 alleles. Interestingly, bam1 caused a stronger suppression of clv3 than bam2 (Table 1).
TABLE 2.
Phenotypic interactions of bam1 and bam2 mutations with null and dominant-negative clv1 alleles
| Genotype | Mean carpels/flower | SEa | nb | Pc |
|---|---|---|---|---|
| clv1-7 | 3.09 | 0.047 | 300 | |
| clv1-7 bam2-1 | 3.80 | 0.037 | 200 | <0.001 |
| clv1-7 bam2-3 | 3.35 | 0.054 | 200 | <0.001 |
| clv1-7 bam1-1 | 4.62 | 0.069 | 300 | <0.001 |
| clv1-7 bam1-3 | 4.34 | 0.080 | 230 | <0.001 |
| clv1-7 bam1-1 bam2-1 | 7.44 | 0.116 | 100 | <0.001 |
| clv1-11 | 3.60 | 0.042 | 200 | |
| clv1-11 bam2-1 | 4.40 | 0.074 | 100 | <0.001 |
| clv1-11 bam2-3 | 4.26 | 0.045 | 200 | <0.001 |
| clv1-11 bam1-1 | 5.18 | 0.072 | 200 | <0.001 |
| clv1-11 bam1-3 | 5.44 | 0.087 | 179 | <0.001 |
| clv1-11 bam1-1 bam2-1 | 8.65 | 0.198 | 40 | <0.001 |
| clv1-1 | 4.32 | 0.051 | 100 | |
| clv1-1 bam2-1 | 4.29 | 0.056 | 200 | 0.693 |
| clv1-1 bam1-1 | 4.37 | 0.050 | 200 | 0.483 |
| clv1-1 bam1-1 bam2-1 | 7.35 | 0.185 | 60 | <0.001 |
| clv1-4 | 5.45 | 0.077 | 130 | |
| clv1-4 bam2-1 | 6.67 | 0.057 | 300 | <0.001 |
| clv1-4 bam1-1 | 6.41 | 0.076 | 200 | <0.001 |
| clv1-4 bam1-1 bam2-1 | 8.97 | 0.200 | 30 | <0.001 |
Standard error.
Number of flowers evaluated.
P-value calculated using Student's t-test comparing clv1 bam1, clv1 bam2, or clv1 bam1 bam2 to the respective clv1 single mutant.
To assess the effect of removing all three receptor kinases on meristem development, bam1-1 bam2-1 clv1-11 and bam1-1 bam2-1 clv1-7 triple-mutant plants were generated. In both cases the bam1 bam2 clv1 triple mutants exhibited rather extreme phenotypes, with the mean number of carpels per flower >8.6 for the clv1-11 combination (Table 2).
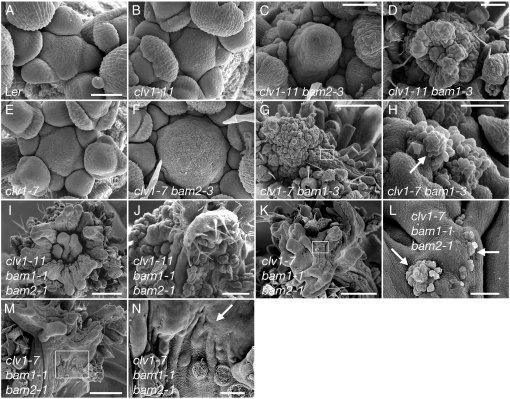
To determine if these unexpected interactions were specific to the flower meristem, we examined shoot apical meristem size and organization in clv1, bam1 clv1, bam2 clv1, and bam1 bam2 clv1 triple-mutant plants. In each case, bam mutations enhanced the shoot meristem defects of clv1-7 and clv1-11, in accordance with the flower meristem enhancement. Namely, bam1 clv1 were more enhanced compared to clv1 than bam2 clv1, and the bam1 bam2 clv1 triple mutant exhibited extreme shoot meristem defects (Figure 1). Indeed, the bam1 bam2 clv1 triple mutants exhibited novel phenotypes that are discussed below.
Figure 1.—
bam1 and bam2 enhance meristem enlargement by clv1 and function synergistically with clv1 resulting in novel phenotypes. Scanning electron micrographs of Ler (A), clv1-11 (B), clv1-11 bam2-3 (C), clv1-11 bam1-3 (D), clv1-7 (E), clv1-7 bam2-3 (F), clv1-7 bam1-3 (G and H), clv1-11 bam1-1 bam2-1 (I and J), clv1-7 bam1-1 bam2-1 (K–N). Older flowers were removed for clarity. H, L, and N correspond to the boxed regions in G, K, and M, respectively. Differentiated cells surrounded by undifferentiated meristem cells are indicated with an arrow (H and L). Lack of flower primordia is indicated by an arrow (N). A, B, E, and F are shown at the same magnification. Bars: 50 μm (A, B, E, F, H, and L); 100 μm (C, D, and N); 250 μm (J); and 500 μm (G, I, K, and M).
These data are not consistent with a model in which BAM genes simply function in a separate pathway antagonistic to the CLV pathway. The observation that BAM genes are required for the weak phenotype of clv1 null alleles suggests that the BAM and CLV loci interact in a complex manner and that BAM and CLV1 genes may act in a functionally redundant manner in the center of the meristem.
Divergent interactions of clv1 dominant-negative alleles:
The apparent functional overlap of the BAM and CLV1 receptors evident from the bam clv1 combinations could occur either within the same cells or within separate cells. Overlap occurring within separate cells would suggest that BAM acts in a complicated fashion on the meristem periphery to regulate meristem development. Overlap within the same cells would suggest that although BAM expression is low at the center of the meristem (DeYoung et al. 2006), functional BAM receptors are still present and acting redundantly with CLV1 in these cells. To distinguish between these possibilities, we combined bam1 and bam2 mutations with the clv1 dominant-negative alleles clv1-1 and clv1-4. These alleles contain missense mutations in the kinase domain and LRR domain, respectively, and result in phenotypes significantly more severe than clv1 null alleles (Clark et al. 1997; Diévart et al. 2003). We have hypothesized that clv1 dominant-negative alleles block the action of receptor kinases with functional overlap.
If the overlapping function of BAM receptors with CLV1 occurs in separate cells, we would expect similar interactions of bam mutants with clv1 null and dominant-negative alleles. If the functional overlap occurs within the same cells, clv1 dominant-negative alleles might partially block BAM1 and/or BAM2 function.
When the bam clv1-1 double mutants were assayed, we observed that neither the bam1 clv1-1 nor the bam2 clv1-1 double mutants exhibited a stronger phenotype than clv1-1 alone (Table 2). This indicates that the genetic interactions of bam1 and bam2 with clv1-1 are different from those with the clv1 null alleles. When determining the genetic interactions with clv1-4, we observed a different result: both bam1 and bam2 were capable of enhancing the clv1-4 allele. Thus, the nature of the dominant-negative effect in the kinase domain missense allele (clv1-1) may be different from the LRR domain missense allele (clv1-4). The LRR domain dominant-negative alleles of clv1 are invariably more severe phenotypically than the kinase domain dominant-negative alleles (Diévart et al. 2003), providing additional evidence of functional differences in dominant-negative action between LRR and kinase mutants.
Neither clv1-1 nor clv1-4 exhibit the most severe clv mutant phenotypes, and CLV3 still functions in these mutant backgrounds (Clark et al. 1995). Consistent with this, the bam1 bam2 clv1-1 and the bam1 bam2 clv1-4 triple-mutant plants were enhanced compared to the corresponding clv1 single mutant, and displayed phenotypes similar to bam1 bam2 clv1 null triple mutants (Table 2).
Novel phenotypes in bam1 bam2 clv triple mutants:
The shoot apical meristems of bam1 bam2 clv1 plants exhibited unique phenotypes not seen in any clv single mutant. These meristems occasionally lost organogenesis in portions of the meristem periphery (Figure 1, M and N). Furthermore, bam1 bam2 clv1 shoot apical meristems, and to a lesser degree, clv1 bam1 shoot apical meristems, developed patches of differentiated cells (in one case, complete leaves) across the normally continuous field of undifferentiated meristem cells (Figure 1, G, H, and J–L). Finally, bam1-1 bam2-1 clv1-11 and bam1-1 bam2-1 clv1-7 flowers developed an enormous indeterminate meristem in the center of each flower (Figure 2Q).
Figure 2.—
clv1 bam1 bam2 plants exhibit novel phenotypes in floral meristem and nonmeristem tissues. Rosettes of Ler (A), clv1-7 (B), clv1-11 (C), bam1-1 bam2-1 (D), clv1-7 bam1-1 bam2-1 (E), and clv1-11 bam1-1 bam2-1 plants 14 days after germination. Gynoecia of Ler (G), clv1-7 (H), and clv1-7 bam1-1 bam2-1 (I and J) plants. Scanning electron micrographs of clv1-7 (K), bam1-1 bam2-1 (L), and clv1-7 bam1-1 bam2-1 (M) young flowers are shown. Some sepals were removed from the clv1-7 (K) and bam1-1 bam2-1 (L) flowers to reveal the inner organs. No organs were removed from the clv1-7 bam1-1 bam2-1 flower (M). clv1-7 bam1-1 bam2-1 plant (right, boxed) beside a sibling plant (left) that is either heterozygous or homozygous wild type at both BAM1 and BAM2 loci (N). Close-up of clv1-7 bam1-1 bam2-1 plant (N, inset). Scanning electron micrographs of clv1-7 (O), bam1-1 bam1-2 (P), and clv1-11 bam1-1 bam1-2 gynoecia are shown. A–F are shown at the same magnification as are G, H, and I. J is a close-up view of a clv1-7 bam1-1 bam2-1 gynoecium. Bars, 100 μm (L and M); 250 μm (K); 500 μm (O–Q). Asterisks indicate attachment site of sepals, petals, and stamens. S, stigmatic tissue; VL, valve-like strips; se, sepal primordium; pe, petal primordium; st, stamen primordium; and ca, carpel primordium.
In addition to the synergistic interactions observed in the shoot meristem, bam1-1 bam2-1 clv1-11 and bam1-1 bam2-1 clv1-7 triple mutants displayed a number of novel phenotypes outside of the meristem (Figure 2). bam1-1 bam2-1 clv1-11 and bam1-1 bam2-1 clv1-7 plants were much smaller than wild-type and bam1-1 bam2-1 plants, even when they were fully grown (Figure 2, E, F, and N; data not shown). bam1-1 bam2-1 clv1-11 and bam1-1 bam2-1 clv1-7 plants had very small leaves and the number of rosette leaves was increased. The stems of the primary inflorescence of clv1-11 bam1-1 bam2-1 and clv1-7 bam1-1 bam2-1 plants were also extremely thick (Figure 2N; see below). The sepals, petals, and stamens of bam1-1 bam2-1 clv1-11 and bam1-1 bam2-1 clv1-7 flowers were sometimes filamentous and generally looked less developed than those of bam1 bam2 flowers (Figure 2M; data not shown). The fruits of bam1-1 bam2-1 clv1-11 and bam1-1 bam2-1 clv1-7 plants were very short, however the pedicels were longer than those of wild type (WT) (Figure 2N).
The replacement of valve tissue by replum tissue (valvelessness) has been previously observed in clv2 mutants (Kayes and Clark 1998). This clv2 phenotype is enhanced by clv1 and clv3 mutants. Valvelessness was also strongly enhanced in the bam1 bam2 clv1 triple mutant, such that every bam1-1 bam2-1 clv1-11 and bam1-1 bam2-1 clv1-7 flower had missing or incomplete valves (Table 3). In addition, large sections of the gynoecia of these plants frequently lacked all valve character. We also observed large valveless regions that developed very thin stripes of valve-like tissue (Figure 2, I and J).
TABLE 3.
bam1 and bam2 affect valvelessness
| Genotype | % valvelessa | nb |
|---|---|---|
| Ler | 0.0 | 100 |
| clv1-7 | 46.3 | 300 |
| clv1-7 bam2-1 | 22.5 | 200 |
| clv1-7 bam1-1 | 95.0 | 300 |
| clv1-7 bam1-1 bam2-1 | 100.0 | 100 |
| clv2-1 | 86.0 | 100 |
| clv2-1 bam2-1 | 45.5 | 200 |
| clv2-1 bam1-1 | 78.5 | 200 |
| clv2-1 bam1-1 bam2-1 | 80.0 | 100 |
Percentage of flowers with at least one missing or incomplete valve. Missing valves were defined as the replacement of a complete valve in the gynoecium by replum tissue. Incomplete valves were defined as valves that occupied <75% of the distance from the receptacle to the stigma.
Number of flowers evaluated.
None of these synergistic shoot meristem or organ phenotypes were seen in either bam1 bam2 clv2 plants or bam1 bam2 clv3 plants (Table 3; Figure 1; data not shown), indicating a specific role for CLV1 outside the meristem. This represents a novel range of developmental pathways regulated by CLV1, and prompts consideration of broad functions for the orthologous rice FON1 and maize thick tassel dwarf1 (td1), both of whose expression appears broader than CLV1 (Suzaki et al. 2004; Bommert et al. 2005).
A defect in internal stem differentiation was observed in bam1 bam2 combinations with all clv mutations. clv mutations normally result in increased vascular bundle number, and rarely vascular bundles located away from the periphery of the stem, presumably a result of the enlarged shoot meristem (L. Doil and S. E. Clark, unpublished data; Figure 3). In various bam1 bam2 clv mutant combinations, a more dramatic patterning defect was observed. Single, large ectopic lignified cells, as well as circular rings of smaller lignified cells were observed in the center of the stem (Figure 3, D–F and H–J). Furthermore, in stronger clv mutant combinations, large portions of the internal stem were composed of poorly vacuolated cells with a mixture of lignified and nonlignified cells (Figure 3, F and I). Stems of plants with this phenotype would occasionally break open during growth, revealing a fiber-like consistency to these abnormally developing stem regions (Figure 3, K and L). What role defects in meristem development and what role defects in later stem differentiation play in this phenotype are unclear.
Figure 3.—
Synergistic vascular patterning defects clv bam1 bam2 mutants. Cross sections of wild-type Ler (A), bam1-1 bam2-1 (B), clv1-1 (C), clv1-1 bam1-1 bam2-1 (D–F), clv3-2 (G), clv3-2 bam1-1 bam2-1 (H), clv2-1 (I), clv2-1 bam1-1 bam2-1 (J) stems stained with phloroglucinol (red) to detect lignified tissues. Scanning electron micrographs of clv1-11 bam1-1 bam2-1 mutants (K and L) show stems that split open during growth to reveal a fiber-like inner structure. Portions of the stem were removed to better visualize this structure. VB, vascular bundle, SM, shoot meristem. Arrows indicate large lignified cells; asterisks indicate rings of smaller lignified cells.
DISCUSSION
A model for BAM function within the meristem:
We investigated the genetic interactions of bam null alleles with various clv mutations. bam1 and bam2 exhibit dramatically different genetic interactions with clv1, clv2, and clv3, resulting in enhancement, no effect, and suppression, respectively. This indicates that the effects of BAM1 and BAM2 on stem cell homeostasis are integral to the CLV signaling pathway and do not represent a separate pathway regulating meristem development.
bam1 bam2 mutations significantly suppressed the stem cell accumulation in clv3-2 flowers, consistent with a requirement for BAM receptors in stem cell maintenance. In contrast, clv2 was unaltered by bam1 bam2 within the meristem. Given previous data suggesting that CLV2 is required in part for CLV1 protein accumulation, we propose that BAM1 and BAM2 function within the meristem is dependent on CLV2. These results suggest that the clv2 phenotype, which is the strongest of any receptor null, would result from the combined reduction of CLV1 and BAM function.
Unexpectedly, when combined with clv1 null alleles, both bam1 and bam2 strongly enhanced the Clv− phenotype, and the triple mutant exhibited very severe defects in stem cell homeostasis. Thus bam mutants have an opposite effect on stem cell accumulation in a clv1 CLV3 background compared to a CLV1 CLV3 or a CLV1 clv3 background, raising the question of how these data can be reconciled.
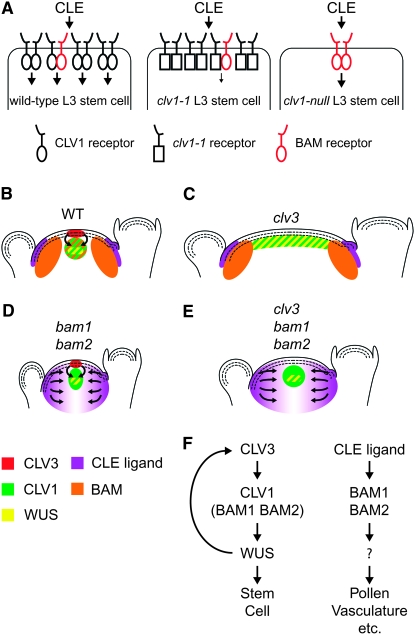
A previous report has indicated that CLV1 likely acts to sequester the ligand CLV3 (Lenhard and Laux 2003). One speculative possibility is that BAM receptors also act to sequester CLE polypeptide ligand(s) (Figure 4, B–E). We have shown that many CLE-containing proteins can replace CLV3 function when expressed within the meristem, and some can act through CLV1 (Ni and Clark 2006). Furthermore, the CLE domain of these proteins appears to be proteolytically released as the active signaling molecule, and this domain is well conserved among Arabidopsis CLE-containing proteins (Cock and McCormick 2001; DeYoung and Clark 2001; Fiers et al. 2004, 2005; Ito et al. 2006; Kondo et al. 2006). Indeed, multiple CLE domains can bind to the CLV1 extracellular domain (Ogawa et al. 2008). Over 30 CLE-containing proteins are expressed in a large number of different tissues, including the meristem periphery, and evidence indicates that these peptides can diffuse over many cell diameters (Lenhard and Laux 2003; Sharma et al. 2003; Fiers et al. 2005). Thus, insulating the meristem center from CLE peptides diffusing from outside the meristem may be critical to maintain the proper feedback between WUS expression and CLV1 signaling (Brand et al. 2000; Schoof et al. 2000).
Figure 4.—
A model for BAM function through signal transduction and ligand sequestration. (A) In wild-type cell layer three (L3) meristem cells, CLV1 is expressed at higher levels than BAM proteins and is the primary transducer of CLE signal. CLV1/CLV1 and CLV1/BAM complexes are consistent with the dominant-negative function of many clv1 alleles and their interaction with bam mutations. In clv1-1 L3 cells, BAM signaling may be blocked by interference of the mutant clv1-1 kinase domain. Residual signaling may come from a partially active BAM/clv1-1 heterodimer and/or the presumably small amount of BAM/BAM homodimers (not shown). In the absence of CLV1 (clv1-null), BAM proteins are available to transduce CLE signal, albeit at a reduced level compared to WT cells. To simplify the diagram, CLV2 and other components have not been included. (B) In a wild-type shoot meristem, CLV3 signals primarily through CLV1, but also through BAM1 and BAM2 to regulate the WUS expression domain. BAM1 and BAM2 also function on the flanks of the meristem where they sequester CLV3-related CLE ligands, creating a buffer around the meristem center. (C) In a clv3 apex, WUS repression is lost, leading to an expansion of the WUS expression domain and an enlargement of the meristem, while CLE ligands outside of the meristem are sequestered. (D) In a bam1 bam2 apex, CLE ligands are no longer sequestered, leading to overactivation of CLV1 and reduction in the meristem size, even meristem termination in bam1 bam2 bam3 triple mutants. (E) In a clv3 bam1 bam2 apex, loss of CLV3 is partially balanced through activation of CLV1 by unsequestered CLE ligands resulting in partial WUS expression control. (F) A model depicting genetic pathways through which CLV1, BAM1, and BAM2 function.
We suggest a model in which one of the functions of BAM receptors on the meristem periphery is to sequester CLE ligands and insulate the meristem center from extraneous signals (Figure 4, B–E). In the absence of BAM receptors (i.e., in bam mutants), ectopic CLE signal would diffuse to the meristem center. This signal would not be subjected to the feedback WUS/CLV3 loop that normally regulates CLV3 expression and could lead to overactivation of CLV1 and meristem reduction or termination. Furthermore, this ectopic CLE signal should be able to replace, in part, CLV3 function. Finally, the impact of this ectopic CLE signal resulting from bam mutations would only have an impact in a CLV1 background. Thus, the model explains the reduced meristem size of bam mutants, the bam suppression of clv3, as well as the requirement of CLV1 for meristem reduction in bam mutant backgrounds. While consistent with the genetic data, this model is quite speculative. We have no evidence of which CLE peptide might bind BAM receptors and whether binding alone, as opposed to binding and signaling, is sufficient for this proposed BAM activity. One possible test as CLE expression data become available would be to inactivate candidate CLEs expressed on the meristem flanks in a bam mutant background. Another possible test would be to determine if CLE-binding, but nonfunctional BAM receptors expressed specifically on the meristem flanks could carry out BAM function there. These experiments are complicated by the exquisite sensitivity of the meristem center to BAM expression (see below), and the tendency of nonfunctional receptor kinases to act as dominant-negative isoforms (Williams et al. 1997; Shpak et al. 2003; Diévart et al. 2003, 2006).
The enhancement of clv1 null alleles by bam mutations could indicate that while BAM receptors are at best expressed at low levels within the meristem center, they are redundant with CLV1 there (Figure 4A). clv1 dominant-negative alleles might be so effective in blocking signaling because there would theoretically be much more CLV1 than BAM receptors present in the meristem center. Redundant BAM/CLV1 function in the meristem center would explain the observation that single bam mutations had no effect on the dominant-negative clv1-1 allele. Interestingly, the LRR missense allele clv1-4 did not display the same interaction with bam mutants, raising the possibility that its mechanism of dominant-negative action may differ from that of the kinase-domain missense mutants.
CLV1/BAM synergy:
Despite previous extensive phenotypic and expression analysis suggesting CLV1 function is meristem specific, genetic analysis with bam mutants suggested otherwise. bam1 bam2 clv1 triple mutants exhibited many phenotypes not seen in any other genetic combination. These included strong dwarfism, filamentous floral organs, and loss of recognizable valve tissue. None of these phenotypes were seen in bam1 bam2 clv2 or bam1 bam2 clv3 plants, indicating that the phenotypes were not indirect consequences of defects in meristem development. This suggests that CLV1 plays a role in regulating the development of many organs in parallel with both BAM1 and BAM2.
In addition, synergistic interactions were seen in the development of clv1 bam1 bam2 shoot meristems. Here, we observed differentiated cells, or even organs in the center of the meristem, and the occasional loss of organogenesis on the flanks of the meristem. These phenotypes are reminiscent of the effects of corona mutants and PLL1 overexpression, both of which enhance clv mutants, leading to a loss of organogenesis at the meristem periphery, massive stem cell accumulation, and predicted eventual loss of meristem identity resulting in differentiation of the entire apex (Green et al. 2005; Song et al. 2006). Alternatively, the loss of organogenesis might reflect a function for BAM1 and BAM2 on the periphery of the meristem, where the corresponding genes are expressed at relatively high levels. Determining the precise developmental roles for BAM1 and BAM2 in the meristem periphery as well as in the various plant organs is an essential next step in uncovering the function of these broad-ranging receptors.
Genetic background effects:
One potential complication of our analysis is that the bam1 alleles were isolated in the Columbia (Col) background, while all of the clv alleles were isolated or backcrossed into the Landsberg er (Ler) background (Koornneef et al. 1983; Clark et al. 1993, 1995; Diévart et al. 2003; DeYoung et al. 2006). We have shown previously that crosses between clv alleles in the Ler and Col backgrounds can have subtle, but measurable effects on the meristem phenotype (Diévart et al. 2003). Two lines of evidence, however, indicate that background effects are not the source of the genetic interactions that we observed. First, the effects caused by genetic background are much smaller in magnitude than those observed here. Second, and most importantly, bam2-1 and bam2-3 alleles were isolated in the Ler background, and they exhibit the same type of interactions with each clv allele as observed for bam1.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01GM62962) (to S.E.C.). B.J.D. was supported in part by the National Institutes of Health Cellular Biotechnology Training Program (5 T32 GM08353).
References
- Bommert, P., C. Lunde, J. Nardmann, E. Vollbrecht, M. Running et al., 2005. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 132 1235–1245. [DOI] [PubMed] [Google Scholar]
- Brand, U., J. C. Fletcher, M. Hobe, E. M. Meyerowitz and R. Simon, 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289 617–619. [DOI] [PubMed] [Google Scholar]
- Byrne, M. E., C. A. Kidner and R. A. Martienssen, 2003. Plant stem cells: divergent pathways and common themes in shoots and roots. Curr. Opin. Genet. Dev. 13 551–557. [DOI] [PubMed] [Google Scholar]
- Carles, C. C., and J. C. Fletcher, 2003. Shoot apical meristem maintenance: the art of a dynamic balance. Trends Plant Sci. 8 394–401. [DOI] [PubMed] [Google Scholar]
- Clark, S. E., 1997. Organ formation at the vegetative shoot meristem. Plant Cell 9 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S. E., M. P. Running and E. M. Meyerowitz, 1993. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S. E., M. P. Running and E. M. Meyerowitz, 1995. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121 2057–2067. [Google Scholar]
- Clark, S. E., R. W. Williams and E. M. Meyerowitz, 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585. [DOI] [PubMed] [Google Scholar]
- Cock, J. M., and S. McCormick, 2001. A large family of genes that share homology with CLAVATA3. Plant Physiol. 126 939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung, B. J., K. L. Bickle, K. J. Schrage, P. Muskett, K. Patel et al., 2006. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45 1–16. [DOI] [PubMed] [Google Scholar]
- DeYoung, B. J., and S. E. Clark, 2001. Signaling through the CLAVATA1 receptor complex. Plant Mol. Biol. 46 505–513. [DOI] [PubMed] [Google Scholar]
- Diévart, A., M. Dalal, F. E. Tax, A. D. Lacey, A. Huttly et al., 2003. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diévart, A., M. J. Hymes, J. Li and S. E. Clark, 2006. Brassinosteroid-independent function of BRI1/CLV1 chimeric receptors. Funct. Plant Biol. 33 723–730. [DOI] [PubMed] [Google Scholar]
- Fiers, M., E. Golemiec, J. Xu, L. van der Geest, R. Heidstra et al., 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17 2542–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers, M., G. Hause, K. Boutilier, E. Casamitjana-Martinez, D. Weijers et al., 2004. Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327 37–49. [DOI] [PubMed] [Google Scholar]
- Green, K. A., M. J. Prigge, R. B. Katzman and S. E. Clark, 2005. CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt, R., and T. Laux, 2003. Stem cell regulation in the shoot meristem. J. Cell Sci. 116 1659–1666. [DOI] [PubMed] [Google Scholar]
- Hord, C. L., C. Chen, B. J. DeYoung, S. E. Clark and H. Ma, 2006. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, Y., I. Nakanomyo, H. Motose, K. Iwamoto, S. Sawa et al., 2006. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313 842–845. [DOI] [PubMed] [Google Scholar]
- Jeong, S., A. E. Trotochaud and S. E. Clark, 1999. The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes, J. M., and S. E. Clark, 1998. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125 3843–3851. [DOI] [PubMed] [Google Scholar]
- Kondo, T., S. Sawa, A. Kinoshita, S. Mizuno, T. Kakimoto et al., 2006. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313 845–848. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., J. van Eden, C. J. Hanhart, P. Stam, F. J. Braaksma et al., 1983. Linkage map of Arabidopsis thaliana. J. Hered. 74 265–272. [Google Scholar]
- Laux, T., K. F. Mayer, J. Berger and G. Jurgens, 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., and T. Laux, 2003. Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130 3163–3173. [DOI] [PubMed] [Google Scholar]
- Mayer, K. F., H. Schoof, A. Haecker, M. Lenhard, G. Jurgens et al., 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- Ni, J., and S. E. Clark, 2006. Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, M., H. Shinohara, Y. Sakagami and Y. Matsubayashi, 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319 294. [DOI] [PubMed] [Google Scholar]
- Prigge, M. J., D. Otsuga, J. M. Alonso, J. R. Ecker, G. N. Drews et al., 2005. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof, H., M. Lenhard, A. Haecker, K. F. Mayer, G. Jurgens et al., 2000. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644. [DOI] [PubMed] [Google Scholar]
- Sharma, V. K., J. Ramirez and J. C. Fletcher, 2003. The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol. Biol. 51 415–425. [DOI] [PubMed] [Google Scholar]
- Shpak, E. D., M. B. Lakeman and K. U. Torii, 2003. Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S.-K., and S. E. Clark, 2005. POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev. Biol. 285 272–284. [DOI] [PubMed] [Google Scholar]
- Song, S. K., M. M. Lee and S. E. Clark, 2006. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 133 4691–4698. [DOI] [PubMed] [Google Scholar]
- Suzaki, T., M. Sato, M. Ashikari, M. Miyoshi, Y. Nagato et al., 2004. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131 5649–5657. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A. E., T. Hao, W. Guang, Z. Yang and S. E. Clark, 1999. The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. W., J. M. Wilson and E. M. Meyerowitz, 1997. A possible role for kinase-associated protein phosphatase in the arabidopsis CLAVATA1 signaling pathway. Proc. Natl. Acad. Sci. USA 94 10467–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L. P., A. K. Miller and S. E. Clark, 2003. POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr. Biol. 13 179–188. [DOI] [PubMed] [Google Scholar]
- Yu, L. P., E. J. Simon, A. E. Trotochaud and S. E. Clark, 2000. POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development 127 1661–1670. [DOI] [PubMed] [Google Scholar]